Recovery of Novel Sequence Variants in Chemically Mutagenized Seed and Vegetatively Propagated Coffea arabica L.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mutagenesis
2.2. DNA Extraction and ddRADseq
2.3. Mutation Discovery and Analysis
2.4. Functional Analysis of Missense Changes and Evaluation of Allele Ratios
3. Results
3.1. Novel Single-Nucleotide Variants Recovered in All Samples
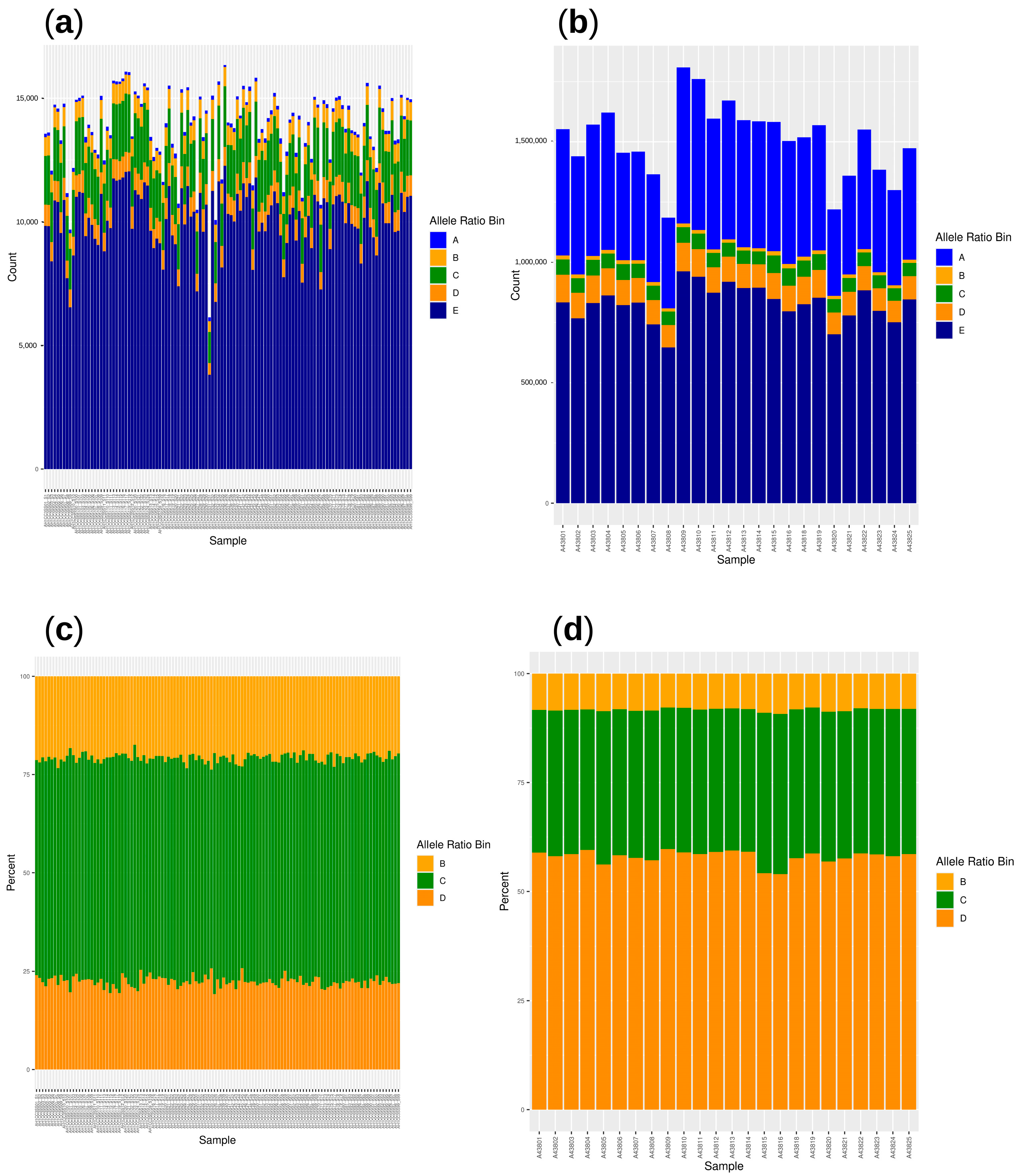
3.2. Ratio of Reads Supporting Reference and Alternative Alleles
3.3. Predicted Effect of Novel Variants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heckman, M.A.; Weil, J.; Gonzalez de Mejia, E. Caffeine (1, 3, 7-trimethylxanthine) in foods: A comprehensive review on consumption, functionality, safety, and regulatory matters. J. Food Sci. 2010, 75, R77–R87. [Google Scholar] [CrossRef] [PubMed]
- Quadra, G.R.; Paranaíba, J.R.; Vilas-Boas, J.; Roland, F.; Amado, A.M.; Barros, N.; Júnio, P.; Dias, R.; Cardoso, S.J. A global trend of caffeine consumption over time and related-environmental impacts. Environ. Pollut. 2020, 256, 113343. [Google Scholar] [CrossRef] [PubMed]
- Vegro, C.L.R.; de Almeida, L.F. Chapter 1—Global coffee market: Socio-economic and cultural dynamics. In Coffee Consumption and Industry Strategies in Brazil; de Almeida, L.F., Spers, E.E., Eds.; Woodhead Publishing: Sawston, CA, USA, 2020; pp. 3–19. [Google Scholar]
- Torga, G.N.; Spers, E.E. Chapter 2—Perspectives of global coffee demand. In Coffee Consumption and Industry Strategies in Brazil; de Almeida, L.F., Spers, E.E., Eds.; Woodhead Publishing: Cambridge, UK, 2020; pp. 21–49. [Google Scholar]
- Castellana, F.; De Nucci, S.; De Pergola, G.; Di Chito, M.; Lisco, G.; Triggiani, V.; Sardone, R.; Zupo, R. Trends in Coffee and Tea Consumption during the COVID-19 Pandemic. Foods 2021, 10, 2458. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.D.; Barkla, B.J.; Rose, T.J.; Liu, L. Does Coffee Have Terroir and How Should It Be Assessed? Foods 2022, 11, 1907. [Google Scholar] [CrossRef] [PubMed]
- Scholz, M.B.d.S.; Kitzberger, C.S.G.; Prudencio, S.H.; Silva, R.S.D.S.F. The typicity of coffees from different terroirs determined by groups of physico-chemical and sensory variables and multiple factor analysis. Food Res. Int. 2018, 114, 72–80. [Google Scholar] [CrossRef]
- Scalabrin, S.; Toniutti, L.; Di Gaspero, G.; Scaglione, D.; Magris, G.; Vidotto, M.; Pinosio, S.; Cattonaro, F.; Magni, F.; Jurman, I.; et al. A single polyploidization event at the origin of the tetraploid genome of Coffea arabica is responsible for the extremely low genetic variation in wild and cultivated germplasm. Sci. Rep. 2020, 10, 4642. [Google Scholar] [CrossRef]
- Scalabrin, S.; Magris, G.; Liva, M.; Vitulo, N.; Vidotto, M.; Scaglione, D.; Terra, L.D.; Ruosi, M.R.; Navarini, L.; Pellegrino, G.; et al. A chromosome-scale assembly reveals chromosomal aberrations and exchanges generating genetic diversity in Coffea arabica germplasm. Nat. Commun. 2024, 15, 463. [Google Scholar] [CrossRef]
- Aisala, H.; Kärkkäinen, E.; Jokinen, I.; Seppänen-Laakso, T.; Rischer, H. Proof of Concept for Cell Culture-Based Coffee. J. Agric. Food Chem. 2023, 71, 18478–18488. [Google Scholar] [CrossRef]
- Avila-Victor, C.M.; Ordaz-Chaparro, V.M.; Arjona-Suárez, E.d.J.; Iracheta-Donjuan, L.; Gómez-Merino, F.C.; Robledo-Paz, A. In Vitro Mass Propagation of Coffee Plants (Coffea arabica L. var. Colombia) through Indirect Somatic Embryogenesis. Plants 2023, 12, 1237. [Google Scholar] [CrossRef]
- Wintgens, J.N.; Zamarripa, C.A. Coffee Propagation. In Coffee: Growing, Processing, Sustainable Production; John Wiley & Sons: Hoboken, NJ, USA, 2004; pp. 87–136. [Google Scholar]
- Salojärvi, J.; Rambani, A.; Yu, Z.; Guyot, R.; Strickler, S.; Lepelley, M.; Wang, C.; Rajaraman, S.; Rastas, P.; Zheng, C.; et al. The genome and population genomics of allopolyploid Coffea arabica reveal the diversification history of modern coffee cultivars. Nat. Genet. 2024, 56, 721–731. [Google Scholar] [CrossRef]
- Henry, R.J. Innovations in plant genetics adapting agriculture to climate change. Curr. Opin. Plant Biol. 2020, 56, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Talhinhas, P.; Batista, D.; Diniz, I.; Vieira, A.; Silva, D.N.; Loureiro, A.; Tavares, S.; Pereira, A.P.; Azinheira, H.G.; Guerra-Guimarães, L.; et al. The coffee leaf rust pathogen Hemileia vastatrix: One and a half centuries around the tropics. Mol. Plant Pathol. 2017, 18, 1039–1051. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.J. Artificial Transmutation of the Gene. Science 1927, 66, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Stadler, L.J. Genetic Effects of X-rays in Maize. Proc. Natl. Acad. Sci. USA 1928, 14, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Auerbach, C.; Robson, J.M.; Carr, J.G. The Chemical Production of Mutations. Science 1947, 105, 243–247. [Google Scholar] [CrossRef]
- Jung, C.; Till, B. Mutagenesis and genome editing in crop improvement: Perspectives for the global regulatory landscape. Trends Plant Sci. 2021, 26, 1258–1269. [Google Scholar] [CrossRef]
- McCallum, C.M.; Comai, L.; Greene, E.A.; Henikoff, S. Targeted screening for induced mutations. Nat. Biotechnol. 2000, 18, 455–457. [Google Scholar] [CrossRef]
- Szurman-Zubrzycka, M.; Kurowska, M.; Till, B.J.; Szarejko, I. Is it the end of TILLING era in plant science? Front. Plant Sci. 2023, 14, 1160695. [Google Scholar] [CrossRef]
- Jankowicz-Cieslak, J.; Hofinger, B.J.; Jarc, L.; Junttila, S.; Galik, B.; Gyenesei, A.; Ingelbrecht, I.L.; Till, B.J. Spectrum and Density of Gamma and X-ray Induced Mutations in a Non-Model Rice Cultivar. Plants 2022, 11, 3232. [Google Scholar] [CrossRef]
- Du, Y.; Feng, Z.; Wang, J.; Jin, W.; Wang, Z.; Guo, T.; Chen, Y.; Feng, H.; Yu, L.; Li, W.; et al. Frequency and Spectrum of Mutations Induced by Gamma Rays Revealed by Phenotype Screening and Whole-Genome Re-Sequencing in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 654. [Google Scholar] [CrossRef]
- Li, S.; Zheng, Y.; Cui, H.; Fu, H.; Shu, Q.; Huang, J. Frequency and type of inheritable mutations induced by γ rays in rice as revealed by whole genome sequencing. J. Zhejiang Univ. Sci. B 2016, 17, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Till, B.J.; Datta, S.; Jankowicz-Cieslak, J. TILLING: The Next Generation. In Plant Genetics and Molecular Biology; Varshney, R.K., Pandey, M.K., Chitikineni, A., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 139–160. [Google Scholar]
- Till, B.J.; Reynolds, S.H.; Weil, C.; Springer, N.; Burtner, C.; Young, K.; Bowers, E.; Codomo, C.A.; Enns, L.C.; Odden, A.R.; et al. Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol. 2004, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Jankowicz-Cieslak, J.; Huynh, O.A.; Brozynska, M.; Nakitandwe, J.; Till, B.J. Induction, rapid fixation and retention of mutations in vegetatively propagated banana. Plant Biotechnol. J. 2012, 10, 1056–1066. [Google Scholar] [CrossRef] [PubMed]
- Krasileva, K.V.; Vasquez-Gross, H.A.; Howell, T.; Bailey, P.; Paraiso, F.; Clissold, L.; Simmonds, J.; Ramirez-Gonzalez, R.H.; Wang, X.; Borrill, P.; et al. Uncovering hidden variation in polyploid wheat. Proc. Natl. Acad. Sci. USA 2017, 114, E913–E921. [Google Scholar] [CrossRef] [PubMed]
- Jankowicz-Cieslak, J.; Till, B.J. Chemical Mutagenesis of Seed and Vegetatively Propagated Plants Using EMS. Curr. Protoc. Plant Biol. 2016, 1, 617–635. [Google Scholar] [CrossRef]
- Bolívar-González, A.; Valdez-Melara, M.; Gatica-Arias, A. Responses of Arabica coffee (Coffea arabica L. var. Catuaí) cell suspensions to chemically induced mutagenesis and salinity stress under in vitro culture conditions. Vitro Cell. Dev. Biol. Plant 2018, 54, 576–589. [Google Scholar] [CrossRef]
- Vargas-Segura, C.; López-Gamboa, E.; Araya-Valverde, E.; Valdez-Melara, M.; Gatica-Arias, A. Sensitivity of Seeds to Chemical Mutagens, Detection of DNA Polymorphisms and Agro-Metrical Traits in M1 Generation of Coffee (Coffea arabica L.). J. Crop Sci. Biotechnol. 2019, 22, 451–464. [Google Scholar] [CrossRef]
- Rojas-Chacón, J.A.; Echeverría-Beirute, F.; Till, B.J.; Gatica-Arias, A. Enhancing coffee diversity: Insights into the impact of sodium azide mutagenesis on quantitative and qualitative traits in Coffea arabica L. Sci. Hortic. 2024, 330, 113043. [Google Scholar] [CrossRef]
- Rojas-Chacón, J.A.; Echeverría-Beirute, F.; Till, B.J.; Gatica-Arias, A. Assessment of Hemileia vastatrix resistance in chemically mutagenized Coffea arabica L. leaf discs and the emergence of a novel resistance scale. J. Plant Pathol. 2024, 106, 1093–1106. [Google Scholar] [CrossRef]
- Gatica-Arias, A.; Bolívar-González, A. Chemical Mutagenesis of Embryogenic Cell Suspensions of Coffea arabica L. var. Catuaí Using EMS and NaN3. In Mutation Breeding in Coffee with Special Reference to Leaf Rust: Protocols; Ingelbrecht, I.L.W., Silva, M.D.C.L.D., Jankowicz-Cieslak, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2023; pp. 95–112. [Google Scholar]
- Teixeira, J.B.; Junqueira, C.S.; Pereira, A.J.P.d.C.; Mello, R.I.S.d.; Silva, A.P.D.d.; Mundim, D.A. Multiplicação clonal de café (Coffea arabica L.) via embriogênese somática. Embrapa Documentos 2004, 1–9. Available online: https://ainfo.cnptia.embrapa.br/digital/bitstream/CENARGEN/24709/1/doc121.pdf (accessed on 27 July 2024).
- van Boxtel, J.; Berthouly, M. High frequency somatic embryogenesis from coffee leaves. Plant Cell Tissue Organ Culture 1996, 44, 7–17. [Google Scholar] [CrossRef]
- Gatica-Arias, A.; Bolívar-González, A.; Sánchez-Barrantes, E.; Araya-Valverde, E.; Molina-Bravo, R. High Resolution Melt (HRM) Genotyping for Detection of Induced Mutations in Coffee (Coffea arabica L. var. Catuaí). In Mutation Breeding in Coffee with Special Reference to Leaf Rust: Protocols; Ingelbrecht, I.L.W., Silva, M.D.C.L.D., Jankowicz-Cieslak, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2023; pp. 275–290. [Google Scholar]
- Doyle, J. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Cooke, T.F.; Fischer, C.R.; Wu, P.; Jiang, T.X.; Xie, K.T.; Kuo, J.; Doctorov, E.; Zehnder, A.; Khosla, C.; Chuong, C.M.; et al. Genetic Mapping and Biochemical Basis of Yellow Feather Pigmentation in Budgerigars. Cell 2017, 171, 427–439.e21. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Till, B.J.; Jiménez-Madrigal, J.P.; Gatica-Arias, A. Identification of Novel Induced Mutations in Seed and Vegetatively Propagated Plants from Reduced Representation or Whole Genome Sequencing Data. Methods Mol. Biol. 2024, 2787, 123–139. [Google Scholar]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118, iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Yan, W.; Deng, X.W.; Yang, C.; Tang, X. The Genome-Wide EMS Mutagenesis Bias Correlates with Sequence Context and Chromatin Structure in Rice. Front. Plant Sci. 2021, 12, 579675. [Google Scholar] [CrossRef] [PubMed]
- Gauffier, C.; Lebaron, C.; Moretti, A.; Constant, C.; Moquet, F.; Bonnet, G.; Caranta, C.; Gallois, J.L. A TILLING approach to generate broad-spectrum resistance to potyviruses in tomato is hampered by eIF4E gene redundancy. Plant J. 2016, 85, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, B.; Singh, L.; Wallace, S.; Schoen, A.; Dong, Y.; Tiwari, V.; Rawat, N. Screening of an Ethyl Methane Sulfonate Mutagenized Population of a Wheat Cultivar Susceptible to Fusarium Head Blight Identifies Resistant Variants. Plant Dis. 2021, 105, 3669–3676. [Google Scholar] [CrossRef] [PubMed]
- Desiderio, F.; Torp, A.M.; Valè, G.; Rasmussen, S.K. TILLING in Plant Disease Control. In Plant Pathogen Resistance Biotechnology; John Wiley & Sons: Hoboken, NJ, USA, 2016; pp. 365–384. [Google Scholar]
- Hussain, M.; Iqbal, M.A.; Till, B.J.; Rahman, M.U. Identification of induced mutations in hexaploid wheat genome using exome capture assay. PLoS ONE 2018, 13, e0201918. [Google Scholar] [CrossRef]
- Acevedo-Garcia, J.; Spencer, D.; Thieron, H.; Reinstädler, A.; Hammond-Kosack, K.; Phillips, A.L.; Panstruga, R. Mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnol. J. 2017, 15, 367–378. [Google Scholar] [CrossRef]
- Lunde, C.; Seong, K.; Kumar, R.; Deatker, A.; Chhabra, B.; Wang, M.; Kaur, S.; Song, S.; Palayur, A.; Davies, C.; et al. Wheat Enhanced Disease Resistance EMS-Mutants Include Lesion-mimics with Adult Plant Resistance to Stripe Rust. bioRxiv 2024. bioRxiv:2024.05.10.593581. [Google Scholar]
- Till, B.J.; Reynolds, S.H.; Greene, E.A.; Codomo, C.A.; Enns, L.C.; Johnson, J.E.; Burtner, C.; Odden, A.R.; Young, K.; Taylor, N.E.; et al. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 2003, 13, 524–530. [Google Scholar] [CrossRef]
- Lababidi, S.; Mejlhede, N.; Rasmussen, S.K.; Backes, G.; Al-Said, W.; Baum, M.; Jahoor, A. Identification of barley mutants in the cultivar ‘Lux’ at the Dhn loci through TILLING. Plant Breed 2009, 128, 332–336. [Google Scholar] [CrossRef]
- Olsen, O.; Wang, X.; von Wettstein, D. Sodium azide mutagenesis: Preferential generation of A.T-->G.C transitions in the barley Ant18 gene. Proc. Natl. Acad. Sci. USA 1993, 90, 8043–8047. [Google Scholar] [CrossRef]
- Talamè, V.; Bovina, R.; Sanguineti, M.C.; Tuberosa, R.; Lundqvist, U.; Salvi, S. TILLMore, a resource for the discovery of chemically induced mutants in barley. Plant Biotechnol. J. 2008, 6, 477–485. [Google Scholar] [CrossRef]
- Spinoso-Castillo, J.L.; Escamilla-Prado, E.; Aguilar-Rincón, V.H.; Morales Ramos, V.; De Los Santos, G.G.; Pérez-Rodríguez, P.; Corona-Torres, T. Genetic diversity of coffee (Coffea spp.) in Mexico evaluated by using DArTseq and SNP markers. Genet. Resour. Crop Evol. 2020, 67, 1795–1806. [Google Scholar] [CrossRef]
- Cariou, M.; Duret, L.; Charlat, S. How and how much does RAD-seq bias genetic diversity estimates? BMC Evol. Biol. 2016, 16, 240. [Google Scholar] [CrossRef] [PubMed]
- Bresadola, L.; Link, V.; Buerkle, C.A.; Lexer, C.; Wegmann, D. Estimating and accounting for genotyping errors in RAD-seq experiments. Mol. Ecol. Resour. 2020, 20, 856–870. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.R. Variant calling in polyploids for population and quantitative genetics. Appl. Plant Sci. 2024, 12, e11607. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Till, B.J.; Jiménez-Madrigal, J.P.; Herrera-Estrella, A.; Atriztán-Hernández, K.; Gatica-Arias, A. Recovery of Novel Sequence Variants in Chemically Mutagenized Seed and Vegetatively Propagated Coffea arabica L. Horticulturae 2024, 10, 1077. https://doi.org/10.3390/horticulturae10101077
Till BJ, Jiménez-Madrigal JP, Herrera-Estrella A, Atriztán-Hernández K, Gatica-Arias A. Recovery of Novel Sequence Variants in Chemically Mutagenized Seed and Vegetatively Propagated Coffea arabica L. Horticulturae. 2024; 10(10):1077. https://doi.org/10.3390/horticulturae10101077
Chicago/Turabian StyleTill, Bradley J., José P. Jiménez-Madrigal, Alfredo Herrera-Estrella, Karina Atriztán-Hernández, and Andrés Gatica-Arias. 2024. "Recovery of Novel Sequence Variants in Chemically Mutagenized Seed and Vegetatively Propagated Coffea arabica L." Horticulturae 10, no. 10: 1077. https://doi.org/10.3390/horticulturae10101077
APA StyleTill, B. J., Jiménez-Madrigal, J. P., Herrera-Estrella, A., Atriztán-Hernández, K., & Gatica-Arias, A. (2024). Recovery of Novel Sequence Variants in Chemically Mutagenized Seed and Vegetatively Propagated Coffea arabica L. Horticulturae, 10(10), 1077. https://doi.org/10.3390/horticulturae10101077






