Construction of SNP-PARMS Fingerprints and Analysis of Genetic Diversity in Taro (Colocasia esculenta)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Simplified Genome Resequencing
2.3. Data Processing and SNP Discovery
2.4. SNP Screening and Primer Designing
2.5. Development of PARMS-SNP Markers
2.6. Analysis of Genetic Diversity
2.7. Construction of Taro Fingerprint Spectra
3. Results
3.1. Simplified Genome Resequencing and Reference Genome Alignment
3.2. SNP Diversity Analysis
3.3. Development and Validation of High-Quality PARMS-SNP Markers
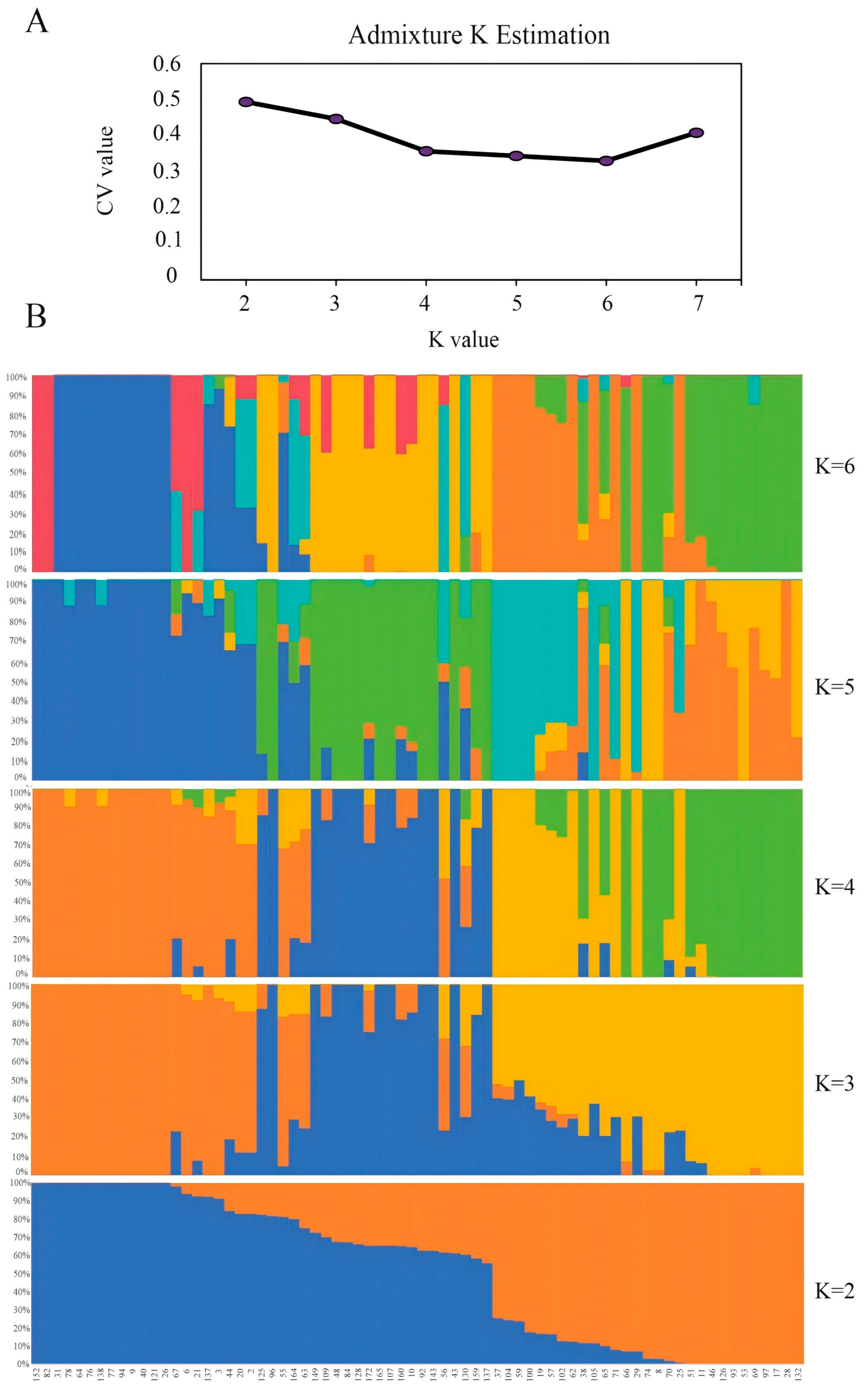
3.4. Analysis of Genetic Diversity Among Taro Resources
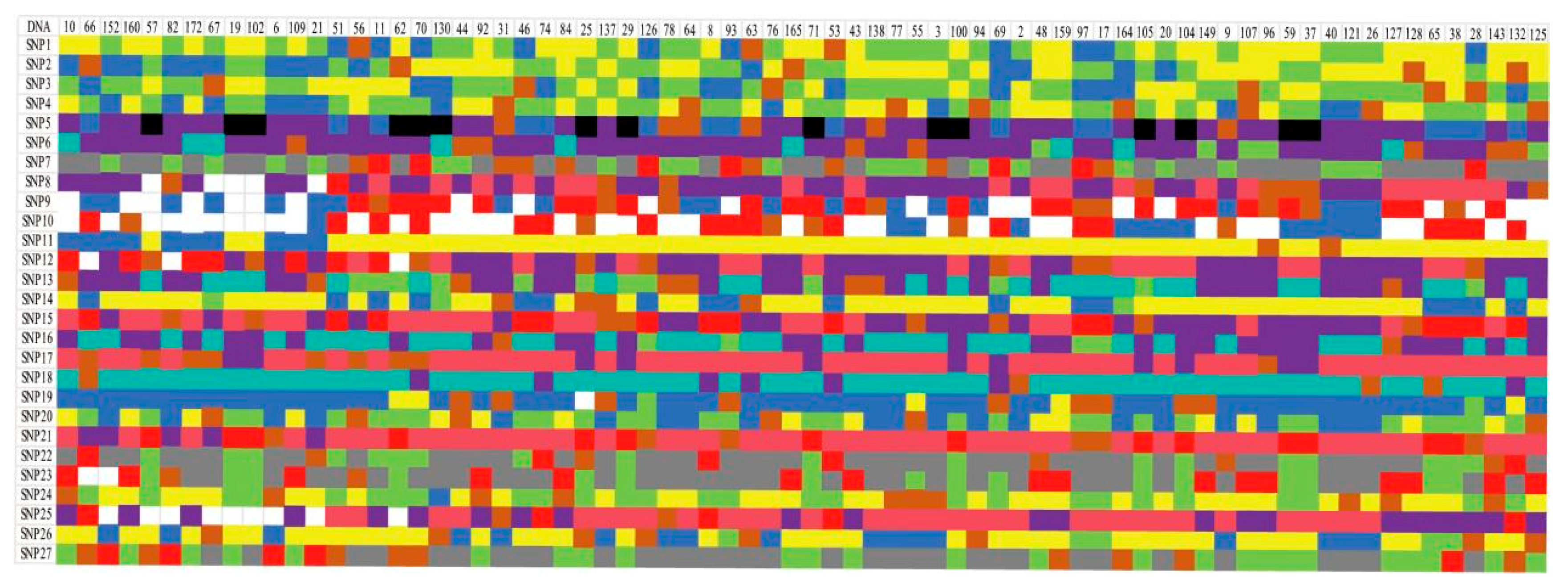
3.5. Construction of PARMS-SNP Fingerprint Maps
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. FAOSTAT: Land Use. 2020. Available online: http://www.fao.org/faostat/en/#data/RL (accessed on 15 June 2024).
- Lebot, V. Tropical Root and Tuber Crops: Cassava, Sweet Potato, Yams and Aroids, 2nd ed.; CABI: Wallingford, UK, 2019. [Google Scholar]
- Wu, W.; Chang, Q.T.; Wang, A. Research progress on taro (Colocasia esculenta) germplasm resources in China. Anhui Agric. Sci. Bull. 2021, 49, 4–7. [Google Scholar]
- Zhang, H.; Teng, C.; Hou, W.; Liu, Y. Construction of core germplasm resources of broad beans using SNP molecular markers. In Proceedings of the 20th Academic Annual Conference of the Chinese Crops Society, Changsha, China, 4 November 2023; Qinghai University: Xining, China, 2023; Volume 1. [Google Scholar]
- Grover, A.; Sharma, P.C. Development and use of molecular markers: Past and present. Crit. Rev. Biotechnol. 2016, 36, 290–302. [Google Scholar] [CrossRef]
- Mammadov, J.; Aggarwal, R.; Buyyarapu, R.; Kumpatla, S. SNP markers and their impact on plant breeding. Int. J. Plant Genom. 2012, 2012, 728398. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Heo, J.H.; Yeon, J.; Jung, J.K.; Shin, I.S.; Sim, S.C. Development of Cost-Effective SNP Markers for Genetic Variation Analysis and Variety Identification in Cultivated Pears (Pyrus spp.). Plants 2024, 13, 2600. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, Z.; Sun, Y.; Huang, X.; Li, F.; Liu, Y.; Zhu, H.; Liu, Z.; Ke, W. Genetic diversity and population structure of eddoe taro in China using genome-wide SNP markers. PeerJ 2020, 8, e10485. [Google Scholar] [CrossRef]
- Pan, R.; Zhu, Q.; Jia, X.; Li, B.; Li, Z.; Xiao, Y.; Luo, S.; Wang, S.; Shan, N.; Sun, J.; et al. Genome-Wide Development of InDel-SSRs and Association Analysis of Important Agronomic Traits of Taro (Colocasia esculenta) in China. Curr. Issues Mol. Biol. 2024, 46, 13347–13363. [Google Scholar] [CrossRef]
- Guo, J.; Wu, T.; Li, M.; Tang, K.; Yao, C.; Li, G.; Luo, W.; Wang, R. Genetic diversity analysis and disease resistance identification of taro germplasm resources based on SSR markers. Chin. J. Agron. 2022, 38, 112–119. [Google Scholar]
- Seki, S.; Kawaguchi, Y.; Chiba, K.; Mikami, Y.; Kizawa, H.; Oya, T.; Mio, F.; Mori, M.; Miyamoto, Y.; Masuda, I.; et al. A functional SNP in CILP, encoding cartilage intermediate layer protein, is associated with susceptibility to lumbar discdisease. Nat. Genet. 2005, 37, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Yao, Z.; Yang, Y.; Wang, Z.; Zou, H.; Zhang, X.; Chen, J.; Fang, B.; Huang, L. Genetic fingerprint construction and genetic diversity analysis of sweet potato (Ipomoea batatas) germplasm resources. BMC Plant Biol. 2023, 23, 355. [Google Scholar] [CrossRef]
- Li, J.; Chang, X.; Huang, Q.; Liu, P.; Zhao, X.; Li, F.; Wang, Y.; Chang, C. Construction of SNP fingerprint and population genetic analysis of honeysuckle germplasm resources in China. Front. Plant Sci. 2023, 14, 1080691. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, W.; Yang, Z.; Luo, C.; Zhang, W.; Xu, F.; Ye, J.; Liao, Y. Genetic diversity analysis and DNA fingerprint construction of Zanthoxylum species based on SSR and iPBS markers. BMC Plant Biol. 2024, 24, 843. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.L.; Liu, G.; Aisa, H.A. Quantitative Determination of Marker Compounds and Fingerprint Analysis of the Seeds of Vernonia anthelmintica. Int. J. Anal. Chem. 2020, 2020, 8859425. [Google Scholar] [CrossRef]
- Li, C.; Tian, Y.; Zhao, C.; Li, S.; Wang, T.; Qiao, B.; Fu, Y. Application of fingerprint combined with quantitative analysis and multivariate chemometric methods in quality evaluation of dandelion (Taraxacum mongolicum). R. Soc. Open Sci. 2021, 8, 210614. [Google Scholar] [CrossRef]
- Guo, R.; Xia, X.; Chen, J.; An, Y.; Mi, X.; Li, R.; Zhang, C.; Chen, M.; Wei, C.; Liu, S. Genetic relationship analysis and molecular fingerprint identification of the tea germplasms from Guangxi Province, China. Breed. Sci. 2021, 71, 584–593. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, H.; Xiang, X.; Yang, A.; Feng, Q.; Dai, P.; Li, Y.; Jiang, X.; Liu, G.; Zhang, X. Construction of a SNP Fingerprinting Database and Population Genetic Analysis of Cigar Tobacco Germplasm Resources in China. Front. Plant Sci. 2021, 12, 618133. [Google Scholar] [CrossRef]
- Xing, X.; Hu, T.; Wang, Y.; Li, Y.; Wang, W.; Hu, H.; Wei, Q.; Yan, Y.; Gan, D.; Bao, C.; et al. Construction of SNP fingerprints and genetic diversity analysis of radish (Raphanus sativus L.). Front. Plant Sci. 2024, 15, 1329890. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yin, J.; Jiang, L.; Wang, L.; Han, X.; Guo, W.; Li, C.; Zhou, Y.; Denton, M.; Zhang, P. A high-quality genome of taro (Colocasia esculenta (L.) Schott), one of the world’s oldest crops. Mol. Ecol. Resour. 2021, 21, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Zhu, P.; He, L.; Li, Y.; Huang, W.; Xi, F.; Lin, L.; Zhi, Q.; Zhang, W.; Tang, Y.T.; Geng, C.; et al. Correction: OTG-snpcaller: An Optimized Pipeline Based on TMAP and GATK for SNP Calling from Ion Torrent Data. Electronic 2015, 26, 1932–6203. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from next-generation sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Liu, K.J.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Zhang, Z. Preliminary exploration of horticultural classification of taro. Chin. Veg. 1984. [Google Scholar] [CrossRef]
- Oladimeji, J.J.; Abe, A.; Kumar, P.L.; Agre, P.A.; Ilesanmi, O.J.; Vetukuri, R.R.; Bhattacharjee, R. Extent and patterns of morphological and molecular genetic diversity and population structure of Nigerian Taro cultivars. BMC Plant Biol. 2024, 24, 1077. [Google Scholar] [CrossRef] [PubMed]
- Ebert, A.W.; Waqainabete, L.M. Conserving and Sharing Taro Genetic Resources for the Benefit of Global Taro Cultivation: A Core Contribution of the Centre for Pacific Crops and Trees. Biopreserv. Biobank. 2018, 16, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Tahara, M.; Nguyen, V.X.; Yoshino, H. Isozyme analyses of Asian diploid and triploid taro, Colocasia esculenta (L.) Shott. Aroideana 1999, 22, 72–78. [Google Scholar]
- Fu, X.Q.; Yang, F.X.; Lu, X.K.; Wang, X.G.; Yang, B.X.; Liu, F.J.; Liu, Y.; Peng, J. Molecular identification of variety purity in a cotton hybrid with unknown parentage using DNA-SSR markers. Genet. Mol. Res. 2017, 16, 1–8. [Google Scholar] [CrossRef]



| Sample ID | Source | Ploidy | Classification | Group | Sample ID | Source | Ploidy | Classification | Group |
|---|---|---|---|---|---|---|---|---|---|
| 0 | Hunan | 2× | Corm type | A | 114 | Hunan | 2× | Corm type | A |
| 3 | Hunan | 2× | Corm type | A | 133 | Hunan | 2× | Corm type | A |
| 4 | Hunan | 2× | Corm type | A | 34 | Hunan | 2× | Leaf type | B |
| 11 | Hunan | 2× | Corm type | A | 45 | Hunan | 2× | Leaf type | B |
| 23 | Hunan | 2× | Corm type | A | 60 | Hunan | 2× | Leaf type | B |
| 38 | Hunan | 2× | Corm type | A | 2 | Hunan | 3× | Corm type | C |
| 65 | Hunan | 2× | Corm type | A | 21 | Yunnan | 3× | Corm type | C |
| 69 | Hunan | 2× | Corm type | A | 75 | Hunan | 3× | Corm type | C |
| 70 | Hunan | 2× | Corm type | A | 78 | Hunan | 3× | Corm type | C |
| 71 | Fujian | 2× | Corm type | A | 80 | Hunan | 3× | Corm type | C |
| 77 | Hunan | 2× | Corm type | A | 85 | Hunan | 3× | Corm type | C |
| 79 | Hunan | 2× | Corm type | A | 94 | Hunan | 3× | Corm type | C |
| 83 | Hunan | 2× | Corm type | A | 96 | Jiangxi | 3× | Corm type | C |
| 98 | Hunan | 2× | Corm type | A | 115 | Hunan | 3× | Corm type | C |
| 111 | Hunan | 2× | Corm type | A |
| ID | Chr | Position | Ref | Alt | Sequence of Primer X | Sequence of Primer Y | Sequence of Primer Z |
|---|---|---|---|---|---|---|---|
| SNP1 | LG01 | 142740049 | C | T | GAAGGTGACCAAGTTCATGCTTGACTTCTGTGGATTATAGCTCGC | GAAGGTCGGAGTCAACGGATTTGACTTCTGTGGATTATAGCTCGT | TCCATGCATTAAACTTGCCATTCA |
| SNP2 | LG01 | 208737408 | C | T | GAAGGTGACCAAGTTCATGCTTTTCTTGTAACACCCCGAAAATTC | GAAGGTCGGAGTCAACGGATTTTTCTTGTAACACCCCGAAAATTT | CGATAATGGCACGTATGTTTTTGG |
| SNP3 | LG02 | 64737033 | T | C | GAAGGTGACCAAGTTCATGCTGCTAAAGAATCAAAGTTAACAATAGATATCT | GAAGGTCGGAGTCAACGGATTGCTAAAGAATCAAAGTTAACAATAGATATCC | TCCAAGCAATAGTATGATCGCTGA |
| SNP4 | LG02 | 65417421 | T | C | GAAGGTGACCAAGTTCATGCTCACTAATTTATTATTTAGAAGAATGGCAGTT | GAAGGTCGGAGTCAACGGATTCACTAATTTATTATTTAGAAGAATGGCAGTC | TTGAGCATTGTCAGGTTGAATTCC |
| SNP5 | LG08 | 8979131 | C | G | GAAGGTGACCAAGTTCATGCTGGAAGAGACAGTTCTGATAGAAACAC | GAAGGTCGGAGTCAACGGATTGGAAGAGACAGTTCTGATAGAAACAG | TGCCATAGACCACTACATTCGATT |
| SNP6 | LG01 | 22020255 | G | T | GAAGGTGACCAAGTTCATGCTGGACCATTTAATTATCAAACATATGTAAAATAG | GAAGGTCGGAGTCAACGGATTGGACCATTTAATTATCAAACATATGTAAAATAT | TCTCTCTATCTCTCTCCTCTCTCT |
| SNP7 | LG01 | 48214697 | A | T | GAAGGTGACCAAGTTCATGCTCCCGGGAAGCACAAATATGTATTAA | GAAGGTCGGAGTCAACGGATTCCCGGGAAGCACAAATATGTATTAT | TATTTCCTTCTTCATCCTTGGCCA |
| SNP8 | LG12 | 99501267 | G | A | GAAGGTGACCAAGTTCATGCTCTGAACAGTAGCCACAAGTGACTG | GAAGGTCGGAGTCAACGGATTCTGAACAGTAGCCACAAGTGACTA | GGAGAAACTCATCAAGGGACCTTA |
| SNP9 | LG10 | 109034243 | T | G | GAAGGTGACCAAGTTCATGCTCAATTCAGCTGCTTACACACAAGA | GAAGGTCGGAGTCAACGGATTCAATTCAGCTGCTTACACACAAGC | TCCTTGTTGACGATCCAAGAAGAT |
| SNP10 | LG05 | 165161699 | A | C | GAAGGTGACCAAGTTCATGCTATTTTACCTTTCTTTAAGTAAGCTTTTGA | GAAGGTCGGAGTCAACGGATTATTTTACCTTTCTTTAAGTAAGCTTTTGC | GTCCATTAAACCCCAAACAACGAT |
| SNP11 | LG04 | 144128461 | T | C | GAAGGTGACCAAGTTCATGCTCGATCGACCGATAATGAAAGCCT | GAAGGTCGGAGTCAACGGATTCGATCGACCGATAATGAAAGCCC | GATTGCCTTGATCCACAATCGATT |
| SNP12 | LG14 | 867311 | A | G | GAAGGTGACCAAGTTCATGCTGTAATTACACTGGATTAAAAATATAACCTATGA | GAAGGTCGGAGTCAACGGATTGTAATTACACTGGATTAAAAATATAACCTATGG | GGGAGAAGTTTTGAGAAACAGTGG |
| SNP13 | LG04 | 148250105 | T | G | GAAGGTGACCAAGTTCATGCTGGTTTCCTCGTGTCAAACAGAAT | GAAGGTCGGAGTCAACGGATTGGTTTCCTCGTGTCAAACAGAAG | GTTTTCGGACTGCTCATCTCAAG |
| SNP14 | LG05 | 153062049 | C | T | GAAGGTGACCAAGTTCATGCTGACAACAAGAGGCAGCCTGC | GAAGGTCGGAGTCAACGGATTGACAACAAGAGGCAGCCTGT | TATAGAGTCGTGCTGTCAAAGACC |
| SNP15 | LG11 | 89325049 | T | C | GAAGGTGACCAAGTTCATGCTTCTGTGTGGTTCTCCGGCA | GAAGGTCGGAGTCAACGGATTTCTGTGTGGTTCTCCGGCG | TATGCTCAGGAGATGAAGAAGAGG |
| SNP16 | LG08 | 170119348 | C | A | GAAGGTGACCAAGTTCATGCTCGAATTTGTCAGAGATTGCTTCCC | GAAGGTCGGAGTCAACGGATTCGAATTTGTCAGAGATTGCTTCCA | TCAAAACAAGCTGTAGAGACCTCA |
| SNP17 | LG10 | 7248248 | A | G | GAAGGTGACCAAGTTCATGCTCCTGGCTTTCCTGTTCACGA | GAAGGTCGGAGTCAACGGATTCCTGGCTTTCCTGTTCACGG | AGGTATTTTCCATGATTCCTGGGT |
| SNP18 | LG06 | 130247652 | G | T | GAAGGTGACCAAGTTCATGCTGCACTCATGGAGGGGGCG | GAAGGTCGGAGTCAACGGATTGCACTCATGGAGGGGGCT | AAGTGGGTATGAGAGAAGAACCAC |
| SNP19 | LG08 | 68668940 | G | A | GAAGGTGACCAAGTTCATGCTTTGGTTATTCCATATTTAGATCTATAGATAGAG | GAAGGTCGGAGTCAACGGATTTTGGTTATTCCATATTTAGATCTATAGATAGAA | TAATAGTAGCGGGGCTTTACATCC |
| SNP20 | LG08 | 17307221 | T | C | GAAGGTGACCAAGTTCATGCTGTACACGTCCCTCTGCTTCAT | GAAGGTCGGAGTCAACGGATTGTACACGTCCCTCTGCTTCAC | TCAAGCTCTCACTGTGATGGTTTA |
| SNP21 | LG14 | 102781792 | T | C | GAAGGTGACCAAGTTCATGCTAGATCTCTTCACTTGTCAAGGATAAATAT | GAAGGTCGGAGTCAACGGATTAGATCTCTTCACTTGTCAAGGATAAATAC | AGCAGACAGATCAAGCTCTGAAAT |
| SNP22 | LG13 | 51666285 | T | A | GAAGGTGACCAAGTTCATGCTTCACAACTGGATACACTTTCAAAGT | GAAGGTCGGAGTCAACGGATTTCACAACTGGATACACTTTCAAAGA | GGAAAGTCTTGTTAAGGCACACTC |
| SNP23 | LG01 | 206486904 | T | A | GAAGGTGACCAAGTTCATGCTATGCCTACAATGCACACCTTCTCT | GAAGGTCGGAGTCAACGGATTATGCCTACAATGCACACCTTCTCA | TCTCAATCATCTCCTCCAGCAAAA |
| SNP24 | LG04 | 64252159 | T | C | GAAGGTGACCAAGTTCATGCTTAGGTCTCCATGTTCAAGAGGAAT | GAAGGTCGGAGTCAACGGATTTAGGTCTCCATGTTCAAGAGGAAC | AATTTGGATTACCACGTGTTGCTT |
| SNP25 | LG02 | 185181757 | G | A | GAAGGTGACCAAGTTCATGCTTTGGGAAAGGGCTGACAAGG | GAAGGTCGGAGTCAACGGATTTTGGGAAAGGGCTGACAAGA | CATCCACATCGAGAAGATTTTCGG |
| SNP26 | LG12 | 83510795 | A | G | GAAGGTGACCAAGTTCATGCTAGCTAAATAGTGAGTCAAAGGATCA | GAAGGTCGGAGTCAACGGATTAGCTAAATAGTGAGTCAAAGGATCG | CCTTACATCACAGTGCTTCACAAG |
| SNP27 | LG01 | 179908017 | A | T | GAAGGTGACCAAGTTCATGCTGAAGGCCTTGCTGAGAAGAGA | GAAGGTCGGAGTCAACGGATTGAAGGCCTTGCTGAGAAGAGT | GTGGACATAAACCTTGTGAAGAGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, S.; Chen, T.; Li, Q.; Wang, X.; Yang, J.; Wang, D. Construction of SNP-PARMS Fingerprints and Analysis of Genetic Diversity in Taro (Colocasia esculenta). Horticulturae 2025, 11, 1224. https://doi.org/10.3390/horticulturae11101224
Wu S, Chen T, Li Q, Wang X, Yang J, Wang D. Construction of SNP-PARMS Fingerprints and Analysis of Genetic Diversity in Taro (Colocasia esculenta). Horticulturae. 2025; 11(10):1224. https://doi.org/10.3390/horticulturae11101224
Chicago/Turabian StyleWu, Shuanghua, Tianxin Chen, Qian Li, Xin Wang, Jianguo Yang, and Duanhua Wang. 2025. "Construction of SNP-PARMS Fingerprints and Analysis of Genetic Diversity in Taro (Colocasia esculenta)" Horticulturae 11, no. 10: 1224. https://doi.org/10.3390/horticulturae11101224
APA StyleWu, S., Chen, T., Li, Q., Wang, X., Yang, J., & Wang, D. (2025). Construction of SNP-PARMS Fingerprints and Analysis of Genetic Diversity in Taro (Colocasia esculenta). Horticulturae, 11(10), 1224. https://doi.org/10.3390/horticulturae11101224






