RsNAC134 Regulates Taproot Skin Color via Positive Regulation of the Chlorophyll Degradation Pathway in Radish (Raphanus sativus)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Gene Expression Analysis
2.3. Vector Construction and Genetic Transformation
2.4. Subcellular Localization
2.5. Transactivation Assay
2.6. Dual-LUC Assay
2.7. Y1H Assay
2.8. Determination of Chlorophyll
2.9. Tobacco Transient Expression
2.10. Phylogenetic Tree Analysis
2.11. Statistical Analysis
3. Results
3.1. Differential Expression of RsNACs in ‘55’ and ‘QZ-16’
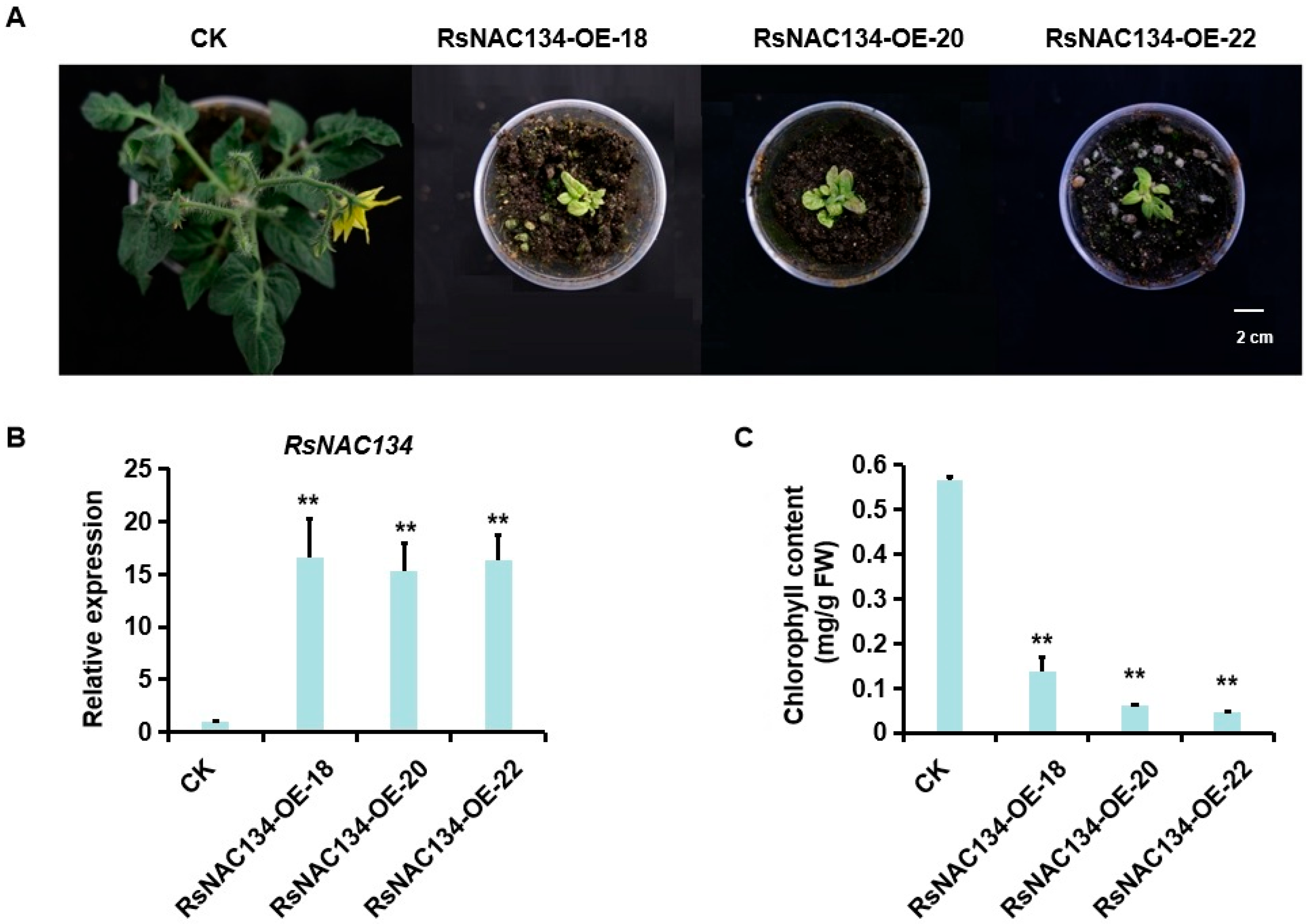
3.2. RsNAC134 Overexpression Caused Chlorosis in Tomatoes
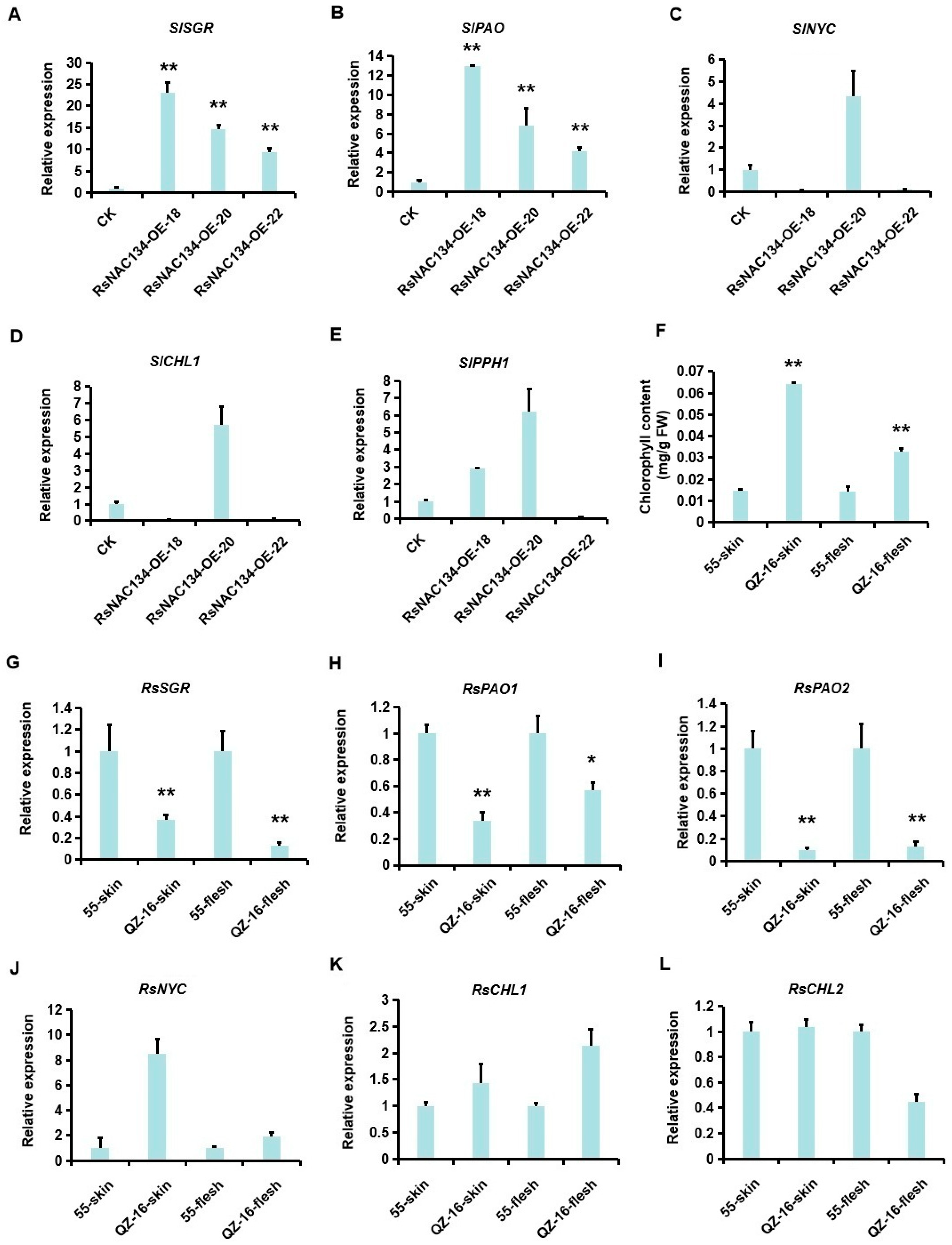
3.3. RsNAC134 Regulates the Expression of Chlorophyll Degradation Genes
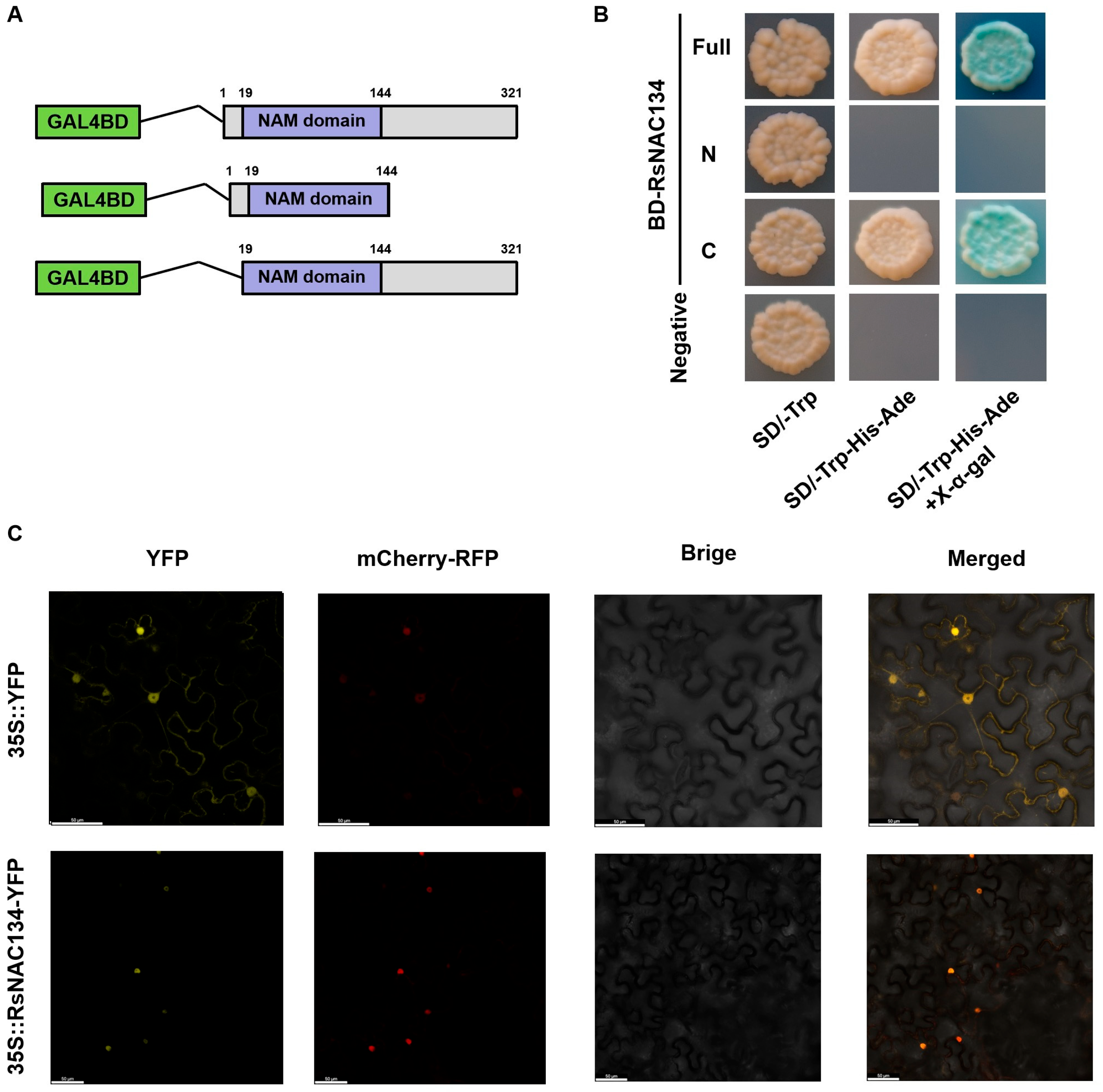
3.4. RsNAC134 Is a Transcriptional Activator That Localized in the Nucleus
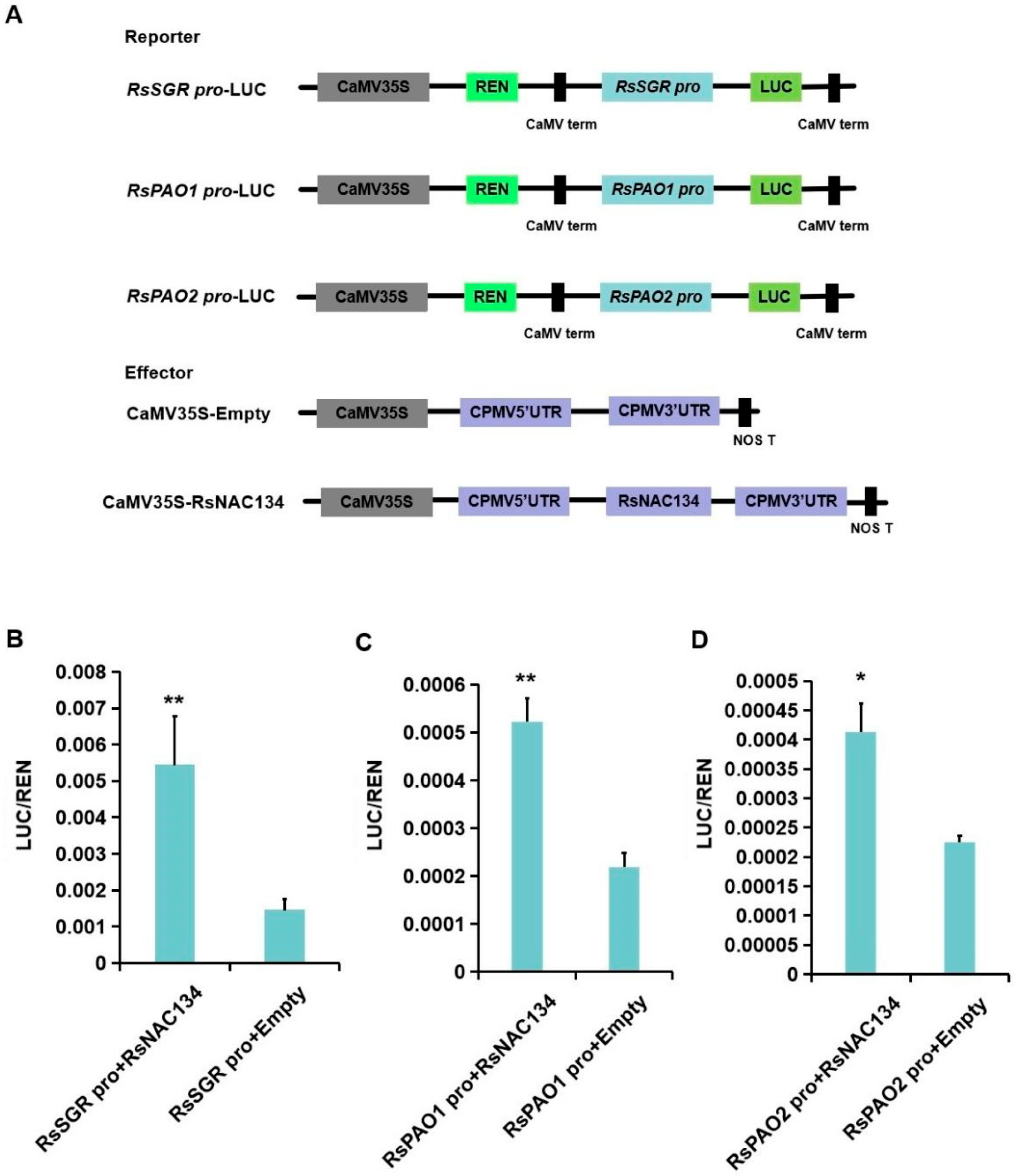
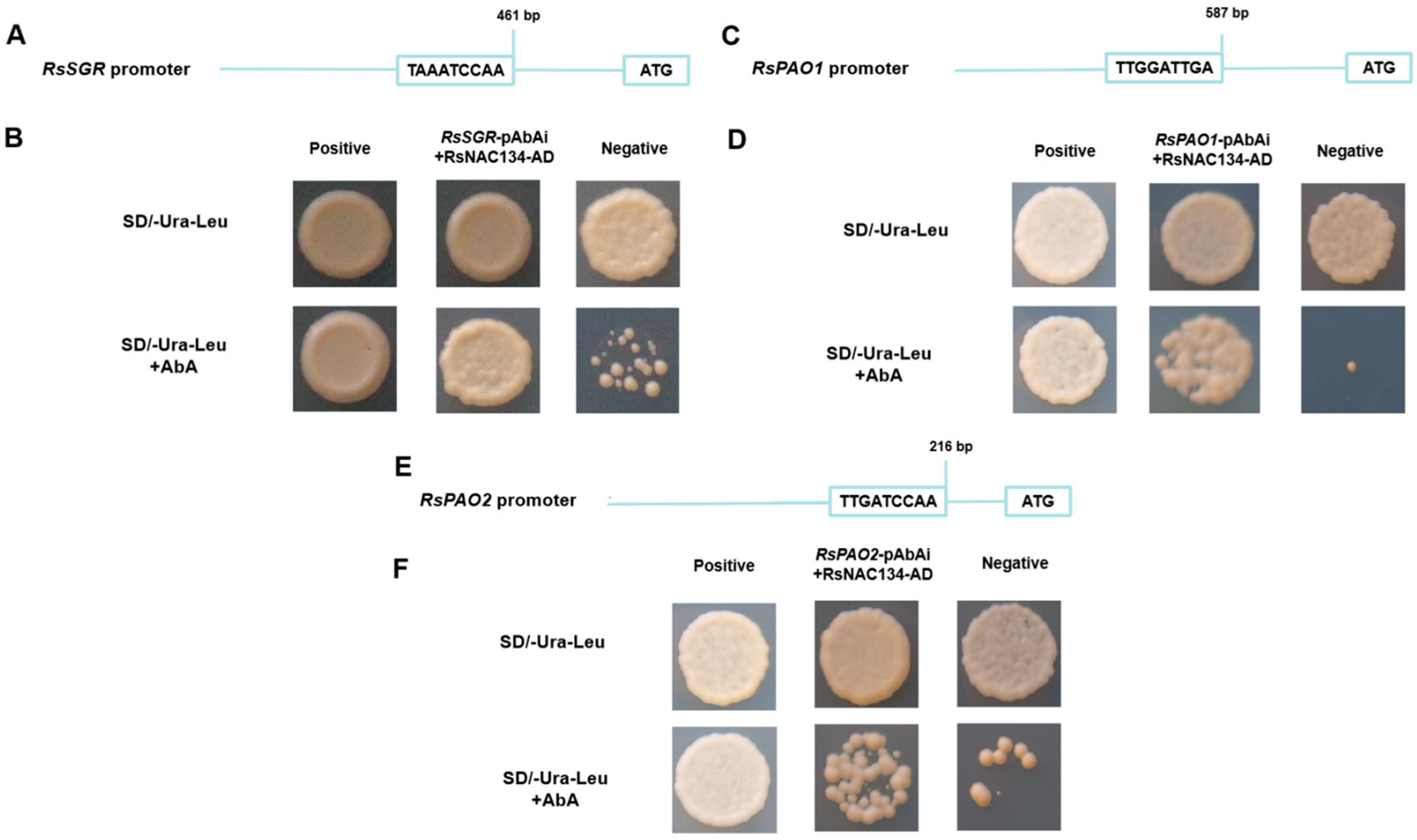
3.5. RsNAC134 Regulated RsSGR and RsPAO Expression by Directly Binding to Their Promoters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sakuraba, Y.; Schelbert, S.; Park, S.Y.; Han, S.H.; Lee, B.D.; Andrès, C.B.; Kessler, F.; Hörtensteiner, S.; Paek, N.C. STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex Ⅱ for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell 2012, 24, 507–518. [Google Scholar] [CrossRef]
- Shimoda, Y.; Ito, H.; Tanaka, A. Arabidopsis STAY-GREEN, Mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 2016, 28, 2147–2160. [Google Scholar] [CrossRef]
- Hörtensteiner, S. Update on the biochemistry of chlorophyll breakdown. Plant Mol. Biol. 2013, 82, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Armstead, I.; Donnison, I.; Aubry, S.; Harper, J.; Hörtensteiner, S.; James, C.; Mani, J.; Moffet, M.; Ougham, H.; Roberts, L.; et al. From crop to model to crop: Identifying the genetic basis of the staygreen mutation in the Lolium/Festuca forage and amenity grasses. New Phytol. 2006, 172, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Yu, J.W.; Park, J.S.; Li, J.; Yoo, S.C.; Lee, N.Y.; Lee, S.K.; Jeong, S.W.; Seo, H.S.; Koh, H.J.; et al. The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef] [PubMed]
- Armstead, I.; Donnison, I.; Aubry, S.; Harper, J.; Hörtensteiner, S.; James, C.; Mani, J.; Moffet, M.; Ougham, H.; Roberts, L.; et al. Cross-species identification of Mendel’s I locus. Science 2007, 315, 73. [Google Scholar] [CrossRef]
- Ono, K.; Kimura, M.; Matsuura, H.; Tanaka, A.; Ito, H. Jasmonate production through chlorophyll a degradation by Stay-Green in Arabidopsis thaliana. J. Plant Physiol. 2019, 238, 53–62. [Google Scholar] [CrossRef]
- Sato, T.; Shimoda, Y.; Matsuda, K.; Tanaka, A.; Ito, H. Mg-dechelation of chlorophyll a by Stay-Green activates chlorophyll b degradation through expressing Non-Yellow Coloring 1 in Arabidopsis thaliana. J. Plant Physiol. 2018, 222, 94–102. [Google Scholar] [CrossRef]
- Matsuda, K.; Shimoda, Y.; Tanaka, A.; Ito, H. Chlorophyll a is a favorable substrate for chlamydomonas Mg-dechelatase encoded by STAY-GREEN. Plant Physiol. Biochem. 2016, 109, 365–373. [Google Scholar] [CrossRef]
- Chung, D.W.; Pruzinska, A.; Hörtensteiner, S.; Ort, D.R. The role of pheophorbide a oxygenase expression and activity in the canola green seed problem. Plant Physiol. 2006, 142, 88–97. [Google Scholar] [CrossRef]
- Gray, J.; Close, P.S.; Briggs, S.P.; Johal, G.S. A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell 1997, 89, 25–31. [Google Scholar] [CrossRef]
- Li, P.; Ma, Y.; Li, X.; Zhang, L.; Wang, Y.; Wang, N. Cloning and expressional characterization of soybean GmLls1 gene. Chin. Sci. Bull. 2006, 51, 1210–1218. [Google Scholar] [CrossRef]
- Ma, N.; Ma, X.; Li, A.; Cao, X.; Kong, L. Cloning and expression analysis of wheat pheophorbide a oxygenase gene TaPaO. Plant Mol. Biol. Rep. 2012, 30, 1237–1245. [Google Scholar] [CrossRef]
- Spassieva, S.; Hille, J. A lesion mimic phenotype in tomato obtained by isolating and silencing an Lls1 homologue. Plant Sci. 2002, 162, 543–549. [Google Scholar] [CrossRef]
- Tang, Y.; Li, M.; Chen, Y.; Wu, P.; Wu, G.; Jiang, H. Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. J. Plant Physiol. 2011, 168, 1952–1959. [Google Scholar] [CrossRef] [PubMed]
- Hörtensteiner, S. Chlorophyll degradation during senescence. Annu. Rev. Plant Biol. 2006, 57, 55–77. [Google Scholar] [CrossRef]
- Hörtensteiner, S.; Krautler, B. Chlorophyll breakdown in higher plants. Biochim. Biophys. Acta. 2011, 1807, 977–988. [Google Scholar] [CrossRef]
- Xie, Q.; Liang, Y.; Zhang, J.; Zheng, H.; Dong, G.; Qian, Q.; Zuo, J. Involvement of a putative bipartite transit peptide in targeting rice pheophorbide a oxygenase into chloroplasts for chlorophyll degradation during leaf senescence. J. Genet. Genom. 2016, 43, 145–154. [Google Scholar] [CrossRef]
- Chen, W.; Provart, N.J.; Glazebrook, J.; Katagiri, F.; Chang, H.S.; Eulgem, T.; Mauch, F.; Luan, S.; Zou, G.; Whitham, S.A.; et al. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 2002, 14, 559–574. [Google Scholar] [CrossRef]
- Zafar, Z.; Fatima, S.; Bhatti, M.F. Comprehensive analyses of NAC transcription factor family in almond (Prunus dulcis) and their differential gene expression during fruit development. Plants 2021, 10, 2200. [Google Scholar] [CrossRef]
- He, X.J.; Mu, R.L.; Cao, W.H.; Zhang, Z.G.; Zhang, J.S.; Chen, S.Y. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005, 44, 903–916. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, K.; Xu, Z.; El-Kereamy, A.; Casaretto, J.A.; Rothstein, S.J. The Arabidopsis transcription factor ANAC032 represses anthocyanin biosynthesis in response to high sucrose and oxidative and abiotic stresses. Front. Plant Sci. 2016, 7, 1548. [Google Scholar] [CrossRef]
- Zhong, R.; Demura, T.; Ye, Z.H. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 2006, 18, 3158–3170. [Google Scholar] [CrossRef]
- Wu, H.; Fu, B.; Sun, P.; Xiao, C.; Liu, J.H. A NAC transcription factor represses putrescine biosynthesis and affects drought tolerance. Plant Physiol. 2016, 172, 1532–1547. [Google Scholar] [CrossRef]
- Oda-Yamamizo, C.; Mitsuda, N.; Sakamoto, S.; Ogawa, D.; Ohme-Takagi, M.; Ohmiya, A. The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Sci. Rep. 2016, 6, 23609. [Google Scholar] [PubMed]
- Zou, S.C.; Zhuo, M.G.; Abbas, F.; Hu, G.B.; Wang, H.C.; Huang, X.M. Transcription factor LcNAC002 coregulates chlorophyll degradation and anthocyanin biosynthesis in litchi. Plant Physiol. 2023, 192, 1913–1927. [Google Scholar] [CrossRef]
- Weil, R.; Kremen, A. Thinking across and beyond disciplines to make cover crops pay. J. Sci. Food Agric. 2006, 87, 551–557. [Google Scholar] [CrossRef]
- Fan, L.; Wang, Y.; Xu, L.; Tang, M.; Zhang, X.; Ying, J.; Li, C.; Dong, J.; Liu, L. A genome-wide association study uncovers a critical role of the RsPAP2 gene in red-skinned Raphanus sativus L. Hortic. Res. 2020, 7, 164. [Google Scholar] [CrossRef]
- Luo, X.; Xu, L.; Wang, Y.; Dong, J.; Chen, Y.; Tang, M.; Fan, L.; Zhu, Y.; Liu, L. An ultra-high-density genetic map provides insights into genome synteny, recombination landscape and taproot skin colour in radish (Raphanus sativus L.). Plant Biotechnol. J. 2020, 18, 274–286. [Google Scholar] [CrossRef]
- Zhang, S.; Yuan, G.; Peng, Z.; Li, X.; Huang, Y.; Yin, C.; Cui, L.; Xiao, G.; Jiao, Z.; Wang, L.; et al. Chemical composition analysis and transcriptomics reveal the R2R3-MYB genes and phenol oxidases regulating the melanin formation in black radish. Int. J. Biol. Macromol. 2024, 271 Pt 1, 132627. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, Y.; Zhang, X.; Sun, Y.; Wang, H.; Song, J.; Li, X. Transcriptome analyses reveal key genes involved in skin color changes of ‘Xinlimei’ radish taproot. Plant Physiol. Biochem. 2019, 139, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Cui, L.; Chen, W.; Liu, Z.; Yuan, W.; Zhu, F.; Jiao, Z.; Zhang, Z.; Deng, X.; Wang, L.; et al. Comprehensive analysis of NAC transcription factors and their expressions during taproot coloration in radish (Raphanus sativus L.). Sci. Hortic. 2022, 299, 111047. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, X.; Gong, Y.; Xu, L.; Wang, Y.; Liu, L. Evaluation of reference genes for gene expression studies in radish (Raphanus sativus L.) using quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2012, 424, 398–403. [Google Scholar] [CrossRef]
- Jones, B.; Frasse, P.; Olmos, E.; Zegzouti, H.; Li, Z.G.; Latché, A.; Pech, J.C.; Bouzayen, M. Down-regulation of DR12, an auxin-response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. Plant J. 2002, 32, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Tzfira, T.; Tian, G.W.; Lacroix, B.t.; Vyas, S.; Li, J.; Leitner-Dagan, Y.; Krichevsky, A.; Taylor, T.; Vainstein, A.; Citovsky, V. pSAT vectors: A modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol. Biol. 2005, 57, 503–516. [Google Scholar] [CrossRef]
- Zheng, F.; Cui, L.; Li, C.; Xie, Q.; Ai, G.; Wang, J.; Yu, H.; Wang, T.; Zhang, J.; Ye, Z.; et al. Hair interacts with SlZFP8-like to regulate the initiation and elongation of trichomes by modulating SlZFP6 expression in tomato. J. Exp. Bot. 2022, 73, 228–244. [Google Scholar] [CrossRef]
- Chen, W.; Hu, T.; Ye, J.; Wang, B.; Liu, G.; Wang, Y.; Yuan, L.; Li, J.; Li, F.; Ye, Z.; et al. A CCAAT-binding factor, SlNFYA10, negatively regulates ascorbate accumulation by modulating the D-mannose/L-galactose pathway in tomato. Hortic. Res. 2020, 7, 200. [Google Scholar] [CrossRef]
- Wellburn, A.R. The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 1994, 144, 307–313. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, Y.; Wu, X.; Shi, W.; Chen, N.; Pang, Y.; Zhang, L. Sequence and epigenetic variations of R2R3-MYB transcription factors determine the diversity of taproot skin and flesh colors in different cultivated types of radish (Raphanus sativus L.). Theor. Appl. Genet. 2024, 137, 133. [Google Scholar] [CrossRef]
- Kuai, B.; Chen, J.; Hörtensteiner, S. The biochemistry and molecular biology of chlorophyll breakdown. J. Exp. Bot. 2018, 69, 751–767. [Google Scholar] [CrossRef]
- Christ, B.; Egert, A.; Sussenbacher, I.; Krautler, B.; Bartels, D.; Peters, S.; Hörtensteiner, S. Water deficit induces chlorophyll degradation via the ‘PAO/phyllobilin’ pathway in leaves of homoio- (Craterostigma pumilum) and poikilochlorophyllous (Xerophyta viscosa) resurrection plants. Plant Cell Environ. 2014, 37, 2521–2531. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Saito, H.; Fukui, M.; Tanaka, A.; Arakawa, K. Poplar leaf abscission through induced chlorophyll breakdown by Mg-dechelatase. Plant Sci. 2022, 324, 111444. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.J.; Liu, K.K.; Li, D.W.; Arisha, M.H.; Chai, W.G.; Gong, Z.H. Cloning and characterization of the pepper CaPAO gene for defense responses to salt-induced leaf senescence. BMC Biotechnol. 2015, 15, 100. [Google Scholar] [CrossRef] [PubMed]
- Broda, M.; Khan, K.; O’Leary, B.; Pružinská, A.; Lee, C.P.; Millar, A.H.; Van Aken, O. Increased expression of ANAC017 primes for accelerated senescence. Plant Physiol. 2021, 186, 2205–2221. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, J.; Xie, Z.; Gao, J.; Ren, G.; Gao, S.; Zhou, X.; Kuai, B. Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 2015, 84, 597–610. [Google Scholar] [CrossRef]
- Singh, S.; Koyama, H.; Bhati, K.K.; Alok, A. The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. J. Plant Res. 2021, 134, 475–495. [Google Scholar] [CrossRef]
- Tak, H.; Negi, S.; Gupta, A.; Ganapathi, T.R. A stress associated NAC transcription factor MpSNAC67 from banana (Musa x paradisiaca) is involved in regulation of chlorophyll catabolic pathway. Plant Physiol. Biochem. 2018, 132, 61–71. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Y.; Turečková, V.; Xue, G.P.; Fernie, A.R.; Mueller-Roeber, B.; Balazadeh, S. The NAC transcription factor SlNAP2 regulates leaf senescence and fruit yield in tomato. Plant Physiol. 2018, 177, 1286–1302. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.; Yan, C.; Chen, L.; Cui, L.; Yuan, W. RsNAC134 Regulates Taproot Skin Color via Positive Regulation of the Chlorophyll Degradation Pathway in Radish (Raphanus sativus). Horticulturae 2025, 11, 1248. https://doi.org/10.3390/horticulturae11101248
Chen W, Yan C, Chen L, Cui L, Yuan W. RsNAC134 Regulates Taproot Skin Color via Positive Regulation of the Chlorophyll Degradation Pathway in Radish (Raphanus sativus). Horticulturae. 2025; 11(10):1248. https://doi.org/10.3390/horticulturae11101248
Chicago/Turabian StyleChen, Weifang, Chenghuan Yan, Leifu Chen, Lei Cui, and Weiling Yuan. 2025. "RsNAC134 Regulates Taproot Skin Color via Positive Regulation of the Chlorophyll Degradation Pathway in Radish (Raphanus sativus)" Horticulturae 11, no. 10: 1248. https://doi.org/10.3390/horticulturae11101248
APA StyleChen, W., Yan, C., Chen, L., Cui, L., & Yuan, W. (2025). RsNAC134 Regulates Taproot Skin Color via Positive Regulation of the Chlorophyll Degradation Pathway in Radish (Raphanus sativus). Horticulturae, 11(10), 1248. https://doi.org/10.3390/horticulturae11101248




