NGS-Based Multi-Allelic InDel Genotyping and Fingerprinting Facilitate Genetic Discrimination in Grapevine (Vitis vinifera L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Screening of InDels Markers
2.3. InDel Primers Design and Selection
2.4. InDel Genotyping
2.5. Phylogenetic Analyses
3. Results
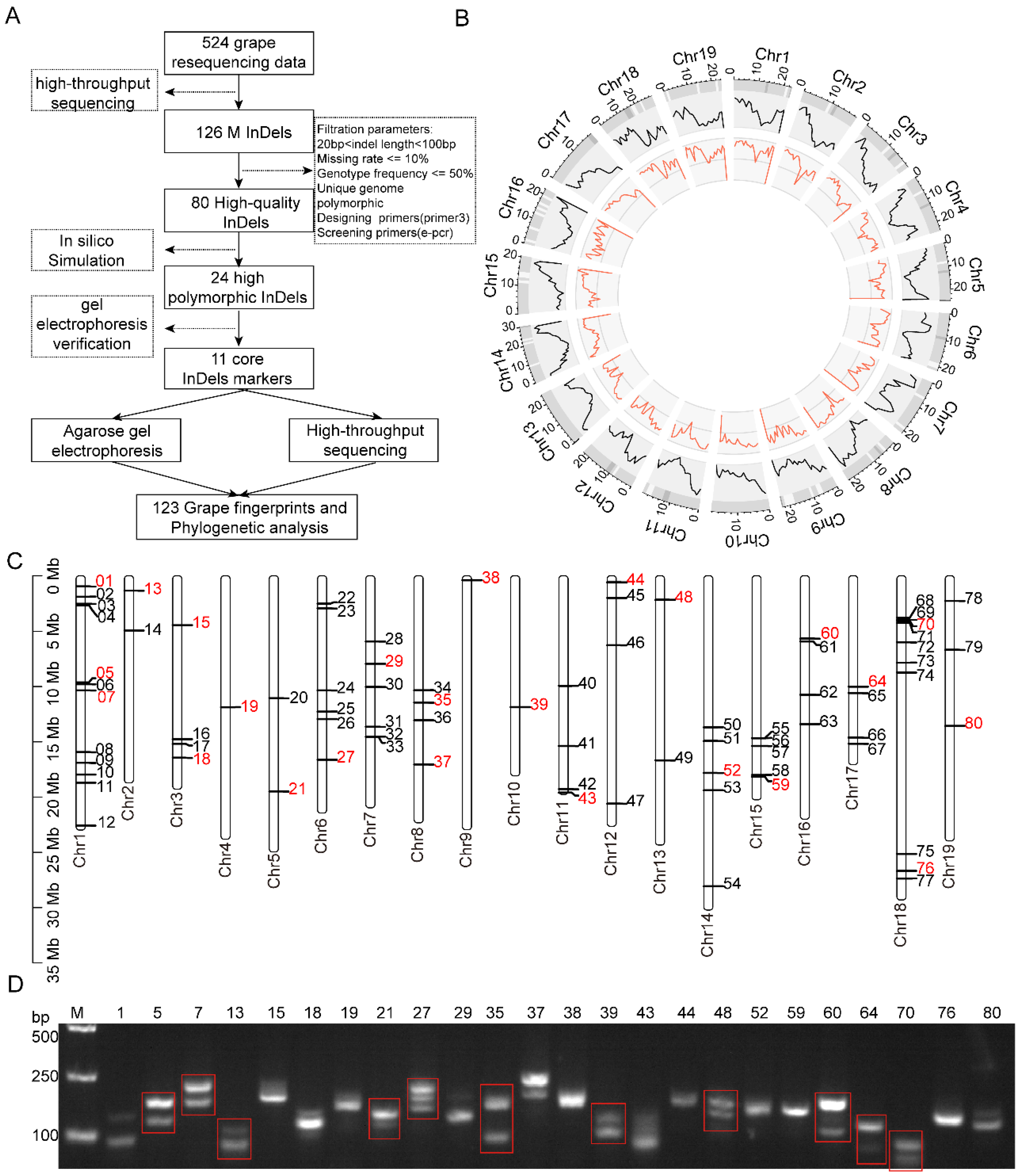
3.1. Screening of InDels Markers
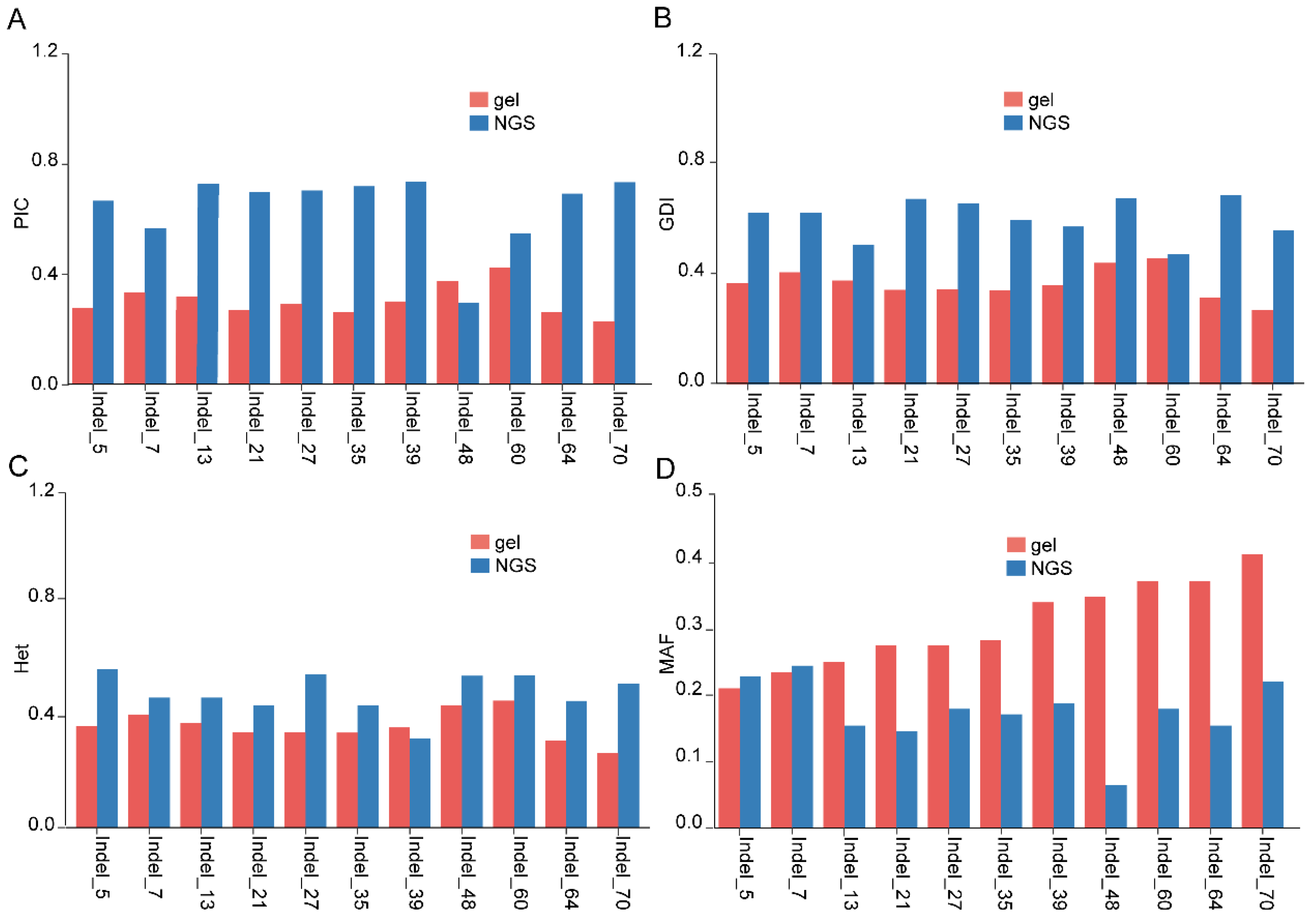
3.2. Genotyping of 123 Grape Lines Using Agarose Gel Electrophoresis
3.3. NGS-Based Genotyping of 123 Grape Lines
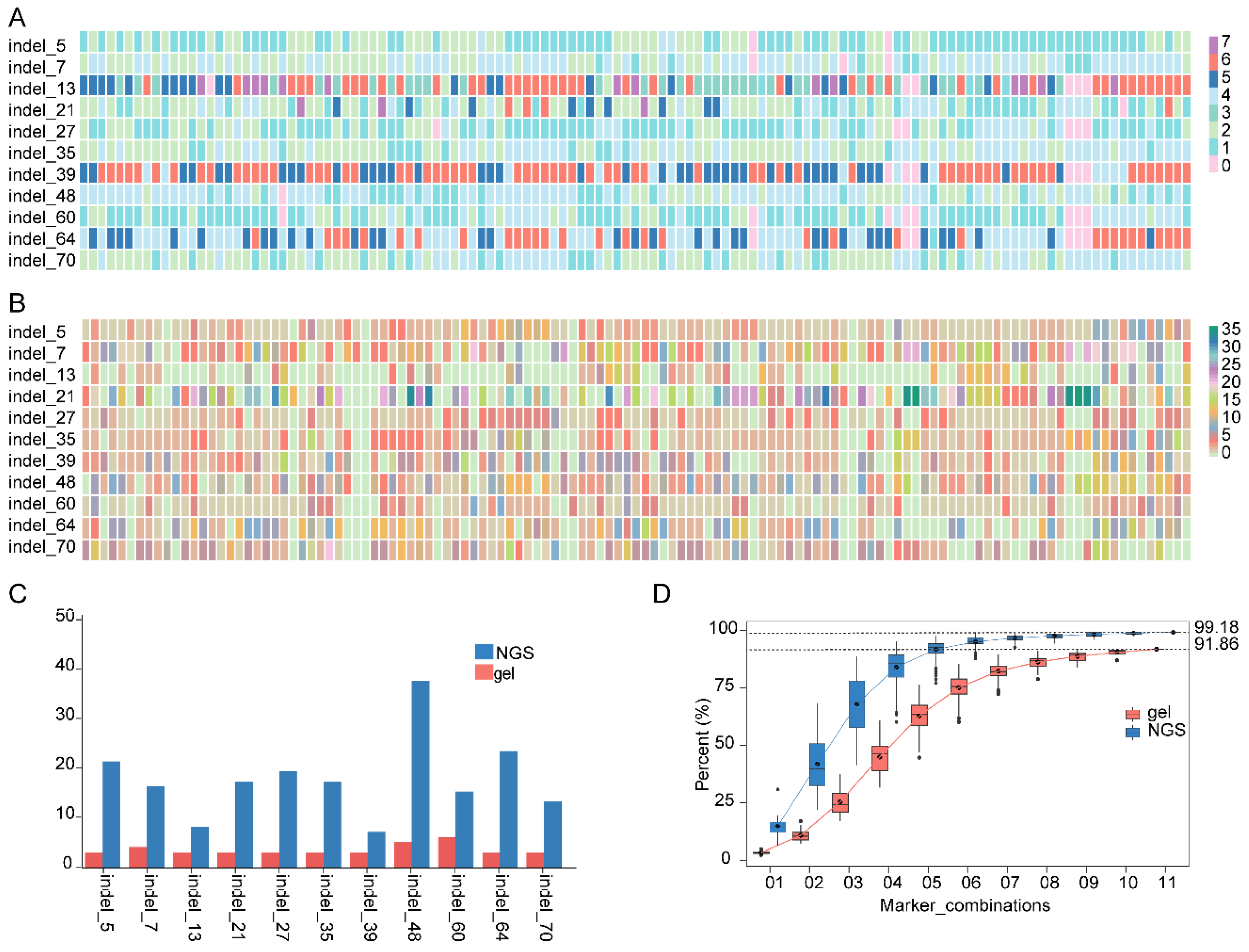
3.4. Discrimination and Fingerprints of 123 Grape Lines
3.5. Phylogenetic Analyses of 123 Grapes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dong, Y.; Duan, S.; Xia, Q.; Liang, Z.; Dong, X.; Margaryan, K.; Musayev, M.; Goryslavets, S.; Zdunić, G.; Bert, P.-F.; et al. Dual domestications and origin of traits in grapevine evolution. Science 2023, 379, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Grassi, F.; Gabriella, D.L. Back to the Origins: Background and Perspectives of Grapevine Domestication. Int. J. Mol. Sci. 2021, 22, 4518. [Google Scholar] [CrossRef] [PubMed]
- Sabir, A.; Tangolar, S.; Büyükalaca, S.; Kafkas, S. Ampelographic and molecular diversity among grapevine (Vitis spp.) cultivars. Czech J. Genet. Plant Breed. 2009, 45, 160–168. [Google Scholar]
- Zhou, Y.; Massonnet, M.; Sanjak, J.S.; Cantu, D.; Gaut, B.S. Evolutionary genomics ofgrape (Vitis vinifera ssp. vinifera) domestication. Proc. Natl. Acad. Sci. USA 2017, 114, 11715–11720. [Google Scholar]
- Xiao, H.; Liu, Z.; Wang, N.; Long, Q.; Cao, S.; Huang, G.; Liu, W.; Peng, Y.; Riaz, S.; Walker, A.M.; et al. Adaptive and maladaptive introgression in grapevine domestication. Proc. Natl. Acad. Sci. USA 2023, 120, e2222041120. [Google Scholar] [CrossRef]
- Martínez, M.C.; Boso, S.; Gago, P.; Muñoz-Organero, G.; De Andrés, M.T.; Gaforio, L.; Cabello, F.; Santiago, J.L. Value of two Spanish live grapevine collections in the resolution of synonyms, homonyms and naming errors. Aust. J. Grape Wine R. 2018, 24, 430–438. [Google Scholar] [CrossRef]
- Tessier, C.; David, J.; This, P.; Boursiquot, J.M.; Charrier, A. Optimization of the choice of molecular markers for varietal identification in Vitis vinifera L. Theor. Appl. Genet. 1999, 98, 171–177. [Google Scholar] [CrossRef]
- Shen, X.; Guo, W.; Zhu, X.; Yuan, Y.; Yu, J.Z.; Kohel, R.J.; Zhang, T. Molecular mapping of QTLs for fiber qualities in three diverse lines in Upland cotton using SSR markers. Mol. Breeding 2005, 15, 169–181. [Google Scholar] [CrossRef]
- Zombardo, A.; Meneghetti, S.; Morreale, G.; Calò, A.; Costacurta, A.; Storchi, P. Study of inter-and intra-varietal genetic variability in grapevine cultivars. Plants 2022, 11, 397. [Google Scholar] [CrossRef] [PubMed]
- Taheri, S.; Abdullah, T.L.; Yusop, M.R.; Hanafi, M.M.; Sahebi, M.; Azizi, P.; Shamshiri, R.R. Mining and development of novel SSR markers using next generation sequencing (NGS) data in plants. Molecules. 2018, 23, 399. [Google Scholar] [CrossRef]
- Lv, Y.; Liu, Y.; Zhao, H. mInDel: A high-throughput and efficient pipeline for genome-wide InDel marker development. BMC Genom. 2016, 17, 290. [Google Scholar] [CrossRef]
- Rimbert, H.; Darrier, B.; Navarro, J.; Kitt, J.; Choulet, F.; Leveugle, M.; Duarte, J.; Rivière, N.; Eversole, K. High throughput SNP discovery and genotyping in hexaploid wheat. PLoS ONE 2018, 13, e0186329. [Google Scholar] [CrossRef]
- Emanuelli, F.; Lorenzi, S.; Grzeskowiak, L.; Catalano, V.; Stefanini, M.; Troggio, M.; Myles, S.; Martinez-Zapater, J.M.; Zyprian, E.; Moreira, F.M.; et al. Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol. 2013, 13, 39. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Fan, X.; Zhang, Y.; Sun, L.; Liu, C.; Jiang, J. Establishment and application of an SNP molecular identification system for grape cultivars. J. Integr. Agric. 2022, 21, 1044–1057. [Google Scholar] [CrossRef]
- Lü, Y.; Cui, X.; Li, R.; Huang, P.; Zong, J.; Yao, D.; Li, G.; Zhang, D.; Yuan, Z. Development of genome-wide insertion/deletion markers in rice based on graphic pipeline platform. J. Integr. Plant Biol. 2015, 57, 980–991. [Google Scholar] [CrossRef]
- Liu, Z.; Zhao, Y.; Zhang, Y.; Xu, L.; Zhou, L.; Yang, W.; Zhao, H.; Zhao, J.; Wang, F. Development of Omni InDel and supporting database for maize. Front. Plant Sci. 2023, 14, 1216505. [Google Scholar] [CrossRef]
- Guan, L.; Cao, K.; Li, Y.; Guo, J.; Xu, Q.; Wang, L. Detection and application of genome-wide variations in peach for association and genetic relationship analysis. BMC Genet. 2019, 20, 101. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shen, F.; Gao, Y.; Wang, K.; Chen, R.; Luo, J.; Yang, L.; Zhang, X.; Qiu, C.; Li, W.; et al. Application of genome-wide insertion/deletion markers on genetic structure analysis and identity signature of Malus accessions. BMC Plant Biol. 2000, 20, 540. [Google Scholar] [CrossRef]
- Liu, X.; Geng, X.; Zhang, H.; Shen, H.; Yang, W. Association and genetic identification of loci for four fruit traits in tomato using InDel markers. Front. Plant Sci. 2017, 8, 1269. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.; Liu, B.; Qiu, Y.; Zhang, X.; Zhang, Z.; Wang, H.; Li, X.; Li, S. Development and application of cucumber InDel markers based on genome re-sequencing. Plant Genet. Res. 2013, 14, 278–283. [Google Scholar]
- Liang, S.; Lin, F.; Qian, Y.; Zhang, T.; Wu, Y.; Qi, Y.; Ren, S.; Ruan, L.; Zhao, H. A cost-effective barcode system for maize genetic discrimination based on bi-allelic InDel markers. Plant Methods 2020, 16, 101. [Google Scholar] [CrossRef] [PubMed]
- This, P.; Lacombe, T.; Thomas, M.R. Historical origins and genetic diversity of wine grapes. Trends Genet. 2006, 22, 511–519. [Google Scholar] [CrossRef]
- Chung, H.Y.; Won, S.Y.; Kim, Y.K.; Kim, J.S. Development of the chloroplast genome-based InDel markers in Niitaka (Pyrus pyrifolia) and its application. Plant Biotechnol. Rep. 2019, 13, 51–61. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics Methods and Protocols; Humana Press: Totowa, NJ, USA, 1999; pp. 365–386. [Google Scholar]
- Schuler, G.D. Sequence mapping by electronic PCR. Genome Res. 1997, 7, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, C.; Jiao, X.; Zhang, H.; Song, L.; Li, Y.; Gao, C.; Wang, K. Hi-TOM: A platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 2019, 62, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Liang, Z.; Duan, S.; Sheng, J.; Zhu, S.; Ni, X.; Shao, J.; Liu, C.; Nick, P.; Du, F.; Fan, P.; et al. Whole-genome resequencing of 472 Vitis accessions for grapevine diversity and demographic history analyses. Nat. Commun. 2019, 10, 1190. [Google Scholar] [CrossRef]
- Myles, S.; Boyko, A.R.; Owens, C.L.; Brown, P.J.; Grassi, F.; Aradhya, M.K.; Prins, B.; Reynolds, A.; Chia, J.-M.; Ware, D.; et al. Genetic structure and domestication history of the grape. Proc. Natl. Acad. Sci. USA 2011, 108, 3530–3535. [Google Scholar] [CrossRef]
- Kvikstad, E.M.; Tyekucheva, S.; Chiaromonte, F.; Makova, K.D. A macaque’s-eye view of human insertions and deletions: Differences in mechanisms. PLoS Comput. Biol. 2007, 3, e176. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Roorkiwal, M.; Kale, S.; Garg, V.; Yadala, R.; Varshney, R.K. InDel markers: An extended marker resource for molecular breeding in chickpea. PLoS ONE 2019, 14, e0213999. [Google Scholar] [CrossRef] [PubMed]
- Adedze, Y.M.N.; Lu, X.; Xia, Y.; Sun, Q.; Nchongboh, C.G.; Alam, M.A.; Liu, M.; Yang, X.; Zhang, W.; Deng, Z.; et al. Agarose-resolvable InDel markers based on whole genome re-sequencing in cucumber. Sci. Rep. 2021, 11, 3872. [Google Scholar] [CrossRef]
- Pan, G.; Li, Z.; Huang, S.; Tao, J.; Shi, Y.; Chen, A.; Li, J.; Tang, H.; Chang, L.; Deng, Y.; et al. Genome-wide development of insertion-deletion (InDel) markers for Cannabis and its uses in genetic structure analysis of Chinese germplasm and sex-linked marker identification. BMC Genom. 2021, 22, 595. [Google Scholar] [CrossRef] [PubMed]
- Lijavetzky, D.; Cabezas, J.; Ibáñez, A.; Rodríguez, V.; Martínez-Zapater, J.M. High throughput SNP discovery and genotyping in grapevine (Vitis vinifera L.) by combining a re-sequencing approach and SNPlex technology. BMC Genom. 2007, 8, 424. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Lyu, M.; Liu, J.; Wu, J.; Wang, Q.; Xie, T.; Li, H.; Chen, R.; Sun, D.; Yang, Y.; et al. Construction of an SNP fingerprinting database and population genetic analysis of 329 cauliflower cultivars. BMC Plant Biol. 2022, 22, 522. [Google Scholar] [CrossRef]
- Shen, Y.; Wang, J.; Shaw, R.K.; Yu, H.; Sheng, X.; Zhao, Z.; Li, S.; Gu, H. Development of GBTS and KASP panels for genetic diversity, population structure, and fingerprinting of a large collection of broccoli (Brassica oleracea L. var. italica) in China. Front. Plant Sci. 2021, 12, 655254. [Google Scholar] [CrossRef]
- Eltaher, S.; Sallam, A.; Belamkar, V.; Emara, H.A.; Nower, A.A.; Salem, K.F.M.; Poland, J.; Baenziger, P.S. Genetic diversity and population structure of F3: 6 Nebraska winter wheat genotypes using genotyping-by-sequencing. Front. Genet. 2018, 9, 76. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, G.; Zhang, N.; Yang, Y.; Jin, Q.; Jiang, J.; Zhang, H.; Guo, Y.; Wang, Q.; Zhang, H.; Wu, J.; et al. NGS-Based Multi-Allelic InDel Genotyping and Fingerprinting Facilitate Genetic Discrimination in Grapevine (Vitis vinifera L.). Horticulturae 2024, 10, 752. https://doi.org/10.3390/horticulturae10070752
Jia G, Zhang N, Yang Y, Jin Q, Jiang J, Zhang H, Guo Y, Wang Q, Zhang H, Wu J, et al. NGS-Based Multi-Allelic InDel Genotyping and Fingerprinting Facilitate Genetic Discrimination in Grapevine (Vitis vinifera L.). Horticulturae. 2024; 10(7):752. https://doi.org/10.3390/horticulturae10070752
Chicago/Turabian StyleJia, Guiying, Na Zhang, Yingxia Yang, Qingdong Jin, Jianfu Jiang, Hong Zhang, Yutong Guo, Qian Wang, He Zhang, Jianjin Wu, and et al. 2024. "NGS-Based Multi-Allelic InDel Genotyping and Fingerprinting Facilitate Genetic Discrimination in Grapevine (Vitis vinifera L.)" Horticulturae 10, no. 7: 752. https://doi.org/10.3390/horticulturae10070752
APA StyleJia, G., Zhang, N., Yang, Y., Jin, Q., Jiang, J., Zhang, H., Guo, Y., Wang, Q., Zhang, H., Wu, J., Chen, R., Huang, J., & Lyu, M. (2024). NGS-Based Multi-Allelic InDel Genotyping and Fingerprinting Facilitate Genetic Discrimination in Grapevine (Vitis vinifera L.). Horticulturae, 10(7), 752. https://doi.org/10.3390/horticulturae10070752





