Abstract
A cytological study was carried out on four Zingiber species from Thailand, namely, Z. chrysostachys, Z. isanense, Z. junceum, and Z. niveum, which are edible and beautiful ornamental plants. They all have somatic chromosomal numbers of 2n = 22. This research contributes to karyological knowledge regarding this species. The somatic chromosomal counts of Z. niveum and Z. isanense are reported for the first time, as are the NFs of all species, which were all discovered to be 44. All four edible and ornamental species had their karyotypes: 16m + 6sm for Z. chrysostachys, 4m + 18sm for Z. isanense, 12m + 10sm for Z. junceum, and 14m + 4sm + 4st for Z. niveum. The dominant characteristics of these four Zingiber species are as follows: Z. chrysostachys has yellow bracts, pale yellow flowers, and a red labellum with white dots; Z. isanensis has red-brown bracts, white flowers, and a white labellum; Z. junceum has green bracts, yellow flowers, and a yellow labellum; and Z. niveum has white bracts, yellow flowers, and a yellow labellum. Additionally, principal component analysis (PCA) of the karyotype formula was used to divide the four Zingiber species into two groups via various points using the chromosome indexes (CIs): Z. niveum (D) with Z. chrysostachys (A), and Z. junceum (C) with Z. isanensis (B). This finding implies that, while being in the same stage, the CIs of these four Zingiber species can be used to distinguish them, revealing their resemblance at unique stages and close relationship. Accordingly, the chromosomal structure, karyotype formulae, and CIs can be used to distinguish these four edibles and ornamental Zingiber species from Thailand.
1. Introduction
The Zingiberaceae family, also referred to as the ginger family, consists of a vast range of flowering plants that are found across tropical and subtropical regions globally, covering Africa, the Americas, and Asia. There are around 57 recognized groups and more than 1900 distinct types of species [1]. Zingiberaceae plants exhibit adaptability to several ecological circumstances, although they are primarily distributed in tropical rainforests and damp habitats. They flourish in tropical and subtropical environments characterized by high temperatures and moisture, where they can benefit from ample precipitation and adequate shelter. However, certain species can also be found in arid regions exposed to direct sunshine. This family consists of both terrestrial and epiphytic species. The Zingiberaceae family is well-known for its exceptional biodiversity, encompassing a variety of plants that are both economically useful and of great importance [2]. These plants exhibit a diverse array of colors, forms, and sizes. Furthermore, they have been employed for diverse applications, and the understanding of their usage has also been transmitted through generations in human societies, specifically in Southeast Asia [1,2]. Thailand has an extensive variety of Zingiberaceae species, making it one of the countries with the highest abundance and diversity of this plant family. Thailand harbors around 29 recognized groups and more than 400 species of Zingiberaceae [1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17]. Thailand’s tropical climate and diversified biosphere provide ideal conditions for the flourishing and spread of Zingiberaceae species. Plants belonging to the ginger family have diverse applications, serving as food and spice sources, medicinal plants, decorative flowers, and cultural symbols. They are involved in traditions, such as offering flowers to monks, and tourism, like the Krachiew flower fields in Chaiyaphum province. Additionally, they are used in the production of cosmetics, dyes, and essential oil extracts and in spas [2]. Thailand is highly significant for understanding the wide variety and range of evolutionary tendencies within this intriguing plant family [6].
The large genus Zingiber belongs to the Zingiberaceae family [2]. Zingiber has been found in China, India, Australia, Southeast Asia, Papua New Guinea, and China [17,18,19]. The medicinal herbs Zingiber officinale (or ginger), Z. montanum, and Z. zerumbet, among others, are important [16]. There are a total of 56 Thai Zingiber species, consistent with 60 taxa [17]. Zingiber, as a result, belongs to one of the most diverse genera in Thailand [17]. There are many uses for the plants in this genus. Many Zingiber species’ rhizomes are often utilized in Thai food as sauces or other components. Other species are also used as food, medicinal plants, ornamental plants, commercial plants, and ritual plants [6]. Zingiber corallinum, Z. myoga, Z. officinale, Z. striolatum, and Z. zerumbet have been the focus of studies on biological and pharmacological activity, but a vast number of phytochemical constituents have since been discovered that are thought to represent genus features [18], suggesting that many more species in this genus should possess the same application potential.
The study of the chromosomes of plants in the ginger family mostly involves studying their chromosome numbers [20,21,22,23,24,25,26]. As for the study of karyotypes and ideograms, it was found that there are very few studies [23,24,25]. This may be due to the small size of chromosomes in the plant family, and Evaluating the distribution of chromosomes is a difficult assignment. This could explain the low number of studies conducted on the chromosomes of plants within the ginger family [13,14,15]. Moreover, it has been found that there are very few chromosome studies on the Zingiber genus [26,27,28,29,30].
Notwithstanding, previous cytogenetic studies on Zingiber showed that Zingiber chrysostachys [22], Z. corallinum [11], Z. junceum [23], Z. ligulatum [25], Z. mekongense [23], Z. mioga [28], Z. macrostachyum [25], Z. montanum [10,29], Z. officinale [15,25,29], Z. ottensii [22,29], Z. parishii subsp. phuphanense [25], Z. purpureum [22,25], Z. roseum [25], Z. rubens [23], Z. wightianum [25], Z. aff. wrayi [15], Z. zerumbet [22,23,25], have a chromosomal count of 2n = 22.
Additionally, karyotype studies have reported the formulae of a few Zingiber species, i.e., Zingiber montanum (2a + 18m + 2sm) [10], Zingiber officinale [31], and Z. ligulatum (10m + 12sm), as well as Z. parishii subsp. phuphanense (16m + 6sm) [25]. Zingiber genus ideogram investigations have been limited to three species: Zingiber ligulatum [25], Z. montanum [10], and Z. parishii subsp. phuphanense [25].
Research team discovered four species of Zingiber, namely, Z. chrysostachys, Z. isanensis, Z. junceum, and Z. niveum, while collecting samples from the Zingiberaceae family in the northeastern part of Thailand. Additionally, villagers utilized these plants in a variety of ways; for example, young inflorescences, leaves, and rhizomes can all be eaten as vegetables or used in local food, including as ingredients in local soups and salads. The rhizomes, pseudostems, and young inflorescences of the four Zingiber species mentioned above were traded at the local market. Furthermore, villagers cultivated them in their gardens for ornamental use, as the inflorescences and pseudostems of all the plants are attractive [2,6,16]. Therefore, each of the four Zingiber plants is widely sought after for both food and ornamental uses and can be readily purchased in the regions where they are native. These forest plants were discovered to be extensively cultivated and propagated within the community for the purpose of harvesting flowers and rhizomes, which are then sold in the local market or sent to other provinces. These four species of Zingiber plants are economically significant at the community level in Northeastern Thailand and have been integral to the local people’s way of life since ancient times [16,17,23].
Moreover, they are consumed locally. Zingiber junceum and Z. isanense are listed as least-concern (LC) species on the IUCN Red List [19]; however, Zingiber chrysostachys and Z. niveum are listed as endangered (EN). All four species are classified as rare in Thailand according to the World Checklist of Selected Plant Families [1]. Researchers are interested in researching the chromosomal information of these plants due to the aforementioned reasons. The purpose of this research is to provide fundamental data for the subsequent advancement of these plants as widely consumed indigenous food crops. The goal of this research is to advocate for the cultivation of ornamental plants and flowers as profitable agricultural commodities. Hence, knowledge of chromosome information holds great significance for future advancements.
Several experts have recognized the significance of chromosomal information in plant systematics and evolution. Chromosome morphology can help us comprehend taxonomic relationships at the general and subgenomic levels. Chromosomal morphologies and structures serve as the basis for taxonomy [20,21]. Previous investigations have clarified the chromosomal structures of very few Zingiber species [22,23,24], although there is still a lack of knowledge of karyology. The chromosomal numbers of these four edibles and ornamental Zingiber species, namely, Z. chrysostachys, Z. isanensis, Z. junceum, and Z. niveum, have been recorded in a few publications. However, their karyological aspects have never been studied before. As a result, the purpose of this study was to provide more information regarding their chromosome numbers, the Fundamental Number (NF: number of chromosome arms), and karyotype forms.
2. Materials and Methods
2.1. Sample Collection
Zingiber chrysostachys (coll. No. Saensouk 3900), Z. niveum (coll. No. Saensouk 3901), Z. isanense (coll. No. Saensouk 3902), and Z. junceum (coll. No. Saensouk 3903) were obtained from natural settings in different provinces in the northeastern part of Thailand. Zingiber chrysostachys was discovered in a villager’s garden (a deciduous dipterocarp forest) in Nakhon Phanom Province, which is located in the upper region of the northeastern part from Thailand. Z. niveum and Z. isanense were collected from a deciduous dipterocarp forest (a villager’s garden) in Ubon Ratchathani Province, which is located in the lower region of Northeast Thailand. Z. junceum was obtained from a villager’s garden (which was like a deciduous dipterocarp forest) in Beung Kan Province, located in the upper region of Northeast Thailand. They were kept alive by being grown in a nursery at Mahasarakham University in Thailand’s Maha Sarakham Province. Figure 1 depicts the blooms of the species investigated in this study.

Figure 1.
Flowers of edible and ornamental Zingiber species in this study: (a) Zingiber chrysostachys, (b) Z. isanense, (c) Z. junceum, and (d) Z. niveum. Scale bars = 1 cm.
2.2. Chromosome Number and Karyotype Study
Root tips from four Zingiber species, which are both edible and ornamental, were prepared with paradichlorobenzene at 4 °C for 6 h before being fixed in ethanol-acetic acid (3:1, v:v) at room temperature for 30 min. The roots were kept at 4 °C in case they needed to be used later. The samples were rinsed in distilled water, hydrolyzed in 1 M of HCl for 5 min at 60 °C, and then washed again in distilled water. They were dyed with 2% aceto-orcein and examined using the squash technique. Observations were made using a light microscope (Zeiss Axiostar Plus: Carl Zeiss Light Microscopy, Göttingen, Germany) operated at 400× magnification [25,26]. The metaphase plates with well-individualized chromosomes were imaged. The chromosomes were counted, and the karyotype formulas were determined from measurements of the metaphase chromosomes apparent in photomicrographs. The chromosome morphology was described using the terminology used [22,24,25,26,27,28,29,30,31,32,33,34,35].
2.3. Statistical Analysis
The average karyotype of all four Zingiber species, which includes measurements of the short arm chromosome, long arm chromosome, total chromosome arms, relative length, and centromeric index, was used as a representation of all the results. The average was calculated by taking into account all the variables, along with one standard deviation (SD). We used calculations, following the methods defined in references [28,29,30,31], to evaluate the similarity among the four Zingiber species. Our study focused on the karyotype formula data and the presence or absence of distinguishing features related to somatic chromosomes. We took into account a somatic chromosomal number of 2n = 22. In addition, we assigned a binary value of 1 or 0 to indicate the morphological traits of the four Zingiber species that were analyzed. A distance matrix was used in the dendrogram (UPGMA) (unweighted pair group method with arithmetic mean) [32,33,34], and principal component analysis (PCA) [22,34,35,36,37,38,39] was performed using SPSS version 29.
3. Results
The Zingiber (family: Zingiberaceae) species Z. chrysostachys, Z. isanense, Z. junceum, and Z. niveum, obtained from several provinces in Thailand for this study (Figure 1), were identified as rare species in Thailand. Two species are considered endangered (Zingiber chrysostachys and Z. niveum), while two are considered least-threatened (Zingiber isanense and Z. junceum). Figure 2 shows photomicrographs of the chromosomes from the four species. Table 1 summarizes the somatic chromosomal counts corresponding to the root tips of four rare Zingiber species. For each species, chromosomal counts were performed on ten metaphase plates. All the species’ chromosomal counts were discovered to be 2n = 22. Figure 3 and Figure 4 show the species’ karyotypes and ideograms, respectively.
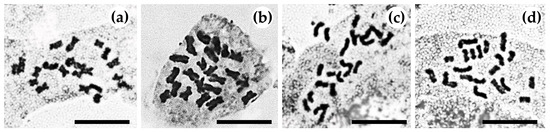
Figure 2.
Photomicrographs of somatic metaphase plate 2n = 22: (a) Zingiber chrysostachys, (b) Z. isanense, (c) Z. junceum, and (d) Z. niveum. Scale bars = 10 µm.

Table 1.
The cytogenetics with conservation status, dominant characteristics, and traditional uses of the four species in the genus Zingiber analyzed in the present study.

Figure 3.
Karyotypes of (a) Zingiber chrysostachys, (b) Z. isanense, (c) Z. junceum, and (d) Z. niveum. Scale bars = 10 µm.
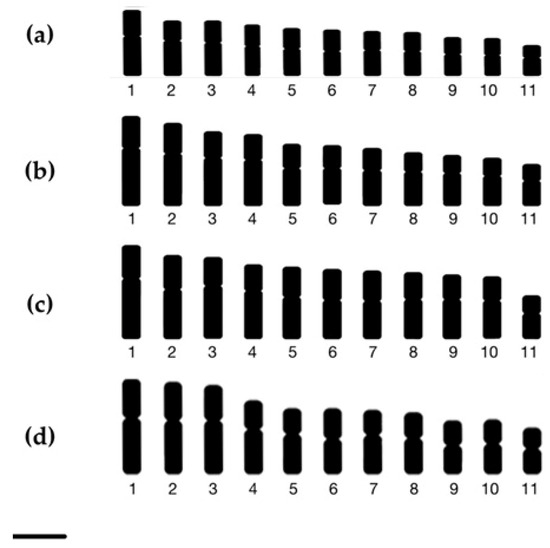
Figure 4.
Ideograms of (a) Zingiber chrysostachys, (b) Z. isanense, (c) Z. junceum, and (d) Z. niveum. Scale bars = 10 µm.
The most important distinguishing characteristics of these four Zingiber species, which can be used for both food and ornamentation, are as follows: Z. chrysostachys has yellow bracts, pale yellow flowers, and a red labellum adorned with white dots. Z. isanensis has red-brown bracts, white flowers, and a white labellum. Z. junceum features green bracts, yellow flowers, and a yellow labellum. Lastly, Z. niveum presents white bracts, yellow flowers, and a yellow labellum.
In the field survey, we recorded the conventional utilization of all four edible and ornamental Zingiber species. The young inflorescences and young leaves of all species are utilized as edible vegetables or local foods. Additionally, young rhizomes of two specific species (Z. junceum and Z. niveum) were eaten as fresh vegetables or as part of the local cuisine. All of these species are utilized for ornamental purposes, namely, as plants for one’s home.
These four species, which can be eaten or used for ornamentation, have a highly beneficial arrangement of chromosomes in each cell. Consequently, in this study, specific counting of chromosomes could be conducted by using intact plant root parts and employing treatments such as chromosomal identifying and technological addition, resulting in good karyotype and ideogram data.
Zingiber chrysostachys has a somatic chromosomal number of 2n = 22 and a fundamental number (NF) of 44 (Figure 2a) (Table 1). The karyotype of Zingiber chrysostachys contained eight pairs of metacentric chromosomes and three pairs of submetacentric chromosomes. The karyotype formula was 16m + 6sm, indicating that the karyotype was symmetrical (Table 1, Figure 3a). Because of the arm ratio, it had a symmetrical karyotype. The short arm length was 0.68 ± 0.04 to 1.40 ± 0.09 µm, the long arm length was 0.98 ± 0.06 to 2.17 ± 0.65 µm, and the total chromosomal length was 1.66 ± 0.11 to 3.57 ± 0.73 µm. The centromeric index (CI) ranged from 0.54 ± 0.06 to 0.64 ± 0.10, and the relative chromosomal length (RL) ranged from 5.91 ± 0.04 to 12.72 ± 0.04% (Table 2). The CI and the average length of chromosomes were used to arrange the chromosomes into an ideogram with size decreasing from left to right (Figure 4a).

Table 2.
Mean lengths of short-arm chromosomes (Ls), long-arm chromosomes (Ll), total chromosome arm (LT), relative length (RL), and centromeric index (CI) of four Zingiber species obtained from 10 metaphase plates.
Zingiber isanense has 2n = 22 somatic chromosomes and an NF = 44 (Figure 2b, Table 1). There were two pairs of metacentric chromosomes and nine pairs of submetacentric chromosomes in the corresponding karyotype. Its arm ratio and karyotype formula of 4m + 18sm place it in the symmetrical karyotype group (Figure 3b). The short arm length was 0.68 ± 0.01 to 1.37 ± 0.05 µm, the long arm length was 1.08 ± 0.01 to 2.45 ± 0.01 µm, and the total chromosomal length was 1.76 ± 0.01 to 3.83 ± 0.06 µm. The RL ranged from 6.00 ± 0.01 to 13.00 ± 0.02%, while the CI ranged from 0.59 ± 0.01 to 0.65 ± 0.01, as shown in Table 2. Figure 4b shows the ideogram that was created.
Zingiber junceum has 2n = 22 chromosomes and an NF = 44 (Figure 2c) (Table 1). Six pairs of metacentric and five pairs of submetacentric chromosomes were discovered. Because of the formula, namely, 12m + 10sm, and the arm ratio of the chromosomes, the karyotype seemed to be symmetrical. The short arm length was 0.66 ± 0.04 to 1.24 ± 0.08 µm, the long arm length was 0.95 ± 0.06 to 2.22 ± 0.65 µm, and the total chromosomal length was 1.61 ± 0.10 to 3.46 ± 0.73 µm. Table 2 shows that the RL ranged from 5.57 ± 0.08 to 12.00 ± 0.16% and that the CI ranged from 0.59 ± 0.10 to 0.64 ± 0.13 (Table 2). Figure 4c shows the ideogram that was created.
The somatic chromosomal number of Zingiber niveum, reported herein for the first time, is 2n = 22, with an NF = 44 (Figure 2d, Table 1). There were seven pairs of metacentric chromosomes, four pairs of submetacentric chromosomes, and four pairs of subtelocentric chromosomes found. 14m + 4sm + 4st was the karyotype formula. The arm ratio revealed that it was a asymmetrical karyotype (Table 1, Figure 3d). The short arm length was 0.86 ± 0.06 to 1.65 ± 0.10 µm, the long arm length was 1.09 ± 0.07 to 2.33 ± 0.68 µm, and the total chromosomal length was 1.94 ± 0.13 to 3.98 ± 0.78 µm. The RL ranged from 6.10 ± 0.12 to 12.50 ± 0.20, and the CI varied from 0.56 ± 0.10 to 0.58 ± 0.10 (Table 2). The ideogram of Zingiber niveum is depicted in Figure 4d.
3.1. The Similarity Index of Four Rare Zingiber Species
The results of a UPMG cluster analysis, which examined the similarity among four species of Zingiber, included Zingiber chrysostachys, Z. isanense, Z. junceum, and Z. niveum. This analysis was based on their karyotype formulas, which represent the chromosome characteristics and 30 morphological characteristics of each species. The UPMG cluster analysis produced a dendrogram, a diagram showing the relationships among these species based on their similarities in terms of chromosome characteristics and morphology. The dendrogram revealed that Zingiber junceum and Z. niveum are the most similar to each other in terms of their karyotype formulas and morphologies; these two species have similar flowers and inflorescences. Following them, Z. chrysostachys showed a moderate level of similarity, while Z. isanense exhibited the least similarity to the other species (Figure 5).
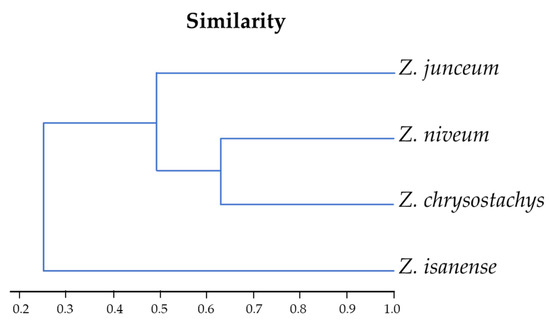
Figure 5.
UPGM cluster analysis dendrogram of four rare Zingiber species based on the distant matrix of karyotype formulars, similarity index, and 30 morphological characteristics with a cophenetic correlation = 0.9686.
The UPMG cluster analysis provided valuable insights into the similarities among four distinct species of Zingiber: Z. chrysostachys, Z. isanense, Z. junceum, and Z. niveum. The analysis yielded karyotype formulas, similarity indices, and 30 morphological characteristics. The cophenetic correlation, which is 0.9686, indicates a high level of similarity among the four Zingiber species, with a 96.86% similarity rate. Crucially, this analysis utilized karyotype formulas as the basis for comparison, offering a glimpse into the unique chromosome characteristics of each species according to their morphologies.
3.2. The PCA Score Plot of CI
The PCA score plot of Zingiber chrysostachys, Z. isanensis, Z. junceum, and Z. niveum, based on CIs, showed that component 1 accounts for 60.25% of the total variance, while component 2 accounts for 37.64%. All four species are in the same stage, which means that their CIs are similar to each other. The CIs can be used to differentiate between different points. This presentation divides the four species into two groups: Zingiber niveum (D) with Z. chrysostachys (A), and Z. junceum (C) with Z. isanensis (B). This indicates that the CIs of each group are closely related (Figure 6). This result suggests that the CIs of these four rare Zingiber species can be employed to differentiate them even if they are in the same stage, signifying their similarity yet distinct stages while remaining closely related.
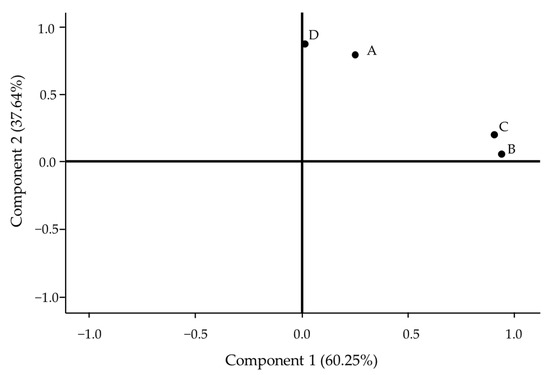
Figure 6.
PCA score plot of (A) Zingiber chrysostachys, (B) Z. isanense, (C) Z. junceum, and (D) Z. niveum based on CI.
4. Discussion
According to the Plants of the World Online database [17], four edible and ornamental species of Zingiber are rare in Thailand: Zingiber chrysostachys, Z. isanensis, Z. junceum, and Z. niveum. Furthermore, two species are classified as endangered (Zingiber chrysostachys and Z. niveum), and two as being of least concern (Zingiber isanensis and Z. junceum), in the International Union for Conservation of Nature and Natural Resources database [16].
Zingiber chrysostachys, Z. isanensis, Z. junceum, and Z. niveum are four rare edible and ornamental Zingiber species discovered in Thailand. They all have the same somatic chromosomal number, 2n = 22 [23,24]. Similarly, it was determined that this species’ haploid chromosome number in Malaysia was n = 11 [23]. The somatic chromosomal counts of Zingiber niveum and Z. isanense are reported herein for the first time. We determined the somatic chromosomal numbers of Zingiber chrysostachys and Z. junceum to be 2n = 22 [23] (Table 1). The NFs of these four Zingiber species were determined to be 44, which aligns with the research results reported by Saensouk and Saensouk [25]. The arm ratio analysis revealed that individuals of three species (Z chrysostachys, Z. isanensis, and Z. junceum) had a karyotype that showed symmetrical characteristics, which corresponds to the findings published by Saensouk and Saensouk [25]. On the other hand, Z. niveum had an asymmetrical karyotype. Nevertheless, the NFs, karyotypes, and ideograms of all four species have been determined for the first time. Therefore, while the chromosome number was consistent across all four species, there were differences in the CIs, RL (%), karyotypes, and ideograms among the species. These differences can be attributed to environmental factors such as the plant’s location, soil composition, water availability, air quality, etc.
The chromosomal structures and karyotype formulae of these four Zingiber species can be used to distinguish them. Researchers can distinguish plants by analyzing their chromosomal and morphological characteristics [36]. The total chromosomal length ranged from 3.46 ± 0.73 µm (Z. junceum) to 3.98 ± 0.78 µm (Z. niveum), which is shorter than the measurements reported in a previous work by Saensouk and Saensouk [25]. This difference in length can be attributed to various environmental conditions, age, or periods of development. The RL content varied from 12.00 ± 0.16% (Z. junceum) to 13.00 ± 0.02% (Z. isanensis), which differs from the earlier findings reported by Saensouk and Saensouk [25]. This difference could be attributed to variations in environmental circumstances, age, or developmental stages. Furthermore, the CI ranged from 0.65 ± 0.01 (Z. isanense) to 0.58 ± 0.10 (Z. niveum), which contrasts with the previous results documented by Saensouk and Saensouk [25]. This disparity can be ascribed to differences in environmental conditions, age, or phases of development.
Karyological data can be employed in cytotaxonomy or karyosystematics since they can represent the genetic links between the species being studied [37]. These features are considered advantageous due to the fact that they are not influenced by environmental circumstances, age, or developmental stages [38]. This research has found that the chromosomal numbers of all four Zingiber species are the same. Nonetheless, banding patterns in karyotypes with identical morphologies can differ significantly [39]. Banding patterns in karyotypes with identical shapes can, however, vary dramatically [39].
The PCA score plot of Zingiber chrysostachys, Z. isanense, Z. junceum, and Z. niveum, based on CI, showed that all four rare species are in the same stage, which means that their CIs are similar to each other. However, this study based on CIs could be divided into two groups, namely, Zingiber niveum (D) with Z. chrysostachys (A) and Z. junceum (C) with Z. isanense (B), which is consistent with [17], who reported the morphological features of leaf blades of Z. niveum (D) and Z. chrysostachys (A) were reported as oblong to lanceolate. On the other hand, the second group, consisting of Z. junceum (C) and Z. isanensis (B), showed narrow lanceolate leaf blade morphology.
Beyond the four rare edible plants belonging to the Zingiber genus, four had distinct karyotype formulas and chromosome structures. However, not all evidence can be sued to differentiate between these four species. The CI is capable of distinguishing between the four species, notwithstanding the fact that the CI values seem to be aligned in the same direction. Additionally, the leaf blade morphologies of these four species can serve as a characteristic with which to differentiate them. According to the findings of this study, it is feasible to categorize all four species of Zingiber using the PCA of the karyotype formula, the CI, and the morphological characteristics of the leaf blade.
Therefore, these data will help in supporting the development of plant breeding for commercial purposes in the future and are expected to generate more income for the community.
5. Conclusions
Cytological research was conducted on four species of Zingiber (Z. chrysostachys, Z. isanensis, Z. junceum, and Z. niveum) that are edible and ornamental, with the aim of obtaining more knowledge of these species. Their somatic chromosomal numbers are all 2n = 22. Our knowledge of these species’ karyology has been increased by this investigation. For the first time, both the somatic chromosomal counts of Zingiber niveum and Z. isanense—in addition to the NFs of all species, all of which were found to be 44—were reported. The karyotypes for all four of these endangered species were made available: 16m + 6sm for Zingiber chrysostachys, 4m + 18sm for Z. isanense, 12m + 10sm for Z. junceum, and 14m + 4sm + 4st for Z. niveum. The karyotypes are symmetrical for each individual. Chromosome architecture and karyotype formulae were used to distinguish four rare Zingiber species from Thailand.
The CIs can be used to categorize these into various points. The four species were divided into two groups in this presentation: Zingiber niveum (D) and Z. chrysostachys (A), and Z. junceum (C) and Z. isanensis (B). This suggests a close relationship between the CIs of each group (Figure 5). This finding implies that, while in the same stage, the CIs of these four rare Zingiber species can be used to distinguish them, indicating their resemblance at unique stages while still being closely related.
The information obtained from this study helps promote the conservation of plants in the areas that are their habitats, which will lead to the conservation of these rare plants and their habitats. Consequently, the genetics of these plants persist, allowing for their propagation and development into ornamental plants with economic value, which could potentially boost a community’s income.
Author Contributions
P.S. (Piyaporn Saensouk) and S.S., methodology; S.S., formal analysis; R.S., data curation; P.S. (Piyaporn Saensouk), S.S., R.S., D.M. and P.S. (Phetlasy Souladeth), writing—original draft preparation; S.S., writing—review and editing; S.S., funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Mahasarakham University.
Data Availability Statement
All data produced and examined are available in this article.
Acknowledgments
This research was financially supported by Mahasarakham University. We are grateful to the Walai Rukhavej Botanical Research Institute, Mahasarakham University, Diversity of Family Zingibeaceae and Vascular Plant of Its Applications Research Unit, and Mahasarakham University, for their facilities during this study. I would like to thank Jolyon Dodgson for conducting language editing and providing suggestions regarding how to improve the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- POWO. Plant of the World Online, Facilitated by the Royal Botanic Gardens, Kew. Available online: http://www.plantsoftheworldonline.org/ (accessed on 12 February 2023).
- Larsen, K.; Larsen, S.S. Ginger of Thailand; Queen Sirikit Botanic Garden, The Botanical Garden Organization: Chiang Mai, Thailand, 2006. [Google Scholar]
- Kress, W.J.; Prince, L.M.; Williams, J.K. The phylogeny and a new classifcation of the gingers (Zingiberaceae): Evidence from molecular data. Am. J. Bot. 2002, 89, 1682–1696. [Google Scholar] [CrossRef] [PubMed]
- Mood, J.D.; Ardiyani, M.; Veldkamp, J.F.; Mandáková, T.; Prince, L.M.; de Boer, H.J. Nomenclatural changes in Zingiberaceae: Haplochorema is reduced to Boesenbergia. Gard. Bull. Singap. 2020, 72, 77–95. [Google Scholar] [CrossRef] [PubMed]
- Aung, M.M.; Tanaka, N.; Miyake, N. Two gingers, Zingiber orbiculatum and Z. flavomaculosum (Zingiberaceae), newly recorded from Myanmar. Bull. Natl. Mus. Nat. Sci. 2015, 41, 107–112. [Google Scholar]
- Saensouk, S.; Saensouk, P.; Pasorn, P.; Chantaranothai, P. Diversity, Traditional Uses and New Record of Zingiberaceae in Nam Nao National Park, Petchabun Province, Thailand. Agric Nat. Resour. 2016, 50, 445–453. [Google Scholar]
- Triboun, T.; Chantaranothai, P.; Larsen, K. Taxonomic changes regarding three species of Zingiber (Zingiberaceae) from Thailand. Acta Phytotax. Sin. 2007, 45, 403–404. [Google Scholar]
- Ikeda, H.; Nam, B.M.; Yamamoto, N.; Funakoshi, H.; Takano, A.; Im, H.T. Chromosome number of myoga ginger (Zingiber mioga: Zingiberaceae) in Korea. Korean J. Plant Taxon. 2021, 51, 100–102. [Google Scholar] [CrossRef]
- Khumkratok, S.; Boontiang, K.; Chutichudet, P.; Pramaul, P. Geographical distributions and ecology of ornamental Curcuma (Zingiberaceae) in Northeastern Thailand. Pak. J. Biol. Sci. 2012, 15, 929–939. [Google Scholar] [CrossRef][Green Version]
- Saensouk, P.; Saensouk, S.; Phechphakdee, T.; Ragsasilp, A. Cytogenetic study in seven species of Zingiberaceae family from Bueng Kan Province, Thailand. Biodiversitas 2023, 24, 68–77. [Google Scholar] [CrossRef]
- Chen, Z.Y. Evolutionary patterns in cytology and pollen structure of Asian Zingiberaceae. In Tropical Forests; Holm-Nielsen, B., Nielsen, I.C., Balslev, H., Eds.; Academic Press Limited: New York, NY, USA, 1989; pp. 185–191. [Google Scholar]
- Lin, Y.-C.; Chao, C.-T.; Chang, C.-Y.; Tseng, Y.-H. Taxonomic revision of Zingiber (Zingiberaceae) of Taiwan. Eur. J. Taxon. 2022, 839, 74–102. [Google Scholar] [CrossRef]
- Eksomtramage, L.; Boontum, K. Chromosome counts of Zingiberaceae. Songklanakarin J. Sci. Technol. 1995, 17, 291–297. [Google Scholar]
- Eksomtramage, L.; Sirirugsa, P.; Sawangchote, P.; Jornead, S.; Saknimit, T.; Leeratiwong, C. Chromosome numbers of some monocot species from Ton-Nga-Chang Wildlife Sanctuary, Southern Thailand. Thai For. Bull. (Bot.) 2001, 29, 63–71. [Google Scholar]
- Eksomtramage, L.; Sirirugsa, P.; Jivanit, P.; Maknoi, C. Chromosome counts of some Zingiberaceous species from Thailand. Songklanakarin J. Sci. Technol. 2002, 24, 311–319. [Google Scholar]
- Ragsasilp, A.; Saensouk, P.; Saensouk, S. Ginger family from Bueng Kan Province, Thailand: Diversity, conservation status, and traditional uses. Biodiversitas 2022, 23, 2739–2752. [Google Scholar] [CrossRef]
- Triboun, P.; Larsen, K.; Chantaranothai, P. A key to the genus Zingiber (Zingiberaceae) in Thailand with descriptions of 10 new taxa. Thai J. Bot. 2003, 6, 53–77. [Google Scholar]
- Deng, M.; Yun, X.; Ren, S.; Qing, Z.; Luo, F. Plants of the Genus Zingiber: A review of their ethnomedicine, phytochemistry and pharmacology. Molecules 2022, 27, 2826. [Google Scholar] [CrossRef] [PubMed]
- IUCN. The IUCN Red List of Threatened Species; Version 2022-2; International Union for Conservation of Nature and Natural Resources: Gland, Switzerland, 2023. [Google Scholar]
- Chaiyasut, K. Cytogenetics and Cytotaxonomy of the Family Zephyranthes; Department of Botany, Faculty of Science, Chulalongkorn University: Bangkok, Thailand, 1989. [Google Scholar]
- Stebbins, G.L. Chromosomal Evolution in Higher Plants; Addison-Wesley Pub. Co.: San Francisco, CA, USA, 1971. [Google Scholar]
- Beltran, I.C.; Kiew, K.Y. Cytotaxonomic studies in the Zingiberaceae. Notes R. Bot. Gard. Edinb. 1984, 41, 541–559. [Google Scholar]
- Saensouk, S.; Chantaranothai, P. The Family Zingiberaceae in Phu Phan National Park. In Proceedings of the 3rd Symposium on the Family Zingiberaceae, Khon Kaen, Thailand, 7–12 July 2002; pp. 16–25. [Google Scholar]
- Saenprom, K.; Saensouk, S.; Saensouk, P.; Senakun, C. Karyomorphological analysis of four species of Zingiberaceae from Thailand. Nucleus 2018, 61, 111–120. [Google Scholar] [CrossRef]
- Saensouk, S.; Saensouk, P. New report on karyotype and ideogram of two Zingiber species, Z. ligulatum and Z. parishii subsp. phuphanense from Thailand. Nucleus 2021, 64, 115–121. [Google Scholar] [CrossRef]
- Bhadra, S.; Bandyopadhyay, M. New chromosome number counts and karyotype analyses in three important genera of Zingiberaceae. Nucleus 2016, 59, 35–40. [Google Scholar] [CrossRef]
- Levan, A.; Fredya, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosome. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Morinaga, T.; Fukushina, E.; Kanui, T.; Tamasaki, Y. Chromosome numbers of cultivated plants. Bot. Mag. 1929, 43, 589–594. [Google Scholar] [CrossRef][Green Version]
- Saensouk, S.; Saensouk, P. Chromosome number of some Zingiberaceous in Thailand. KKU Res. J. 2004, 9, 3–9. [Google Scholar]
- Hong, D.Y.; Zhang, S.Z. Observations on chromosomes of some plants from western Sichuan. Cathaya 1990, 2, 191–197. [Google Scholar]
- Das, A.B.; Rai, S.; Das, P. Estimation of 4C DNA and karyotype analysis in ginger (Zingiber officinale Rosc.) II. Cytologia 1998, 63, 133–139. [Google Scholar] [CrossRef]
- Senavongse, R.; Saensouk, S.; Saensouk, P. Karyological study in three native species of genus Alocasia (Araceae) in the northeast of Thailand. Nucleus 2020, 63, 81–85. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Sengthong, A.; Saensouk, S.; Saensouk, P.; Souladeth, P. Cytogenetic Study of Five Varieties of Callisia repens (Jacq.) L. (Commelinaceae) from Laos. Horticulturae 2023, 9, 1050. [Google Scholar] [CrossRef]
- Bro, R.; Smilde, A.K. Principal component analysis. Anal. Methods 2014, 6, 2812–2831. [Google Scholar] [CrossRef]
- Santhosh, B. Cytological and Palynological Studies on the Family Apocynaceae; Department of Botany, University of Kerala: Thiruvananthapuram, India, 1999. [Google Scholar]
- Dobigny, G.; Ducroz, J.F.; Robinson, T.J.; Volobouev, V. Cytogenetics and Cladistics. Syst. Biol. 2004, 53, 470–484. [Google Scholar] [CrossRef]
- Guerra, M. Chromosome numbers in plant taxonomy: Concepts and implications. Cytogenet. Genome Res. 2008, 120, 339–350. [Google Scholar] [CrossRef]
- Dobigny, G.; Aniskin, V.; Volobouev, V. Explosive chromosome evolution and speciation in the gerbil genus Taterillus (Rodentia, Gerbillinae): A case of two new cryptic species. Cytogenet. Cell Genet. 2002, 96, 117–124. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).