Abstract
Guava (Psidium guajava L.), a perennial species native to tropical regions of the Americas, holds significant economic value and plays an important role in the global fruit industry. Although several reference genomes have been published, population-level genomic studies remain limited, hindering genetic improvement efforts. In this study, we conducted whole genome re-sequencing of 62 guava accessions, primarily from Southern China and Brazil. A total of 4,887,006 high-quality SNPs and 731,469 InDels were identified for population genomic analyses. Phylogenetic and population structure analyses revealed subgroupings that largely corresponded to geographic origins. The data indicated that extensive hybridization between accessions from Brazil and or within China has contributed to the development of many dominant commercial varieties. Genetic diversity analyses showed that Brazilian accessions exhibited higher nucleotide diversity and more rapid linkage disequilibrium decay than those from China. Environmental factors and artificial selection likely imposed selective pressures that shaped guava’s adaptability and agronomic traits. A preliminary genome-wide association study (GWAS) identified PgMYB4 as a candidate gene potentially associated with fruit flesh color. These findings provide novel insights into the genetic diversity, population history, and domestication of guava, and lay a valuable foundation for future breeding and improvement strategies.
1. Introduction
Psidium guajava, commonly known as guava, is a perennial plant species in the genus Psidium, family Myrtaceae. Native to tropical regions of the Americas, including Mexico, Peru, Brazil, and the West Indies, guava is widely valued for its applications in the food, healthcare, and pharmaceutical industries [1,2,3]. It holds substantial economic importance and plays a vital role in the global fruit crop sector. Guava was introduced to China approximately 800 years ago and is now extensively cultivated in tropical and subtropical regions, particularly favored in the Southern coastal areas of China.
However, several challenges currently hinder the development of the guava industry. First, due to local growers often naming cultivars based on morphological traits or geographic origins, varietal nomenclature has become highly inconsistent. This leads to frequent confusion, with different genotypes sharing the same name (homonyms) or identical genotypes bearing different names (synonyms) [4]. Second, the large-scale planting of a few elite cultivars selected for specific traits has led to global genetic uniformity. For example, around 70% of guava cultivated in Brazil belongs to the cultivar ‘Paluma’ [5], while in Pakistan, guava production is dominated by two main types: ‘Gola’ (round fruit) and ‘Surahi’ (pear-shaped fruit) [6]. In China, ‘Zhenzhu’ has long been the dominant cultivar. This genetic bottleneck and reduced germplasm diversity limit breeding progress and increase susceptibility to pests, diseases, and environmental stressors [7].
To address these issues, accurate cultivar identification, diversification of breeding resources, and enrichment of guava germplasm pools have become key objectives for geneticists and breeders [7]. Simple sequence repeat (SSR) markers have been widely used to assess genetic diversity in guava populations, with studies conducted in Cuba, the United States, and India demonstrating their effectiveness [8,9,10]. Arévalo-Marín et al. applied SSR markers to evaluate genetic diversity, population structure, and dissemination patterns among guava accessions from Central and South America, suggesting that the Amazon region may be the center of origin for guava [11].
With the advancement of sequencing technologies, single-nucleotide polymorphisms (SNPs) derived from whole genome re-sequencing have become powerful tools for genetic research in horticultural crops such as grapevine, Brassica napus (rapeseed), and plum [12,13,14]. These approaches offer high-resolution insights into genetic parameters and enable the identification of genomic regions under selection. Despite the release of the first chromosome-scale guava genome in 2021 [15], few studies have applied high-throughput sequencing to population-level analyses in guava [16]. Notably, comprehensive population genetic studies in Asian guava germplasm remain scarce, limiting our understanding of its domestication and breeding potential in this region.
In this study, to better explore the genetic diversity and enrich the genomic resources of guava, we performed whole genome re-sequencing of 60 guava accessions preserved at the Fruit Tree Research Institute in Guangzhou, Guangdong Province, China, along with two accessions of a related species, P. cattleianum (commonly known as strawberry guava). The panel includes traditional landraces and commercial cultivars from China, as well as germplasm introduced from Brazil. We investigated the genetic diversity and population structure of these accessions to elucidate the genetic background and potential selection signals in guava. Furthermore, we conducted a genome-wide association study (GWAS) to identify candidate genes and variants associated with the pink flesh trait. These results are expected to accelerate genomic research in guava and enhance our understanding of its population history, genetic diversity, and breeding potential.
2. Materials and Methods
2.1. Plant Materials and Sequencing
A total of 62 guava accessions (Table S1), including 60 Psidium guajava and two Psidium cattleianum accessions, were cultivated at the Fruit Tree Research Institute, Guangzhou, China. Young leaves were collected for genomic DNA extraction using the M5 CTAB Plant gDNA Extraction Kit (Mei5 Biotechnology Co., Ltd., Beijing, China). DNA quality was checked using NanoDrop Ultra Microvolume UV-Vis Spectrophotometers (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 1% agarose gel. The extracted DNA was used for whole genome re-sequencing on the Illumina NovaSeq 6000 Sequencing System (Illumina, Inc., San Diego, CA, USA), generating 150 bp paired-end reads.
2.2. Mapping and Variant Calling
Low-quality reads and adapter sequences were removed using Trimmomatic (v0.39) [17] after quality check using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/, accessed on 6 May 2024). The cleaned reads were aligned to the guava reference genome [15] using BWA (v2.2.1) [18]. Mapping rate and coverage were calculated with SAMtools (v1.19.2) [19]. SNPs and small InDels were identified using the GATK (v4.5.0) pipeline [20]. SNPs were filtered using the following parameters: QD < 2.0 || MQ < 40.0 || FS > 60.0 || SOR > 3.0 || MQRankSum < −12.5 || ReadPosRankSum < −8.0. InDels were filtered using: QD < 2.0 || FS > 200.0 || SOR > 10.0 || MQRankSum < −12.5 || ReadPosRankSum < −8.0. The aforementioned parameters are the filtering criteria recommended by GATK (https://gatk.broadinstitute.org/hc/en-us (accessed on 21 September 2025)). SNPs and InDels with a sample missing rate < 0.5 and minor allele frequency (MAF) ≥ 0.05 were retained for downstream analyses.
2.3. Phylogenetic and Population Structure Analysis
A maximum likelihood (ML) tree was constructed using all SNP sites and the GTR model in FastTree (v2.1.3) [21]. Principal component analysis (PCA) was performed using PLINK (v1.90) [22]. ADMIXTURE (v1.3.0) was used to infer population structure and determine the optimal number of subpopulations (K) based on cross-validation error [23]. Linkage disequilibrium (LD) decay was analyzed using PopLDdecay (v3.41) by calculating the squared correlation coefficient (r2) for each group as well as the whole population [24].
2.4. Population Selection Analysis
Two parameters were used to detect candidate genomic regions under selection between populations. The first was nucleotide diversity (π) [25,26,27], where the π-ratio (πPOP3/πPOP1) was used to assess diversity differences between populations. The second was the fixation index (FST) [28]. Both metrics were calculated using VCFtools (v0.1.16) with a sliding window size of 100,000 bp and step size of 10,000 bp [29]. Regions falling in the top 5% of both π-ratio and FST and overlapping were defined as candidate selective regions. Genes (along with their 2 kb upstream and downstream flanking sequences) located within these regions were considered candidate genes. GO enrichment analysis of these genes was conducted using the R package clusterProfiler (v4.2.2) [30].
2.5. Transcriptome Analysis
Transcriptome data were obtained from pink-fleshed guava (‘Meiyin’) and white-fleshed guava (‘Zhenzhu’) previously published and available in the NCBI database (PRJNA820889) [31]. Raw sequencing data were quality-controlled using Fastp (v0.23.4) [32] and aligned to the guava reference genome using HISAT2 (v2.2.1) [33]. The resulting alignment files were sorted and converted with SAMtools (v1.19.2) [19]. Gene expression levels were quantified using StringTie (v2.2.1) [34], and differential expression analysis was performed using DESeq2 (v1.40.2) [35].
2.6. Genome-Wide Association Study
SNPs and InDels with MAF ≥ 0.05 and missing rate ≤ 0.1 were selected for GWAS analysis, yielding 4,406,090 SNPs and 2,539,071 indels, respectively. A preliminary GWAS analysis was performed on a set of accessions with flesh color phenotype available (Table S2). To minimize complications from color intensity variations, only pink-colored and white-colored accessions were used for GWAS, as they represent the largest groups. GWAS was conducted using a linear mixed model implemented in EMMAX (v20120210) [36]. The genome-wide significance threshold was set using the Bonferroni correction: −log10(1/n), where n is the total number of variants used in the analysis, to control the type I error rate. Regional linkage disequilibrium and haplotype blocks were visualized using LDBlockShow (v1.40) [37]. Candidate gene annotation was refined manually using IGV-GSAman (v0.6.83) in combination with transcriptome alignment results [38]. Gene functions were annotated using the UniProt database (https://www.uniprot.org/) [39].
2.7. RNA Extraction and qRT-PCR
Total RNA was prepared using the FastPure Universal Plant Total RNA Isolation Kit (Vazyme Co., Ltd., Nanjing, China). NanoDrop and gel electrophoresis (1% agarose gel) were used for quality and quantity checks. cDNA synthesis was performed using the Hifiar III 1st SrandcDNA Synthesis SuperMix for qPCR (gDNA digest plus) (Yeasen Biotechnology Co., Ltd., Shanghai, China). Primers were designed using Primer Premier 5 (Premier Biosoft, Palo Alto, CA, USA). Primer sequences are as follows: gsa.evm.model.ctg29.542_F: 5′-GTCTTTGATTGCTGGGAGATTG-3′, gsa.evm.model.ctg29.542_R: 5′-GCTGCTGATAGGGAGGGCTTA-3′. Tubulin (PgTUB1) served as the reference gene [40]. All biological samples were analyzed in triplicate using iTaq™ universal SYBR Green Supermix (Bio-Rad Laboratories, Inc., Hercules, CA, USA) in the LightCycler480 system (Roche Group, Basel, Switzerland).
3. Results
3.1. Sequencing and Variant Identification
We re-sequenced a total of 62 guava accessions (Table S1) maintained at the Fruit Tree Research Institute in Guangzhou, Guangdong Province, China. They comprised 32 accessions collected from Southern China, 28 accessions originating from Brazil, and two accessions of Psidium cattleianum (strawberry guava). A total of 335 Gb of sequencing data was generated, with an average sequencing depth of 23.43× (ranging from 17.00× to 34.62×). The re-sequencing reads were aligned to the ‘New Age’ reference genome, achieving an average mapping rate of 96.97% (Table S1). After variant calling with the GATK pipeline and filtering, a total of 4,887,006 SNPs and 731,469 InDels of high quality were obtained, which are distributed across all chromosomes (Figure 1). Approximately 64.40% of SNPs were located in intergenic regions, while 5.03% were located within coding sequences (CDS) (Table 1). Similarly, 73.6% of InDels were found in intergenic regions, and 2.9% occurred within CDS.
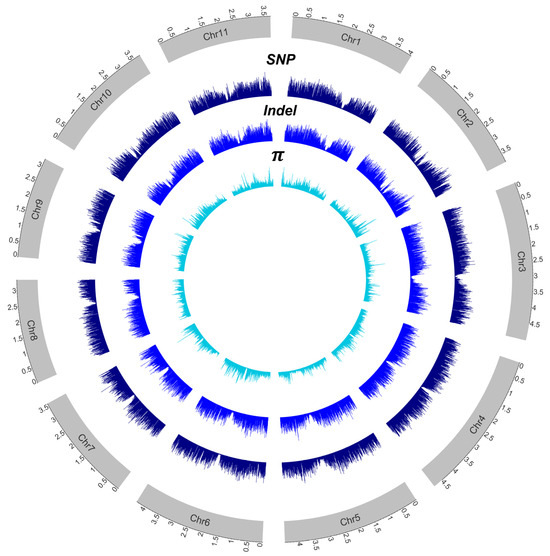
Figure 1.
A circos plot illustrating the density of identified variants and genetic diversity (π). π was calculated using all 62 samples. The unit is in Mb.

Table 1.
Annotation of identified SNPs and InDels using ANNOVAR (v2020-06-07).
3.2. Phylogeny, Population Structure, and Kinship Relationships
A phylogenetic tree was constructed using two P. cattleianum accessions as outgroups, revealing three major clades: POP1, POP2, and POP3 (Figure 2B). This clustering aligned with the geographic origin of the accessions. All POP1 accessions were collected from China, whereas those from Brazil were distributed across POP2 and POP3, indicating distinct genetic backgrounds. This inference was further supported by principal component analysis (PCA), where the accessions from Brazil (represented by red dots) formed two distinct clusters (Figure 2A).
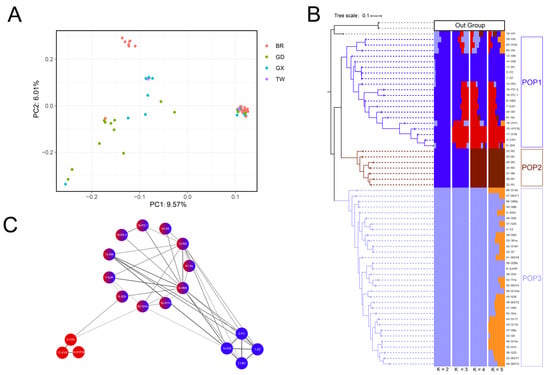
Figure 2.
Phylogeny, population structure, and kinship analysis of guava germplasm accessions. (A) Principal component analysis (PCA), in which the color of each dot represents the recorded geographic origin of the accession, including Brazil (BR), Guangdong (GD), Guangxi (GX), and Taiwan (TW), China. (B) Phylogenetic tree and population structure analysis. Based on the phylogenetic relationships, population structure, and geographic origins, the guava accessions were grouped into three sub-populations (POP1, POP2, and POP3). (C) Identity-by-descent (IBD) network of accessions within POP1. The network, including blue, red, and admixed (red-blue) accessions, was visualized using Cytoscape (v3.10.3). The color intensity of the connecting edges indicates IBD values, with darker lines representing closer genetic relationships.
Population structure analysis identified the optimal number of sub-populations as K = 4 (Figure 2B). Across K values from 2 to 5, POP3 accessions consistently separated from the rest, suggesting a distinct genetic lineage. At K = 3, Chinese accessions (POP1) split into two subgroups, with many individuals showing mixed ancestry. Notably, POP1 samples appeared scattered on the PCA plot, indicating internal diversity. At K = 4, POP2 was clearly distinguished from the blue genetic component within POP1. Interestingly, three accessions with a blue genetic background, 1-ZZ, 2-FC, and 14-DW, were bred in Taiwan and recently introduced to Guangdong, implying their potential use as parental lines in breeding programs.
To further investigate genetic relationships, identity-by-descent (IBD) analysis of the 60 guava accessions was visualized using a pedigree network constructed with Cytoscape (v3.10.3). The admixed individuals (red-blue) in POP1 showed kinship connections to both red and blue individuals, whereas no direct kinship was observed between the red and blue individuals themselves (Figure 2C). This suggests that admixed individuals likely arose from recent hybridization events, and several are, in fact, widely cultivated commercial varieties in China. Additionally, the pedigree network indicated introgression of the light blue genetic background (POP3) into POP1, exemplified by the cultivated variety ‘Xiguahong’ (Figure S1).
3.3. Genetic Diversity, Population Differentiation, and Selection
Nucleotide diversity (π) followed the order POP1 > POP3 > POP2, indicating that POP1 harbors the highest genetic diversity (Figure 3A). FST analysis showed the order FST (POP3/POP2) > FST (POP3/POP1) > FST (POP2/POP1), indicating the highest level of genetic differentiation between POP2 and POP3. In contrast, POP1 and POP2 exhibited lower differentiation, consistent with the results from the population structure analysis. Linkage disequilibrium (LD) analysis revealed that LD decay occurred most rapidly in POP1, followed by POP2 and then POP3 (Figure 3B). The r2 decayed to half of its maximum value at approximately 60 kb (r2 = 0.39) for POP1, 368 kb (r2 = 0.43) for POP2, and 454 kb (r2 = 0.43) for POP3. Moreover, the nucleotide diversity of POP1 (π = 3.08 × 10−3) is lower than that reported in some other sequenced perennial fruit trees, such as Pyrus (π = 5.5 × 10−3) [41], but higher than in others, including Prunus persica (π = 1.1 × 10−3) [42], Malus domestica (π = 1.1 × 10−3) [43], and cultivated Eriobotrya japonica (π = 1.015 × 10−3) [44]. These results suggest that the current guava germplasm maintains a moderate level of genetic diversity still with a certain level of potential for future breeding and genetic improvement.
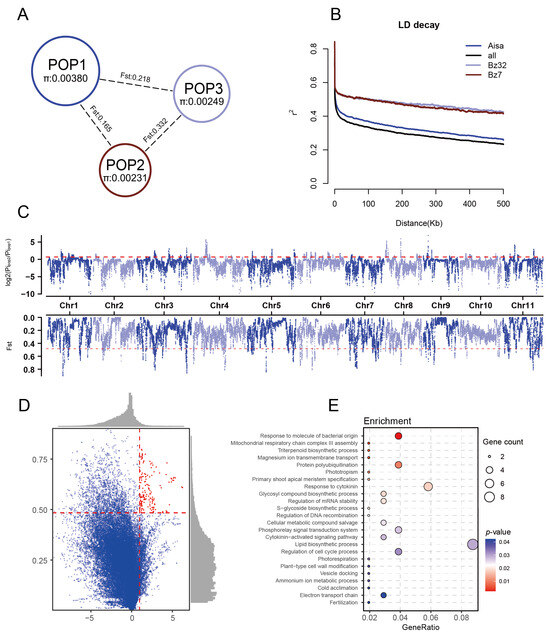
Figure 3.
Population diversity and selective sweep analysis of guava. (A) Nucleotide diversity (π) within each of the three populations and pairwise genetic differentiation (FST) between them. (B) LD decay patterns in the three populations and the entire guava panel. (C) Distribution of FST and π-ratio signals between POP1 and POP3. Red dashed lines indicate the top 5% thresholds. (D) Two-dimensional plot of FST versus π-ratio between POP1 and POP3. Red dots represent genomic regions falling within the top 5% of both metrics, defined as candidate selective sweep regions. (E) Functional enrichment analysis of genes located within the identified candidate selective sweep regions.
Selective sweep analysis was conducted by comparing FST and π-ratio values between POP1 and POP3, considering their sample sizes and geographic representation from China and Brazil, respectively. A total of 2207 genomic regions were identified as candidate selective sweep regions, based on the top 5% of values from both metrics. Among these, only 162 regions were shared between the two methods (Figure 3D). These overlapping regions, located on chromosomes 1, 3, 4, and 8, collectively spanned approximately 0.65% of the genome and were considered as putative selective sweep regions between POP1 and POP3. Within these regions, 249 candidate genes were identified. Gene enrichment analysis revealed that these genes were significantly associated with several biological processes (Table S3), including “response to molecule of bacterial origin” (GO:0002237), “triterpenoid biosynthetic process” (GO:0016104), “cold acclimation” (GO:0009631), “response to cytokinin” (GO:0009735), “regulation of cell cycle process” (GO:0010564), “electron transport chain” (GO:0022900), and “glycosyl compound biosynthetic process” (GO:1901659) (Figure 3E). These findings suggest that both geographic environmental factors and artificial selection have imposed selective pressures on guava, potentially shaping its adaptability and developmental traits.
3.4. Preliminary GWAS for Fruit Flesh Color
To investigate the genetic basis underlying fruit flesh color in guava, we conducted a preliminary genome-wide association study (GWAS) applying a linear mixed model implemented in EMMAX, using accessions with either pink or white flesh phenotypes available. The SNP-based GWAS was not presented due to an excessive number of anomalously correlated loci, which obscured meaningful associations. In contrast, the InDel-based GWAS identified 130 loci that exceeded the Bonferroni-corrected significance threshold (Figure 4A), distributed across all chromosomes except chromosome 10. Within 200 kb upstream and downstream of these loci, 946 genes were identified, among which 38 candidate genes were annotated as being involved in flavonoid-related pathways (Tables S4 and S5). The frequencies of variants in these regions were summarized in Table S6. On average, one SNP and one InDel were expected in every 69 bp and 581 bp, respectively, in the CDS regions. In addition, publicly available transcriptome data from a white-fleshed and a pink-fleshed guava accession were incorporated to aid in the identification of candidate loci [31]. Among the genes differentially expressed between the white- and pink-fleshed accession, 423 overlapped with GWAS-identified genes (Table S7), including 13 annotated as being involved in flavonoid-related pathways (Table S5).
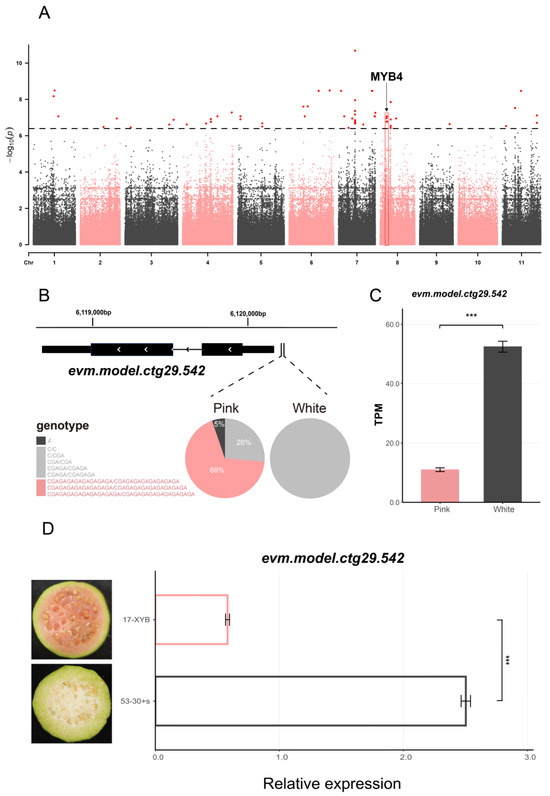
Figure 4.
Genome-wide association analysis of loci associated with pink flesh color in guava. (A) Manhattan plot showing the GWAS results for pink versus white flesh color in guava. The black horizontal line indicates the Bonferroni-corrected significance threshold (−log10(1/n) = 6.40). The red dots represent variants exceeding this threshold. The candidate region is highlighted with a red box, and the arrow indicates the candidate gene within this region. (B) Genotypic distribution of the candidate gene and its associated upstream insertion variants. A total of 22 pink-fleshed and 10 white-fleshed accessions are included for calculation. (C) Expression levels of the candidate gene evm.model.ctg29.542 based on published transcriptome data comparing the pink-fleshed (‘Meiyin’) and white-fleshed (‘Zhenzhu’) guava accession. Asterisks indicate statistical significance (‘***’ for p < 0.001). (D) qRT-PCR of evm.model.ctg29.542 in the plink-fleshed (17-XYB) and white-fleshed (53-30+s) guava, with fruit transverse sections. Asterisks indicate statistical significance (‘***’ for p < 0.001).
To further characterize these 13 genes, BLAST (v2.2.31) searches were performed against the UniProt database. One gene, evm.model.ctg29.542 (Figure S2), showed the best match to MYB4, a known transcriptional repressor of flavonoid biosynthetic genes that reduces flavonoid accumulation in plants. According to the transcriptomic analysis, evm.model.ctg29.542 (PgMYB4) was significantly down-regulated (p < 0.01) in the pink-fleshed accession (Figure 4C). qRT-PCR results from white-flesh sample 17-XYB and pink-flesh sample 53-30+s (p < 0.001) further validated the differential expression (Figure 4D). Further sequence analysis identified 0–20 bp insertions located 38 bp upstream of the start codon of this gene. All white-fleshed individuals carried either short insertions (≤7 bp) or no insertion, while 69% of red-fleshed individuals harbored a long insertion haplotype (≥17 bp) (Figure 4B). These findings suggest that the upstream insertion may affect the expression of evm.model.ctg29.542, potentially contributing to the regulation of flesh color in guava, which warrants further investigation.
4. Discussion
Guava is an important fruit crop in many tropical and subtropical regions. Accurate characterization and classification of genetic variation among guava germplasm are crucial for its conservation, effective utilization, and the development of improved cultivars with desirable traits. However, genetic and genomic resources for guava remain limited [7]. Although the reference genomes have become available, population-level genomic studies are still scarce [15,16]. In this study, we analyzed the genome-wide diversity of 60 guava accessions conserved in Guangzhou, China, through whole genome re-sequencing. Based on these data, we explored phylogenetic relationships, population genomic parameters, and identified loci potentially associated with flesh color. The findings of this study would enhance our understanding of guava’s population history and genetic diversity, and provide a foundation for future genetic improvement efforts.
The phylogenetic and population structure analyses of the 60 guava accessions largely corresponded with their geographic origins. Although the accessions from Brazil included in this study were limited in number, they were sufficient to reveal substantial genetic divergence from the accessions of China (Figure 2). Further population classification indicated that germplasm in China comprises at least two distinct ancestral components. Based on historical records and structure analysis, one group (represented in red) likely corresponds to traditional landraces from Southern China (e.g., Guangdong Province), while the other group (in blue) may represent modern cultivars introduced from Taiwan, China (Figure 2B). These two groups, along with their hybrids, constitute a significant portion of the guava cultivars currently grown in China. Moreover, some Brazilian germplasm introduced into China has been used in breeding programs to develop new cultivars, such as ‘Xiguahong’. Kinship analysis further supports the observation that Chinese breeders are actively hybridizing Brazilian accessions with local germplasm to enhance genetic diversity and improve agronomic traits.
Genetic differentiation can be driven by divergent selection pressures acting on different populations [44]. By comparing the two sub-populations of Chinese and Brazilian origin, we investigated potential genomic signatures shaped by geographic isolation and artificial selection. Preliminary observations revealed candidate selective sweep regions between the two populations harboring genes associated with “response to molecule of bacterial origin” that may play a role in disease resistance, including genes evm.model.ctg30.654 (Hypersensitivity-Related 4), evm.model.ctg30.655 (Hypersensitivity-Related 4), evm.model.ctg9.1333 (Pre-mRNA PROCESSING factor 8A), and evm.model.ctg9.1385 (WRKY transcription factor 40), along with 44 additional genes potentially involved in environmental adaptation and plant growth (Table S3). While the functional divergence of these genes remains undefined, the reported candidate gene set establishes a robust foundation for subsequent genetic dissection and functional validation, which may play pivotal roles in regulating economically important traits.
Kwee and Chong (1990) reported that deep pink flesh is a desirable trait in commercially cultivated guava varieties [45]. Subsequent studies have demonstrated that red-fleshed guavas possess stronger antioxidant activity compared to their white-fleshed counterparts [46,47]. These findings underscore the nutritional and commercial value of flesh coloration, highlighting the importance of elucidating its genetic basis to facilitate targeted breeding for this trait in guava.
Based on high-coverage whole genome re-sequencing data, our GWAS analysis using InDels, in combination with previous metabolomic and transcriptomic studies in guava, enabled the identification of potential loci and a candidate gene associated with pink flesh coloration. Through rigorous gene annotation, we identified PgMYB4 as a promising candidate. MYB4 has been reported as a negative regulator of the phenylpropanoid pathway in Petunia × hybrida, where it suppresses the expression of CINNAMATE-4-HYDROXYLASE (PhC4H1 and PhC4H2), a key enzyme in flavonoid precursor biosynthesis [48]. In Arabidopsis thaliana, MYB4 also interferes with flavonoid biosynthesis via multiple mechanisms, including repression of the MBW complex’s transcriptional activity, inhibition of Arogenate Dehydratase 6 (ADT6), and downregulation of key pathway genes such as 4-coumarate/coenzyme A ligase (4CL) and Flavonol Synthase (FLS) [49,50]. Further experimental validation is needed to confirm the functional role of PgMYB4 in regulating flesh pigmentation in guava. It is noteworthy that the PgMYB4 promoter variant explains only about 69% of the pink-fleshed accessions, suggesting that additional loci identified in the GWAS may also contribute to fruit flesh color. We also acknowledge that a limitation of the GWAS analysis is the relatively small number of accessions. However, as one of the first whole genome re-sequencing studies in guava, we believe our analysis provides meaningful signals and a valuable resource for the guava research community.
In summary, we present a comprehensive analysis of genome-wide diversity in a guava germplasm collection, providing a theoretical foundation for its conservation and future utilization. This study also contributes to the dissection of the genetic basis of complex agronomic traits. The identified selective signals, candidate genes, and associated variants constitute valuable genomic resources for the genetic improvement of guava through breeding. It is anticipated that the germplasm panel and associated genomic resources developed in this study will also facilitate the dissection of genetic mechanisms underlying other important traits in guava.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae11101194/s1, Figure S1: Analysis of linkage disequilibrium blocks for variants surrounding the region harboring evm.model.ctg29.542, Figure S2: Genetic pedigree network of the 60 guava samples, Table S1: Sequencing Statistics and Alignment Rate, Table S2: Samples used for GWAS analysis on fruit flesh color, Table S3: GO enrichment analysis for genes in candidate selective sweep regions, Table S4: Genes located within the 200 kb region of the significant GWAS loci, Table S5: Differential gene expression analysis for the candidate flavonoid-related genes, Table S6: Summary of frequencies of variants located within the candidate regions, Table S7: Differentially expressed genes obtained by comparing the white-fleshed and pink-fleshed accession.
Author Contributions
Conceptualization, J.C. and Z.P.; methodology, J.H. and X.Y.; formal analysis, J.H., X.Y., C.Z., Z.P. and J.C.; investigation, J.H., X.Y., C.Z., Z.P. and J.C.; resources, J.C. and Z.P.; data curation, J.H. and X.Y.; writing—original draft preparation, J.H., X.Y., Z.P. and J.C.; writing—review and editing, Z.P. and J.C.; visualization, J.H. and X.Y.; supervision, Z.P. and J.C.; project administration, Z.P. and J.C.; funding acquisition, J.C. and Z.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Guangdong Modern Agricultural Industry Technology System Innovation Team Construction Project—Rare and Premium Fruit Industry Technology System (2024CXTD09).
Data Availability Statement
The raw sequencing data have been deposited in the Genome Sequence Archive in the National Genomics Data Center, China National Center for Bioinformation under accession number CRA028095.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gutiérrez, R.M.P.; Mitchell, S.; Solis, R.V. Psidium guajava: A review of its traditional uses, phytochemistry and pharmacology. J. Ethnopharmacol. 2008, 117, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Hiwale, S. Sustainable Horticulture in Semiarid Dry Lands; Springer: New Delhi, India, 2015; pp. 1–393. [Google Scholar]
- Landrum, L.R. Psidium guajava L.: Taxonomy, relatives and possible origin. In Guava: Botany, Production and Uses; CABI Publishing: Oxfordshire, UK, 2021; pp. 1–21. [Google Scholar] [CrossRef]
- Kareem, A.; Jaskani, M.J.; Mehmood, A.; Khan, I.A.; Awan, F.S.; Sajid, M.W. Morpho-genetic profiling and phylogenetic relationship of guava (Psidium guajava L.) As genetic resources in Pakistan. Rev. Bras. Frutic. 2018, 40, e-069. [Google Scholar] [CrossRef]
- Pereira, F.M.; Kavati, R. Contribuição da pesquisa científica Brasileira no desenvolvimento de algumas frutíferas de clima subtropical1. J. Ethnopharmacol. 2008, 117, 27. [Google Scholar]
- Mehmood, A.; Jaskani, M.J.; Khan, I.A.; Ahmad, S.; Ahmad, R.; Luo, S.; Ahmad, N.M. Genetic diversity of Pakistani guava (Psidium guajava L.) Germplasm and its implications for conservation and breeding. Sci. Hortic. 2014, 172, 221–232. [Google Scholar] [CrossRef]
- Vasugi, C.; Chaturvedi, K.; Vishwakarma, P.K. Guava. In Fruit and Nut Crops; Rajasekharan, P.E., Rao, V.R., Eds.; Springer Nature: Singapore, 2023; Volumes 1–27. [Google Scholar]
- Sitther, V.; Zhang, D.; Harris, D.L.; Yadav, A.K.; Zee, F.T.; Meinhardt, L.W.; Dhekney, S.A. Genetic characterization of guava (Psidium guajava L.) Germplasm in the United States using microsatellite markers. Genet. Resour. Crop Evol. 2014, 61, 829–839. [Google Scholar] [CrossRef]
- Kherwar, D.; Usha, K.; Mithra, S.V.A.; Singh, B. Microsatellite (SRR) marker assisted assessment of population structure and genetic diversity for morpho-physiological traits in guava (Psidium guajava L.). J. Plant Biochem. Biotechnol. 2018, 27, 284–292. [Google Scholar] [CrossRef]
- Valdés-Infante Herrero, J.; Rodríguez, N.N.; Becker, D.; Velázquez, B.; Sourd, D.; Espinosa, G.; Rohde, W. Microsatellite characterization of guava (Psidium guajava L.) Germplasm collection in Cuba. Cultiv. Trop. 2007, 28, 61–67. [Google Scholar]
- Arévalo-Marín, E.; Casas, A.; Alvarado-Sizzo, H.; Ruiz-Sanchez, E.; Castellanos-Morales, G.; Jardón-Barbolla, L.; Fermin, G.; Padilla-Ramírez, J.S.; Clement, C.R. Genetic analyses and dispersal patterns unveil the Amazonian origin of guava domestication. Sci. Rep. 2024, 14, 15755. [Google Scholar] [CrossRef]
- Peng, W.; Liang, F.; Chen, Z.; Gong, Z.; Zhang, M.; Wei, R.; Li, H.; Zhang, T.; Pan, F.; Yang, X.; et al. Genomic signals of divergence and hybridization between a wild grape (Vitis adenoclada) and domesticated grape (‘shine muscat’). Fruit Res. 2024, 4, e028. [Google Scholar] [CrossRef]
- Lu, K.; Wei, L.; Li, X.; Wang, Y.; Wu, J.; Liu, M.; Zhang, C.; Chen, Z.; Xiao, Z.; Jian, H.; et al. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 2019, 10, 1154. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Shen, F.; Chen, Y.; Cao, K.; Wang, L. Chromosome-scale genome assembly and population genomics provide insights into the adaptation, domestication, and flavonoid metabolism of Chinese plum. Plant J. 2021, 108, 1174–1192. [Google Scholar] [CrossRef]
- Feng, C.; Feng, C.; Lin, X.; Liu, S.; Li, Y.; Kang, M. A chromosome-level genome assembly provides insights into ascorbic acid accumulation and fruit softening in guava (Psidium guajava). Plant Biotechnol. J. 2021, 19, 717–730. [Google Scholar] [CrossRef]
- Mittal, A.; Thakur, S.; Sharma, A.; Boora, R.S.; Arora, N.K.; Singh, D.; Singh Gill, M.I.; Dhillon, G.S.; Chhuneja, P.; Yadav, I.S.; et al. Guava cv. Allahabad safeda chromosome scale assembly and comparative genomics decodes breeders’ choice marker trait association for pink pulp colour. bioRxiv 2024. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and samtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Mckenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A mapreduce framework for analyzing next-generation dna sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. Fasttree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. Plink: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.; Xu, J.; He, W.; Yang, T. Poplddecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Tajima, F. Evolutionary relationship of DNA sequences in finite populations. Genetics 1983, 105, 437–460. [Google Scholar] [CrossRef]
- Luikart, G.; England, P.R.; Tallmon, D.; Jordan, S.; Taberlet, P. The power and promise of population genomics: From genotyping to genome typing. Nat. Rev. Genet. 2003, 4, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Schlötterer, C. Hitchhiking mapping—Functional genomics from the population genetics perspective. Trends Genet. 2003, 19, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, M.A. Adaptation and speciation: What can FST tell us? Trends Ecol. Evol. 2005, 20, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; Depristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and vcftools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. Clusterprofiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Zheng, B.; Zhao, Q.; Wu, H.; Ma, X.; Xu, W.; Li, L.; Liang, Q.; Wang, S. Metabolomics and transcriptomics analyses reveal the potential molecular mechanisms of flavonoids and carotenoids in guava pulp with different colors. Sci. Hortic. 2022, 305, 111384. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one fastq preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with hisat2 and hisat-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.; Mendell, J.T.; Salzberg, S.L. Stringtie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef]
- Dong, S.; He, W.; Ji, J.; Zhang, C.; Guo, Y.; Yang, T. Ldblockshow: A fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files. Brief. Bioinform. 2021, 22, bbaa227. [Google Scholar] [CrossRef]
- Chen, C.; Li, J.; Feng, J.; Liu, B.; Feng, L.; Yu, X.; Li, G.; Zhai, J.; Meyers, B.C.; Xia, R. Srnaanno—A database repository of uniformly annotated small rnas in plants. Hortic. Res. 2021, 8, 45. [Google Scholar] [CrossRef]
- The, U.C. Uniprot: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef]
- Kumar, S.; Muthukumar, M.; Bajpai, A.; Kushwaha, A.K.; Ahmad, I.; Bajpai, Y.; Singh, A.; Damodaran, T.; Trivedi, M. Selection and validation of stable reference genes in guava (Psidium guajava L.) For reliable and consistent gene expression analysis. Electron. J. Biotechnol. 2025, 75, 49–56. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Y.; Xu, J.; Korban, S.S.; Fei, Z.; Tao, S.; Ming, R.; Tai, S.; Khan, A.M.; Postman, J.D.; et al. Diversification and independent domestication of Asian and European pears. Genome Biol. 2018, 19, 77. [Google Scholar] [CrossRef]
- Li, Y.; Cao, K.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Zhao, P.; Guo, J.; Ding, T.; Guan, L.; et al. Genomic analyses of an extensive collection of wild and cultivated accessions provide new insights into peach breeding history. Genome Biol. 2019, 20, 36. [Google Scholar] [CrossRef]
- Duan, N.; Bai, Y.; Sun, H.; Wang, N.; Ma, Y.; Li, M.; Wang, X.; Jiao, C.; Legall, N.; Mao, L.; et al. Genome re-sequencing reveals the history of apple and supports a two-stage model for fruit enlargement. Nat. Commun. 2017, 8, 249. [Google Scholar] [CrossRef]
- Wang, Y.; Paterson, A.H. Loquat (Eriobotrya japonica (Thunb.) Lindl) population genomics suggests a two-staged domestication and identifies genes showing convergence/parallel selective sweeps with apple or peach. Plant J. 2021, 106, 942–952. [Google Scholar] [CrossRef]
- Kwee, L.T.; Chong, K.K. Guava in Malaysia: Production, Pests, and Diseases; Tropical Press: Singapore, 1990. [Google Scholar]
- Corrêa, L.C.; Santos, C.A.F.; Vianello, F.; Lima, G.P.P. Antioxidant content in guava (Psidium guajava) and araçá (Psidium spp.) Germplasm from different Brazilian regions. Plant Genet. Resour. 2011, 9, 384–391. [Google Scholar] [CrossRef]
- Mahattanatawee, K.; Manthey, J.A.; Luzio, G.; Talcott, S.T.; Goodner, K.; Baldwin, E.A. Total antioxidant activity and fiber content of select Florida-grown tropical fruits. J. Agric. Food. Chem. 2006, 54, 7355–7363. [Google Scholar] [CrossRef]
- Colquhoun, T.A.; Kim, J.Y.; Wedde, A.E.; Levin, L.A.; Schmitt, K.C.; Schuurink, R.C.; Clark, D.G. Phmyb4 fine-tunes the floral volatile signature of Petunia × hybrida through phc4h. J. Exp. Bot. 2011, 62, 1133–1143. [Google Scholar] [CrossRef]
- Wang, X.; Wu, J.; Guan, M.; Zhao, C.; Geng, P.; Zhao, Q. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J. 2020, 101, 637–652. [Google Scholar] [CrossRef]
- Banerjee, S.; Agarwal, P.; Choudhury, S.R.; Roy, S. MYB4, a member of R2R3-subfamily of MYB transcription factor functions as a repressor of key genes involved in flavonoid biosynthesis and repair of UV-B induced DNA double strand breaks in Arabidopsis. Plant Physiol. Biochem. 2024, 211, 108698. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).