Establishment of a Protocol for the Characterization of Secreted Biomolecules in Somatic Embryogenic Cultures of Olea europaea L.
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Isolation of Secretome and Extracellular Metabolome from Conditioned Medium
2.3. Analysis of the Secretome Purity Using a Cytosolic Biomarker
2.4. Establishment of a Procedure for Secretome Precipitation and Characterization
2.4.1. Selection of the Most Efficient Method for Secretome Precipitation
2.4.2. Protein Solubilization and Quantification
2.4.3. Analysis of the Secretome Band Patterns by SDS-PAGE
2.5. Analysis of the Secretome by LC-MS/MS
2.6. Analysis of the Extracellular Metabolome by 1H NMR Spectroscopy
3. Results and Discussion
3.1. Selection of the Most Efficient Method for Protein Precipitation
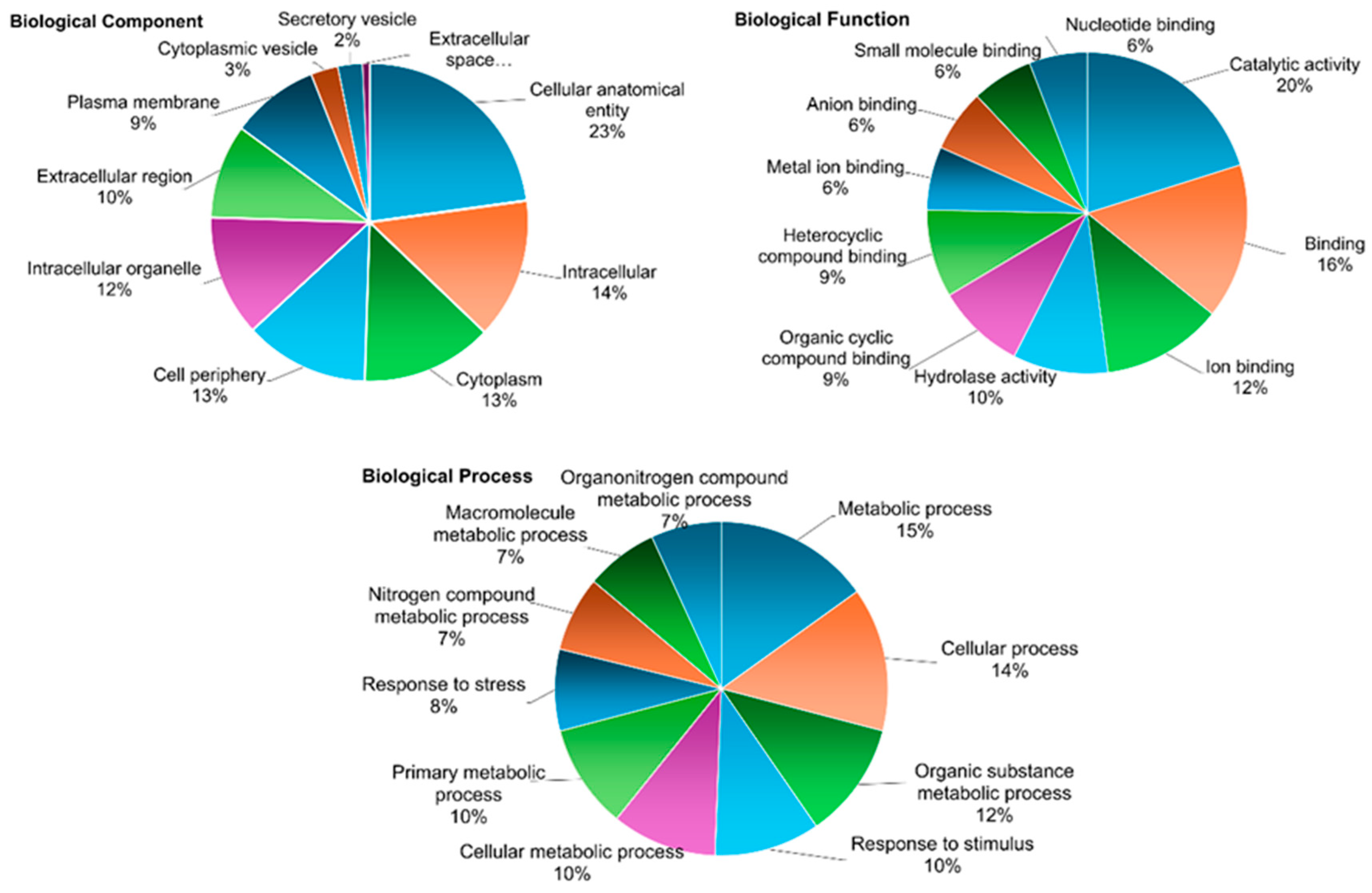
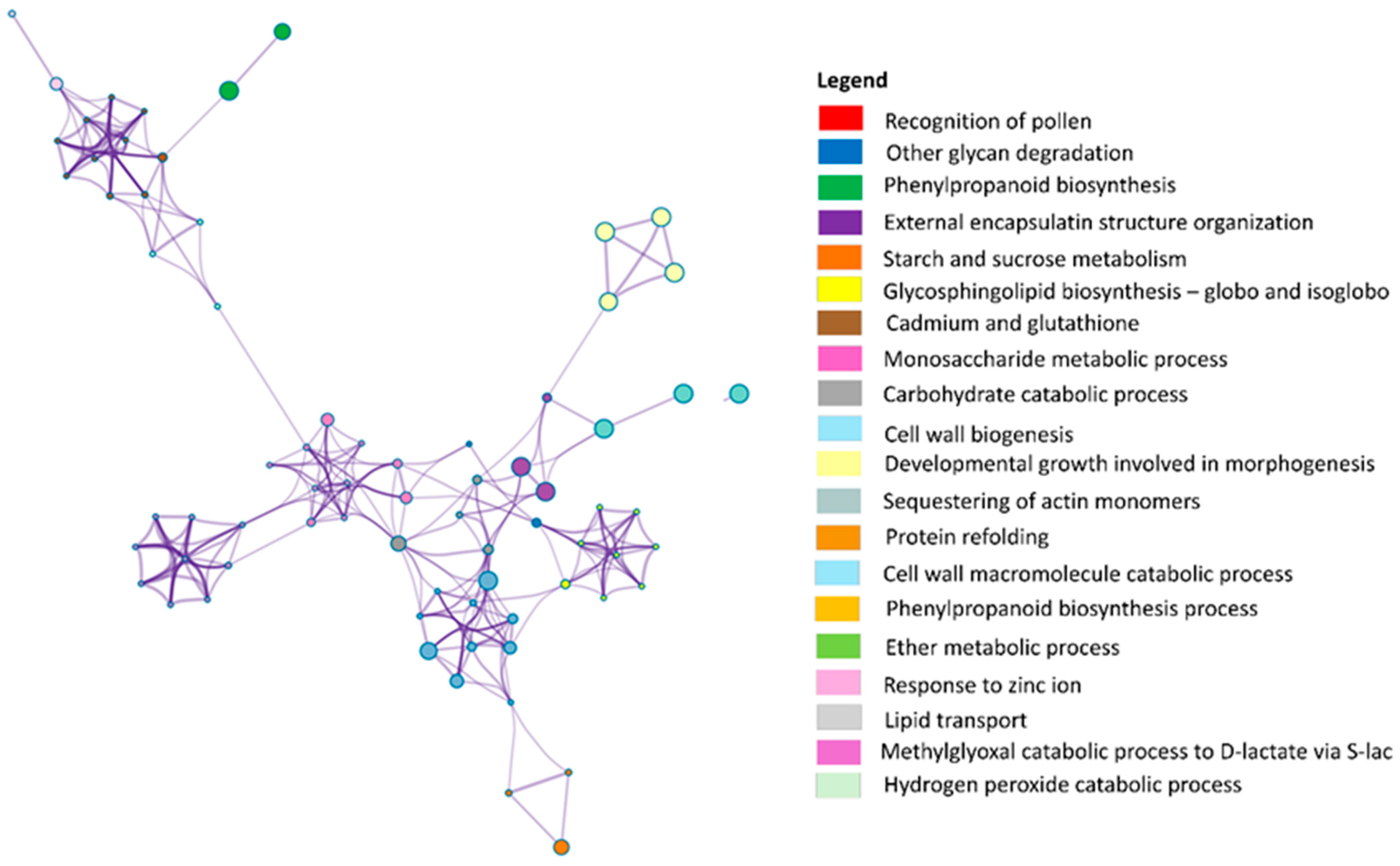
3.2. LC-MS/MS-Based Analysis of Secretome from Embryogenic Calli
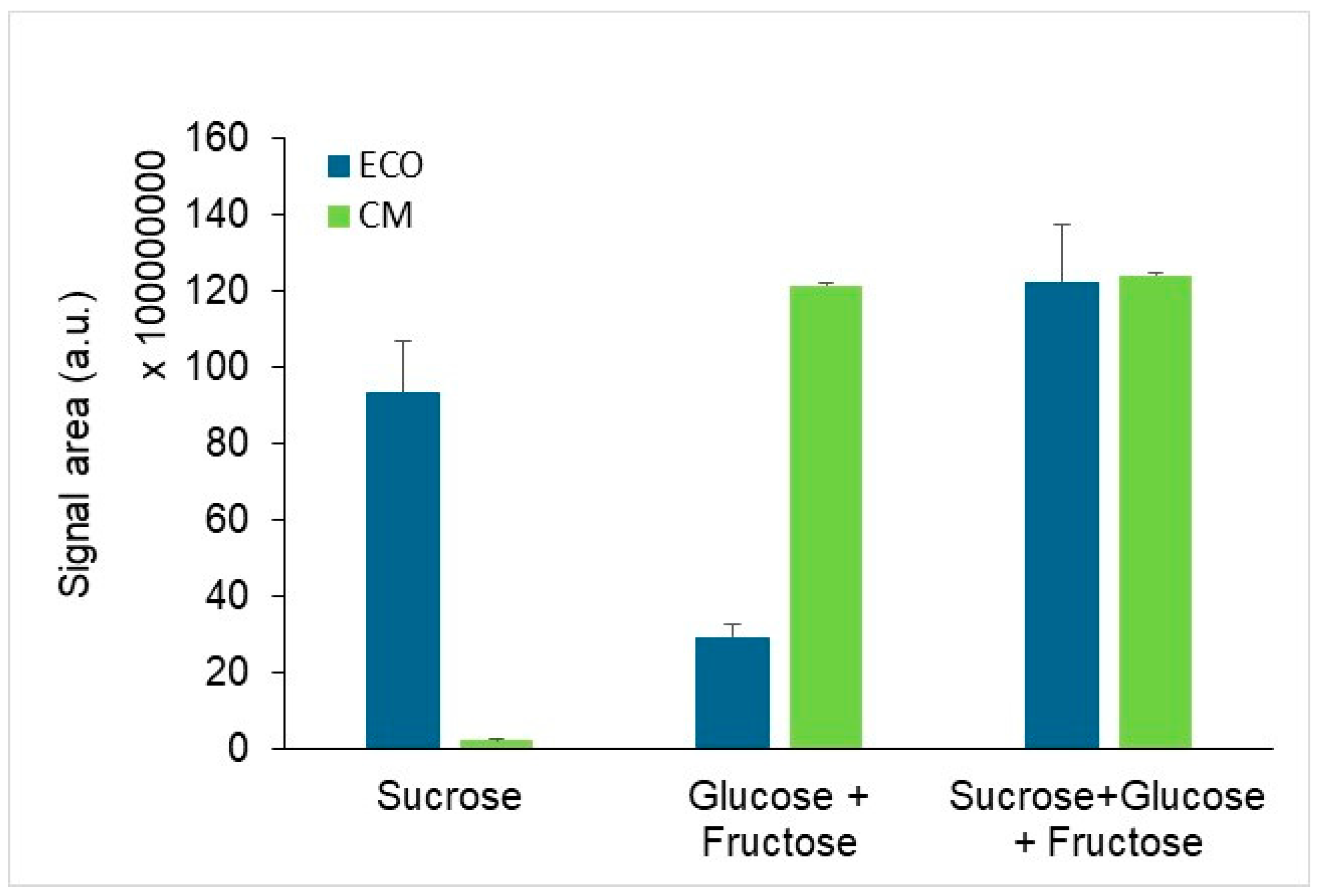
3.3. Detection of Changes in the Exametabolome Composition Through 1H NMR Spectroscopy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SE | Somatic embryogenesis |
| CM | Conditioned medium |
| SDS-PAGE | Sodium dodecyl sulfate–polyacrylamide gel electrophoresis |
| LC-MS/MS | Liquid chromatography coupled to mass spectrometry |
| NMR | Nuclear Magnetic Resonance |
| OMc | Olive Medium Culture |
| ECO | Embryogenesis Cyclic Olive |
| MDH | Malate dehydrogenase |
| NAD | Nicotinamide adenine dinucleotide |
| TCA | Trichloroacetic acid |
| AMY | α-amylase |
| POX | Peroxidase |
Appendix A


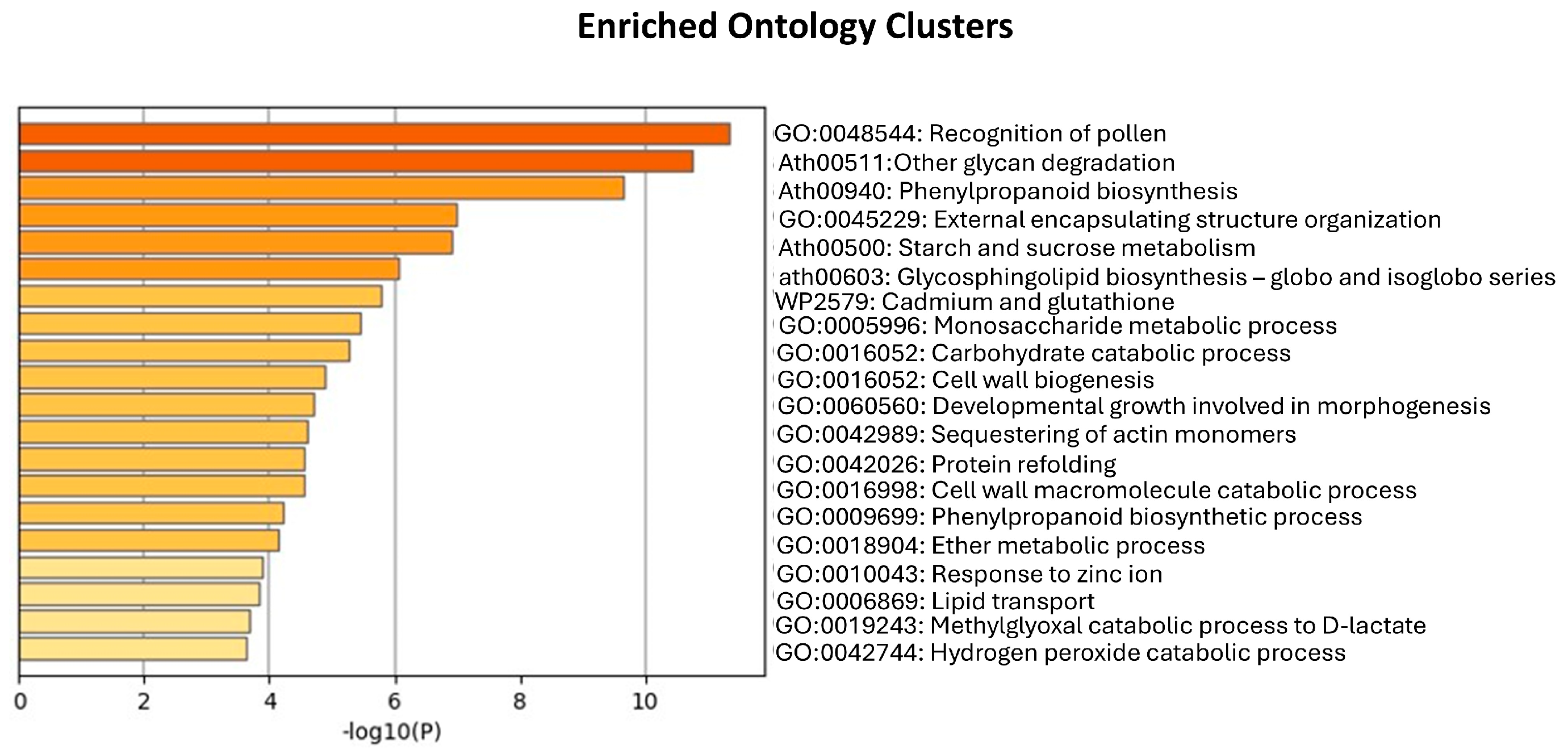
Appendix B
| Precipitation Methods | ||||
|---|---|---|---|---|
| Lyophilized | ||||
| Bands | Acetone | TCA/Acetone | Methanol/Chloroform | p-Value |
| 1 | 291.3 ± 291.3 | 1849.7 ± 464.5 | 381.7 ± 381.7 | 0.052 |
| 2 | 324.7 ± 324.7 | 1807.7 ± 158.4 | 378.3 ± 378.3 | 0.052 |
| 3 | 326.3 ± 326.3 | 1762.7 ± 33.7 | 319.3 ± 319.3 | 0.052 |
| 4 | 409.3 ± 409.3 | 851.7 ± 429.7 | 262.0 ± 262.0 | 0.5 |
| 5 | 456.4 ± 456.3 a | 1598.0 ± 94.92 b | nt | 0.035 |
| 6 | 381.0 ± 381.0 | 607.3 ± 607.33 | 525.3 ± 262.7 | 0.948 |
| Non-lyophilized | ||||
| 1 | 146.0 ± 146.0 | 877.3 ± 114.9 | 547.7 ± 279.2 | 0.120 |
| 2 | 399.7 ± 201.6 | 506.0 ± 253.8 | 291.3 ± 291.3 | 0.794 |
| 3 | nt | 612.3 ± 313.5 | 337.0 ± 337.0 | 0.281 |
| 4 | 184.3 ± 184.3 | 366.7 ± 366.7 | nt | 0.558 |
| 5 | 491.0 ± 249.5 | 522.7 ± 271.3 | 294.0 ± 294.0 | 0.854 |
| 6 | nt | 443.0 ± 443.0 | nt | 0.368 |
References
- Juarez-Escobar, J.; Bojórquez-Velázquez, E.; Elizalde-Contreras, J.M.; Guerrero-Analco, J.A.; Loyola-Vargas, V.M.; Mata-Rosas, M.; Ruiz-May, E. Current Proteomic and Metabolomic Knowledge of Zygotic and Somatic Embryogenesis in Plants. Int. J. Mol. Sci. 2021, 22, 11807. [Google Scholar] [CrossRef] [PubMed]
- Hazubska-Przybył, T.; Ratajczak, E.; Obarska, A.; Pers-Kamczyc, E. Different Roles of Auxins in Somatic Embryogenesis Efficiency in Two Picea Species. Int. J. Mol. Sci. 2020, 21, 3394. [Google Scholar] [CrossRef] [PubMed]
- Vidal, N.; Mallón, R.; Valladares, S.; Meijomín, A.M.; Vieitez, A.M. Regeneration of Transgenic Plants by Agrobacterium-Mediated Transformation of Somatic Embryos of Juvenile and Mature Quercus robur. Plant Cell Rep. 2010, 29, 1411–1422. [Google Scholar] [CrossRef] [PubMed]
- Heringer, A.S.; Santa-Catarina, C.; Silveira, V. Insights from Proteomic Studies into Plant Somatic Embryogenesis. Proteomics 2018, 18, 1700265. [Google Scholar] [CrossRef]
- Guan, Y.; Li, S.-G.; Fan, X.-F.; Su, Z.-H. Application of Somatic Embryogenesis in Woody Plants. Front. Plant Sci. 2016, 7, 938. [Google Scholar] [CrossRef]
- Pais, M.S. Somatic Embryogenesis Induction in Woody Species: The Future After OMICs Data Assessment. Front. Plant Sci. 2019, 10, 240. [Google Scholar] [CrossRef]
- Trabelsi, E.B.; Bouzid, S.; Bouzid, M.; Elloumi, N.; Belfeleh, Z.; Benabdallah, A.; Ghezel, R. In-Vitro Regeneration of Olive Tree by Somatic Embryogenesis. J. Plant Biol. 2003, 46, 173–180. [Google Scholar] [CrossRef]
- Brhadda, N.; Abousalim, A. Loudyi Dou Elmacane Walali Effets du milieu de culture et de la lumière sur l’embryogenèse somatique de l’olivier (Olea europaea L.) cv. Picholine marocaine. Fruits 2003, 58, 167–174. [Google Scholar] [CrossRef]
- Capelo, A.M.; Silva, S.; Brito, G.; Santos, C. Somatic Embryogenesis Induction in Leaves and Petioles of a Mature Wild Olive. Plant Cell Tiss. Organ Cult. 2010, 103, 237–242. [Google Scholar] [CrossRef]
- Oulbi, S.; Belkoura, I.; Loutfi, K. Somatic Embryogenesis from Somatic Explants of a Moroccan Olive (Olea europaea L.) Cultivar, ‘Moroccan Picholine’. Acta Hortic. 2018, 1199, 91–96. [Google Scholar] [CrossRef]
- Mazri, M.A.; Belkoura, I.; Pliego-Alfaro, F.; Belkoura, M. Somatic Embryogenesis from Leaf and Petiole Explants of the Moroccan Olive Cultivar Dahbia. Sci. Hortic. 2012, 159, 88–95. [Google Scholar] [CrossRef]
- Narváez, I.; Martín, C.; Jiménez-Díaz, R.M.; Mercado, J.A.; Pliego-Alfaro, F. Plant Regeneration via Somatic Embryogenesis in Mature Wild Olive Genotypes Resistant to the Defoliating Pathotype of Verticillium dahliae. Front. Plant Sci. 2019, 10, 1471. [Google Scholar] [CrossRef]
- Cardoso, H.G.; Campos, M.C.; Pais, M.S.; Peixe, A. Somatic Embryogenesis in Iberian Grapevine (Vitis vinifera) Cultivars Using Carpels as Initial Explants: Protocol Establishment and Histological Evaluation. J. Agric. Sci. Technol. 2019, 9. [Google Scholar] [CrossRef]
- Pires, R.; Cardoso, H.; Ribeiro, A.; Peixe, A.; Cordeiro, A. Somatic Embryogenesis from Mature Embryos of Olea Europaea L. Cv. ‘Galega Vulgar’ and Long-Term Management of Calli Morphogenic Capacity. Plants 2020, 9, 758. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Romero, C. Somatic Embryogenesis in Olive. Plants 2021, 10, 433. [Google Scholar] [CrossRef]
- Pritsa, T.S.; Voyiatzis, D.G. The in Vitro Morphogenetic Capacity of Olive Embryo Explants at Different Develop-mental Stages, as Affected by L-Glutamine, L-Arginine and 2,4-D. J. Biol. Res. 2004, 1, 55–61. [Google Scholar]
- Mazri, M.A.; Naciri, R.; Belkoura, I. Maturation and Conversion of Somatic Embryos Derived from Seeds of Olive (Olea Europaea L.) Cv. Dahbia: Occurrence of Secondary Embryogenesis and Adventitious Bud Formation. Plants 2020, 9, 1489. [Google Scholar] [CrossRef]
- Titouh, K.; Hadj Moussa, K.; Boufis, N.; Khelifi, L. Impact of Cultural Conditions on Germination of Olive (Olea europaea L.) Somatic Embryos and Plantlets Development from the Algerian Cultivar Chemlal. Adv. Hortic. Sci. 2022, 36, 185–191. [Google Scholar] [CrossRef]
- Cerezo, S.; Mercado, J.A.; Pliego-Alfaro, F. An Efficient Regeneration System via Somatic Embryogenesis in Olive. Plant Cell Tiss. Organ Cult. 2011, 106, 337–344. [Google Scholar] [CrossRef]
- Rugini, E.; Caricato, G. Somatic Embryogenesis and Plant Recovery from Mature Tissues of Olive Cultivars (Olea europaea L.) ‘Canino’ and ‘Moraiolo’. Plant Cell Rep. 1995, 14, 257–260. [Google Scholar] [CrossRef]
- Trabelsi, E.B.; Naija, S.; Elloumi, N.; Belfeleh, Z.; Msellem, M.; Ghezel, R.; Bouzid, S. Somatic Embryogenesis in Cell Suspension Cultures of Olive Olea europaea (L.) ‘Chetoui’. Acta Physiol. Plant 2011, 33, 319–324. [Google Scholar] [CrossRef]
- Toufik, I.; Guenoun, F.; Belkoura, I. Embryogenesis Expression from Somatic Explants of Olive (Olea europaea L.) Cv. Picual. Moroc. J. Biol. 2014, 11, 17–25. [Google Scholar]
- Eigel, L.; Koop, H.-U. Nurse Culture of Individual Cells: Regeneration of Colonies from Single Protoplasts of Nicotiana tabacum, Brassica napus and Hordeum vulgare. J. Plant Physiol. 1989, 134, 577–581. [Google Scholar] [CrossRef]
- Westcott, R.J. Production of Embryogenic Callus from Nonembryonic Explants of Norway Spruce Picea Abies (L.) Karst. Plant Cell Rep. 1994, 14, 47–49. [Google Scholar] [CrossRef]
- Komai, F.; Morohashi, H.; Horita, M. Application of Nurse Culture for Plant Regeneration from Protoplasts of Lilium japonicum Thunb. In Vitro Cell. Dev. Biol. Plant 2006, 42, 252–255. [Google Scholar] [CrossRef]
- Hargreaves, C.; Reeves, C.; Gough, K.; Montalbán, I.A.; Low, C.; Van Ballekom, S.; Dungey, H.S.; Moncaleán, P. Nurse Tissue for Embryo Rescue: Testing New Conifer Somatic Embryogenesis Methods in a F1 Hybrid Pine. Trees 2017, 31, 273–283. [Google Scholar] [CrossRef]
- Dyachok, J.V.; Wiweger, M.; Kenne, L.; Von Arnold, S. Endogenous Nod-Factor-Like Signal Molecules Promote Early Somatic Embryo Development in Norway Spruce. Plant Physiol. 2002, 128, 523–533. [Google Scholar] [CrossRef]
- Pernis, M.; Salaj, T.; Bellová, J.; Danchenko, M.; Baráth, P.; Klubicová, K. Secretome Analysis Revealed That Cell Wall Remodeling and Starch Catabolism Underlie the Early Stages of Somatic Embryogenesis in Pinus nigra. Front. Plant Sci. 2023, 14, 1225424. [Google Scholar] [CrossRef]
- Nic-Can, G.I.; Galaz-Ávalos, R.M.; De-la-Peña, C.; Alcazar-Magaña, A.; Wrobel, K.; Loyola-Vargas, V.M. Somatic Embryogenesis: Identified Factors That Lead to Embryogenic Repression. A Case of Species of the Same Genus. PLoS ONE 2015, 10, e0126414. [Google Scholar] [CrossRef]
- Awada, R.; Campa, C.; Gibault, E.; Déchamp, E.; Georget, F.; Lepelley, M.; Abdallah, C.; Erban, A.; Martinez-Seidel, F.; Kopka, J.; et al. Unravelling the Metabolic and Hormonal Machinery During Key Steps of Somatic Embryogenesis: A Case Study in Coffee. Int. J. Mol. Sci. 2019, 20, 4665. [Google Scholar] [CrossRef]
- Awada, R.; Verdier, D.; Froger, S.; Brulard, E.; De Faria Maraschin, S.; Etienne, H.; Breton, D. An Innovative Automated Active Compound Screening System Allows High-Throughput Optimization of Somatic Embryogenesis in Coffea arabica. Sci. Rep. 2020, 10, 810. [Google Scholar] [CrossRef] [PubMed]
- Ben-Amar, A. Secretome-Derived Cultured Cell System: Overview Towards Extracellular Protein Characterization and Biotechnological Applications. J. Basic Appl. Sci. 2021, 17, 13–24. [Google Scholar] [CrossRef]
- Ngcala, M.G.; Goche, T.; Brown, A.P.; Chivasa, S.; Ngara, R. Heat Stress Triggers Differential Protein Accumulation in the Extracellular Matrix of Sorghum Cell Suspension Cultures. Proteomes 2020, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Egertsdotter, U. Development of Somatic Embryos in Norway Spruce. J. Exp. Bot. 1998, 49, 155–162. [Google Scholar] [CrossRef]
- Couillerot, J.-P.; Windels, D.; Vazquez, F.; Michalski, J.-C.; Hilbert, J.-L.; Blervacq, A.-S. Pretreatments, Condi-tioned Medium and Co-Culture Increase the Incidence of Somatic Embryogenesis of Different Cichorium Species. Plant Sig. Behav 2012, 7, 121–131. [Google Scholar] [CrossRef]
- Guo, H.; Guo, H.; Zhang, L.; Tang, Z.; Yu, X.; Wu, J.; Zeng, F. Metabolome and Transcriptome Association Analysis Reveals Dynamic Regulation of Purine Metabolism and Flavonoid Synthesis in Transdifferentiation during Somatic Embryogenesis in Cotton. Int. J. Mol. Sci. 2019, 20, 2070. [Google Scholar] [CrossRef]
- Gulzar, B.; Mujib, A.; Malik, M.Q.; Sayeed, R.; Mamgain, J.; Ejaz, B. Genes, Proteins and Other Networks Regu-lating Somatic Embryogenesis in Plants. JGEB 2020, 18, 31. [Google Scholar] [CrossRef]
- Duchow, S.; Dahlke, R.I.; Geske, T.; Blaschek, W.; Classen, B. Arabinogalactan-Proteins Stimulate Somatic Embryogenesis and Plant Propagation of Pelargonium Sidoides. Carbohydr. Polym. 2016, 152, 149–155. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, D.; Shi, P.; Htwe, Y.M.; Yu, Q.; Huang, L.; Zhou, H.; Liu, L.; Wang, Y. Cell Wall Lignification May Be Necessary for Somatic Embryogenesis of Areca Palm (Areca catechu). Sci. Hortic. 2023, 307, 111538. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, S.; Wang, J. Transcriptomic and Proteomic Analyses of Embryogenic Tissues in Picea balfouriana Treated with 6-benzylaminopurine. Physiol. Plant. 2015, 154, 95–113. [Google Scholar] [CrossRef]
- Fráterová, L.; Salaj, T.; Matušíková, I.; Salaj, J. The Role of Chitinases and Glucanases in Somatic Embryogenesis of Black Pine and Hybrid Firs. Open Life Sci. 2013, 8, 1172–1182. [Google Scholar] [CrossRef]
- Kreuger, M.; Van Holst, G.-J. Arabinogalactan Proteins Are Essential in Somatic Embryogenesis of Daucus carota L. Planta 1993, 189, 243–248. [Google Scholar] [CrossRef]
- Ben Amar, A.; Cobanov, P.; Ghorbel, A.; Mliki, A.; Reustle, G.M. Involvement of Arabinogalactan Proteins in the Control of Cell Proliferation of Cucurbita pepo Suspension Cultures. Biol. Plant. 2010, 54, 321–324. [Google Scholar] [CrossRef]
- Tchorbadjieva, M.I. Protein Markers for Somatic Embryogenesis. In Somatic Embryogenesis; Mujib, A., Šamaj, J., Eds.; Plant Cell Monographs; Springer: Berlin/Heidelberg, Germany, 2006; Volume 2, pp. 215–233. [Google Scholar]
- Rodríguez-Celma, J.; Ceballos-Laita, L.; Grusak, M.A.; Abadía, J.; López-Millán, A.-F. Plant Fluid Proteomics: Delving into the Xylem Sap, Phloem Sap and Apoplastic Fluid Proteomes. Biophys. Acta Proteins Proteom. 2016, 1864, 991–1002. [Google Scholar] [CrossRef]
- Caeiro, A.; Ventura, F.; Correia, S.; Canhoto, J. Changes of Secondary Metabolites during Tamarillo Somatic Embryogenesis. Biol. Life Sci. Forum 2022, 11, 39. [Google Scholar] [CrossRef]
- Segers, K.; Declerck, S.; Mangelings, D.; Heyden, Y.V.; Eeckhaut, A.V. Analytical Techniques for Metabolomic Studies: A Review. Bioanalysis 2019, 11, 2297–2318. [Google Scholar] [CrossRef]
- Dowlatabadi, R.; Weljie, A.M.; Thorpe, T.A.; Yeung, E.C.; Vogel, H.J. Metabolic Footprinting Study of White Spruce Somatic Embryogenesis Using NMR Spectroscopy. Plant Physiol. Biochem. 2009, 47, 343–350. [Google Scholar] [CrossRef]
- Cañas, L.A.; Benbadis, A. In Vitro Plant Regeneration from Cotyledon Fragments of the Olive Tree (Olea europaea L). Plant Sci. 1988, 54, 65–74. [Google Scholar] [CrossRef]
- Orinos, T.; Mitrakos, K. Rhizogenesis and somatic embryogenesis in calli from wild olive (Olea europaea var. sylvestris (Miller) Lehr) mature zygotic embryos. Plant Cell Tissue Organ Cult. 1991, 27, 183–187. [Google Scholar] [CrossRef]
- Regente, M.; Monzón, G.C.; De La Canal, L. Phospholipids Are Present in Extracellular Fluids of Imbibing Sunflower Seeds and Are Modulated by Hormonal Treatments. J. Exp. Bot. 2008, 59, 553–562. [Google Scholar] [CrossRef]
- Genova, M.L.; Lenaz, G. Functional Role of Mitochondrial Respiratory Supercomplexes. Biochim. Biophys. Acta 2014, 1837, 427–443. [Google Scholar] [CrossRef] [PubMed]
- Diniz, I.; Azinheira, H.; Figueiredo, A.; Gichuru, E.; Oliveira, H.; Guerra-Guimarães, L.; Silva, M.C. Fungal Penetration Associated with Recognition, Signaling and Defence-Related Genes and Peroxidase Activity during the Resistance Response of Coffee to Colletotrichum kahawae. Physiol. Mol. Plant Pathol. 2019, 105, 119–127. [Google Scholar] [CrossRef]
- Loureiro, A.; Guerra-Guimarães, L.; Lidon, F.C.; Bertrand, B.; Silva, M.C.; Várzea, V. Isoenzymatic Characterization of Colletotrichum kahawae Isolates with Different Levels of Aggressiveness. Trop. Plant Pathol. 2011, 36, 287–293. [Google Scholar] [CrossRef]
- Shaw, C.R.; Prasad, R. Starch gel electrophoresis of enzymes—A compilation of recipes. Biochem. Genet. 1970, 4, 297–320. [Google Scholar] [CrossRef]
- Damerval, C.; De Vienne, D.; Zivy, M.; Thiellement, H. Technical Improvements in Two-dimensional Electrophoresis Increase the Level of Genetic Variation Detected in Wheat-seedling Proteins. Electrophoresis 1986, 7, 52–54. [Google Scholar] [CrossRef]
- Wessel, D.; Flügge, U.I. A Method for the Quantitative Recovery of Protein in Dilute Solution in the Presence of Detergents and Lipids. Anal. Biochem. 1984, 138, 141–143. [Google Scholar] [CrossRef]
- Laemmli, U. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Candiano, G.; Bruschi, M.; Musante, L.; Ghiggeri, G.M.; Carnemolla, B.; Orecchia, P.; Zardi, L.; Righetti, P.B. Blue Silver: A Very Sensitive Colloidal Coomassie G-250 Staining for Proteome Analysis. Electrophoresis 2004, 25, 1327–1333. [Google Scholar] [CrossRef]
- Ciordia, S.; Alvarez-Sola, G.; Rullán, M.; Urman, J.M.; Ávila, M.A.; Corrales, F.J. Digging Deeper into Bile Proteome. J. Proteom. 2021, 230, 103984. [Google Scholar] [CrossRef]
- Ciordia, S.; Alvarez-Sola, G.; Rullán, M.; Urman, J.M.; Ávila, M.A.; Corrales, F.J. Bile Processing Protocol for Improved Proteomic Analysis. In Clinical Proteomics. Methods in Molecular Biology; Humana: New York, NY, USA, 2022; Volume 2420. [Google Scholar]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE Database Resources in 2022: A Hub for Mass Spectrometry-Based Proteomics Evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-Throughput Functional Annotation and Data Mining with the Blast2GO Suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- ElNaker, N.A.; Daou, M.; Ochsenkühn, M.A.; Amin, S.A.; Yousef, A.F.; Yousef, L.F. A Metabolomics Approach to Evaluate the Effect of Lyophilization versus Oven Drying on the Chemical Composition of Plant Extracts. Sci. Rep. 2021, 11, 22679. [Google Scholar] [CrossRef]
- Rogulska, O.; Vackova, I.; Prazak, S.; Turnovcova, K.; Kubinova, S.; Bacakova, L.; Jendelova, P.; Petrenko, Y. Storage Conditions Affect the Composition of the Lyophilized Secretome of Multipotent Mesenchymal Stromal Cells. Sci. Rep. 2024, 14, 10243. [Google Scholar] [CrossRef]
- Peng, W.; Chang, M.; Wu, Y.; Zhu, W.; Tong, L.; Zhang, G.; Wang, Q.; Liu, J.; Zhu, X.; Cheng, T.; et al. Lyophilized Powder of Mesenchymal Stem Cell Supernatant Attenuates Acute Lung Injury through the IL-6–p-STAT3–P63–JAG2 Pathway. Stem Cell Res. Ther. 2021, 12, 216. [Google Scholar] [CrossRef]
- Abla, K.K.; Mehanna, M.M. Freeze-Drying: A Flourishing Strategy to Fabricate Stable Pharmaceutical and Biological Products. Int. J. Pharm. 2022, 628, 122233. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Gong, F.; Wang, F. Protein Extraction from Plant Tissues for 2DE and Its Application in Proteomic Analysis. Proteomics 2014, 14, 645–658. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havli, J.; Olsen, J.V.; Mann, M. In-Gel Digestion for Mass Spectrometric Characterization of Proteins and Proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef]
- Niu, L.; Zhang, H.; Wu, Z.; Wang, Y.; Liu, H.; Wu, X.; Wang, W. Modified TCA/Acetone Precipitation of Plant Proteins for Proteomic Analysis. PLoS ONE 2018, 13, e0202238. [Google Scholar] [CrossRef]
- Zhang, Y.; Bottinelli, D.; Lisacek, F.; Luban, J.; Strambio-De-Castillia, C.; Varesio, E.; Hopfgartner, G. Optimization of Human Dendritic Cell Sample Preparation for Mass Spectrometry-Based Proteomic Studies. Anal. Biochem. 2015, 484, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Sharma, I.P.; Kumar, N.; Sharma, A.K. A Rapid Isolation Method of Extracellular Proteins Produced by Pseudomonad Strains. App. Sci. Rep. 2017, 17, 1–3. [Google Scholar] [CrossRef]
- Saad, M.G.; Beyenal, H.; Dong, W.J. Dual Roles of the Conditional Extracellular Vesicles Derived from Pseudomonas Aeruginosa Biofilms: Promoting and Inhibiting Bacterial Biofilm Growth. Biofilm 2024, 7, 100183. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Guimarães, L.; Tenente, R.; Pinheiro, C.; Chaves, I.; Silva, M.D.C.; Cardoso, F.M.H.; Planchon, S.; Barros, D.R.; Renaut, J.; Ricardo, C.P. Proteomic Analysis of Apoplastic Fluid of Coffea Arabica Leaves Highlights Novel Biomarkers for Resistance against Hemileia Vastatrix. Front. Plant Sci. 2015, 6, 478. [Google Scholar] [CrossRef]
- Jaswanthi, N.; Krishna, M.S.R.; Sahitya, U.L.; Suneetha, P. Apoplast Proteomic Analysis Reveals Drought Stress-Responsive Protein Datasets in Chilli (Capsicum annuum L.). Data Brief 2019, 25, 104041. [Google Scholar] [CrossRef]
- Jiang, S.; Pan, L.; Zhou, Q.; Xu, W.; He, F.; Zhang, L.; Gao, H. Analysis of the Apoplast Fluid Proteome during the Induction of Systemic Acquired Resistance in Arabidopsis thaliana. PeerJ 2023, 11, e16324. [Google Scholar] [CrossRef]
- Ribeiro, D.G.; De Almeida, R.F.; Fontes, W.; De Souza Castro, M.; De Sousa, M.V.; Ricart, C.A.O.; Da Cunha, R.N.V.; Lopes, R.; Scherwinski-Pereira, J.E.; Mehta, A. Stress and Cell Cycle Regulation during Somatic Embryogenesis Plays a Key Role in Oil Palm Callus Development. J. Prot. 2019, 192, 137–146. [Google Scholar] [CrossRef]
- Peng, C.; Gao, F.; Tretyakova, I.N.; Nosov, A.M.; Shen, H.; Yang, L. Transcriptomic and Metabolomic Analysis of Korean Pine Cell Lines with Different Somatic Embryogenic Potential. Int. J. Mol. Sci. 2022, 23, 13301. [Google Scholar] [CrossRef]
- Domżalska, L.; Kędracka-Krok, S.; Jankowska, U.; Grzyb, M.; Sobczak, M.; Rybczyński, J.J.; Mikuła, A. Proteomic Analysis of Stipe Explants Reveals Differentially Expressed Proteins Involved in Early Direct Somatic Embryogenesis of the Tree Fern Cyathea Delgadii Sternb. Plant Sci. 2017, 258, 61–76. [Google Scholar] [CrossRef]
- Oulbi, S.; Kohaich, K.; Baaziz, M.; Belkoura, I.; Loutfi, K. Peroxidase Enzyme Fractions as Markers of Somatic Embryogenesis Capacities in Olive (Olea europaea L.). Plants 2021, 10, 901. [Google Scholar] [CrossRef]
- Singh, A.K. Principles of Nanotoxicology. In Engineered Nanoparticles; Elsevier: Amsterdam, The Netherlands, 2016; pp. 171–227. ISBN 978-0-12-801406-6. [Google Scholar]
- Kalluri, R.; LeBleu, V.S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Tancini, B.; Buratta, S.; Delo, F.; Sagini, K.; Chiaradia, E.; Pellegrino, R.M.; Emiliani, C.; Urbanelli, L. Lysosomal Exocytosis: The Extracellular Role of an Intracellular Organelle. Membranes 2020, 10, 406. [Google Scholar] [CrossRef]
- Ibata, K.; Yuzaki, M. Destroy the Old to Build the New: Activity-Dependent Lysosomal Exocytosis in Neurons. Neurosci. Res. 2021, 167, 38–46. [Google Scholar] [CrossRef]
- Su, J.; Song, Y.; Zhu, Z.; Huang, X.; Fan, J.; Qiao, J.; Mao, F. Cell–Cell Communication: New Insights and Clinical Implications. Sig. Transduct. Target Ther. 2024, 9, 196. [Google Scholar] [CrossRef]
- Morel, A.; Trontin, J.-F.; Corbineau, F.; Lomenech, A.-M.; Beaufour, M.; Reymond, I.; Le Metté, C.; Ader, K.; Harvengt, L.; Cadene, M.; et al. Cotyledonary Somatic Embryos of Pinus Pinaster Ait. Most Closely Resemble Fresh, Maturing Cotyledonary Zygotic Embryos: Biological, Carbohydrate and Proteomic Analyses. Planta 2014, 240, 1075–1095. [Google Scholar] [CrossRef]
- Wang, F.-X.; Shang, G.-D.; Wu, L.-Y.; Xu, Z.-G.; Zhao, X.-Y.; Wang, J.-W. Chromatin Accessibility Dynamics and a Hierarchical Transcriptional Regulatory Network Structure for Plant Somatic Embryogenesis. Dev. Cell 2020, 54, 742–757.e8. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, U.C.; Shrivastava, R. Interaction of Viral Proteins with Metal Ions: Role in Maintaining the Structure and Functions of Viruses. FEMS Immunol. Med. Microbiol. 2005, 43, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, C.; Vijayalakshmi, P.; Alex, A.M.; Alothaim, A.S.; Vijayakumar, R.; Umapathy, V.R. Chapter Three—Metalloproteins Structural and Functional Insights into Immunological Patterns. In Advances in Protein Chemistry and Structural Biology; Academic Press: Cambridge, MA, USA, 2024. [Google Scholar]
- Permyakov, E.A. Metal Binding Proteins. Encyclopedia 2021, 1, 261–292. [Google Scholar] [CrossRef]
- Gallego, P.; Martin, L.; Blazquez, A.; Guerra, H.; Villalobos, N. Involvement of Peroxidase Activity in Developing Somatic Embryos of Medicago Arborea L. Identification of an Isozyme Peroxidase as Biochemical Marker of Somatic Embryogenesis. J.Plant Physiol. 2014, 171, 78–84. [Google Scholar] [CrossRef]
- Arnholdt-Schmitt, B.; Ragonezi, C.; Cardoso, H. Do Mitochondria Play a Central Role in Stress-Induced Somatic Embryogenesis? In In Vitro Embryogenesis in Higher Plants; Germana, M.A., Lambardi, M., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2016; Volume 1359, pp. 87–100. [Google Scholar]
- Belmonte, M.F.; Donald, G.; Reid, D.M.; Yeung, E.C.; Stasolla, C. Alterations of the Glutathione Redox State Improve Apical Meristem Structure and Somatic Embryo Quality in White Spruce (Picea glauca). J. Exp. Bot. 2005, 56, 2355–2364. [Google Scholar] [CrossRef]
- Pasternak, T.; Potters, G.; Caubergs, R.; Jansen, M.A.K. Complementary Interactions between Oxidative Stress and Auxins Control Plant Growth Responses at Plant, Organ, and Cellular Level. J. Experim. Bot. 2005, 56, 1991–2001. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ahmad, S.; Ali, A.; Guo, C.; Li, H.; Yu, J.; Zhang, Y.; Gao, X.; Guo, Y. Characterization of Somatic Embryogenesis Receptor-Like Kinase 4 as a Negative Regulator of Leaf Senescence in Arabidopsis. Cells 2019, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhou, J.; Kou, X.; Liu, Y.; Zhao, X.; Qin, G.; Wang, M.; Qian, G.; Li, W.; Huang, Y.; et al. Syntaxin of Plants71 Plays Essential Roles in Plant Development and Stress Response via Regulating pH Homeostasis. Front. Plant Sci. 2023, 14, 1198353. [Google Scholar] [CrossRef]
- Dobrenel, T.; Marchive, C.; Azzopardi, M.; Clément, G.; Moreau, M.; Sormani, R.; Robaglia, C.; Meyer, C. Sugar Metabolism and the Plant Target of Rapamycin Kinase: A Sweet operaTOR? Front. Plant Sci. 2013, 4, 93. [Google Scholar] [CrossRef] [PubMed]
- Ju, L.; Pan, Z.; Zhang, H.; Li, Q.; Liang, J.; Deng, G.; Yu, M.; Long, H. New Insights into the Origin and Evolution of α-Amylase Genes in Green Plants. Sci. Rep. 2019, 9, 4929. [Google Scholar] [CrossRef]
- Damaris, R.N.; Lin, Z.; Yang, P.; He, D. The Rice Alpha-Amylase, Conserved Regulator of Seed Maturation and Germination. Int. J. Mol. Sci. 2019, 20, 450. [Google Scholar] [CrossRef]
- Swaisgood, H.E. Characteristics of Milk. In Fennema’s Food Chemistry; CRC Press: Boca Raton, FL, USA, 2007; pp. 885–921. [Google Scholar]
- Simpson, J.P.; Olson, J.; Dilkes, B.; Chapple, C. Identification of the Tyrosine- and Phenylalanine-Derived Soluble Metabolomes of Sorghum. Front. Plant Sci. 2021, 12, 714164. [Google Scholar] [CrossRef]
- Greenwell, Z.L.; Ruter, J.M. Effect of Glutamine and Arginine on Growth of Hibiscus Moscheutos “in Vitro”. Ornam. Hortic. 2018, 24, 393–399. [Google Scholar] [CrossRef]
- Okumoto, S.; Funck, D.; Trovato, M.; Forlani, G. Editorial: Amino Acids of the Glutamate Family: Functions beyond Primary Metabolism. Front. Plant Sci. 2016, 7, 318. [Google Scholar] [CrossRef]
- Cruzat, V.; Macedo Rogero, M.; Noel Keane, K.; Curi, R.; Newsholme, P. Glutamine: Metabolism and Immune Function, Supplementation and Clinical Translation. Nutrients 2018, 10, 1564. [Google Scholar] [CrossRef]
- Efzueni Rozali, S.; Rashid, K.A.; Mat Taha, R. Micropropagation of an Exotic Ornamental Plant, Calathea crotalifera, for Production of High Quality Plantlets. Sci. World J. 2014, 2014, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Hwang, K.S.; Choi, P.S. Effect of Carbon Sources on Somatic Embryogenesis and Cotyledon Number Variations in Carrot (Daucus carota L.). J. Plant Biotechnol. 2023, 50, 12. [Google Scholar] [CrossRef]
- Wang, G.; Liu, Y.; Gao, Z.; Li, H.; Wang, J. Effects of Amino Acids on Callus Proliferation and Somatic Embryogenesis in Litchi chinensis Cv. ‘Feizixiao’. Horticulturae 2023, 9, 1311. [Google Scholar] [CrossRef]
- Bartos, P.M.C.; Gomes, H.T.; Do Amaral, L.I.V.; Teixeira, J.B.; Scherwinski-Pereira, J.E. Biochemical Events during Somatic Embryogenesis in Coffea arabica L. 3 Biotech 2018, 8, 209. [Google Scholar] [CrossRef] [PubMed]
- Pescador, R.; Kerbauy, G.B.; Kraus, J.E.; De Melo Ferreira, W.; Guerra, M.P.; Figueiredo-Ribeiro, R.D.C.L. Changes in Soluble Carbohydrates and Starch Amounts during Somatic and Zygotic Embryogenesis of Acca sellowiana (Myrtaceae). Vitr. Cell. Dev. Biol. Plant 2008, 44, 289–299. [Google Scholar] [CrossRef]
- Xie, J.; Cai, K.; Hu, H.-X.; Jiang, Y.-L.; Yang, F.; Hu, P.; Cao, D.; Li, W.; Chen, Y.; Zhou, C.; et al. Structural Analysis of the CatalyticMechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase. J. Biol. Biol. Chem. 2016, 291, 25667. [Google Scholar] [CrossRef]



| Method | Lyophilized (µg/mL) | Non-Lyophilized (µg/mL) |
|---|---|---|
| Acetone | 8.93 | nt |
| TCA/Acetone | 54.00 | nt |
| Methanol/Chloroform | nt | nt |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pires, R.; Rodrigues, L.; Santos, F.M.; Duarte, I.F.; Ciordia, S.; Peixe, A.; Cardoso, H. Establishment of a Protocol for the Characterization of Secreted Biomolecules in Somatic Embryogenic Cultures of Olea europaea L. Horticulturae 2025, 11, 331. https://doi.org/10.3390/horticulturae11030331
Pires R, Rodrigues L, Santos FM, Duarte IF, Ciordia S, Peixe A, Cardoso H. Establishment of a Protocol for the Characterization of Secreted Biomolecules in Somatic Embryogenic Cultures of Olea europaea L. Horticulturae. 2025; 11(3):331. https://doi.org/10.3390/horticulturae11030331
Chicago/Turabian StylePires, Rita, Lénia Rodrigues, Fátima Milhano Santos, Iola F. Duarte, Sergio Ciordia, Augusto Peixe, and Hélia Cardoso. 2025. "Establishment of a Protocol for the Characterization of Secreted Biomolecules in Somatic Embryogenic Cultures of Olea europaea L." Horticulturae 11, no. 3: 331. https://doi.org/10.3390/horticulturae11030331
APA StylePires, R., Rodrigues, L., Santos, F. M., Duarte, I. F., Ciordia, S., Peixe, A., & Cardoso, H. (2025). Establishment of a Protocol for the Characterization of Secreted Biomolecules in Somatic Embryogenic Cultures of Olea europaea L. Horticulturae, 11(3), 331. https://doi.org/10.3390/horticulturae11030331












