Expression Analysis and Interaction Protein Screening of CoGI, the Key Factor in Photoperiod Regulation of Flowering in Camellia oleifera Abel
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. RNA Extraction and cDNA Synthesis
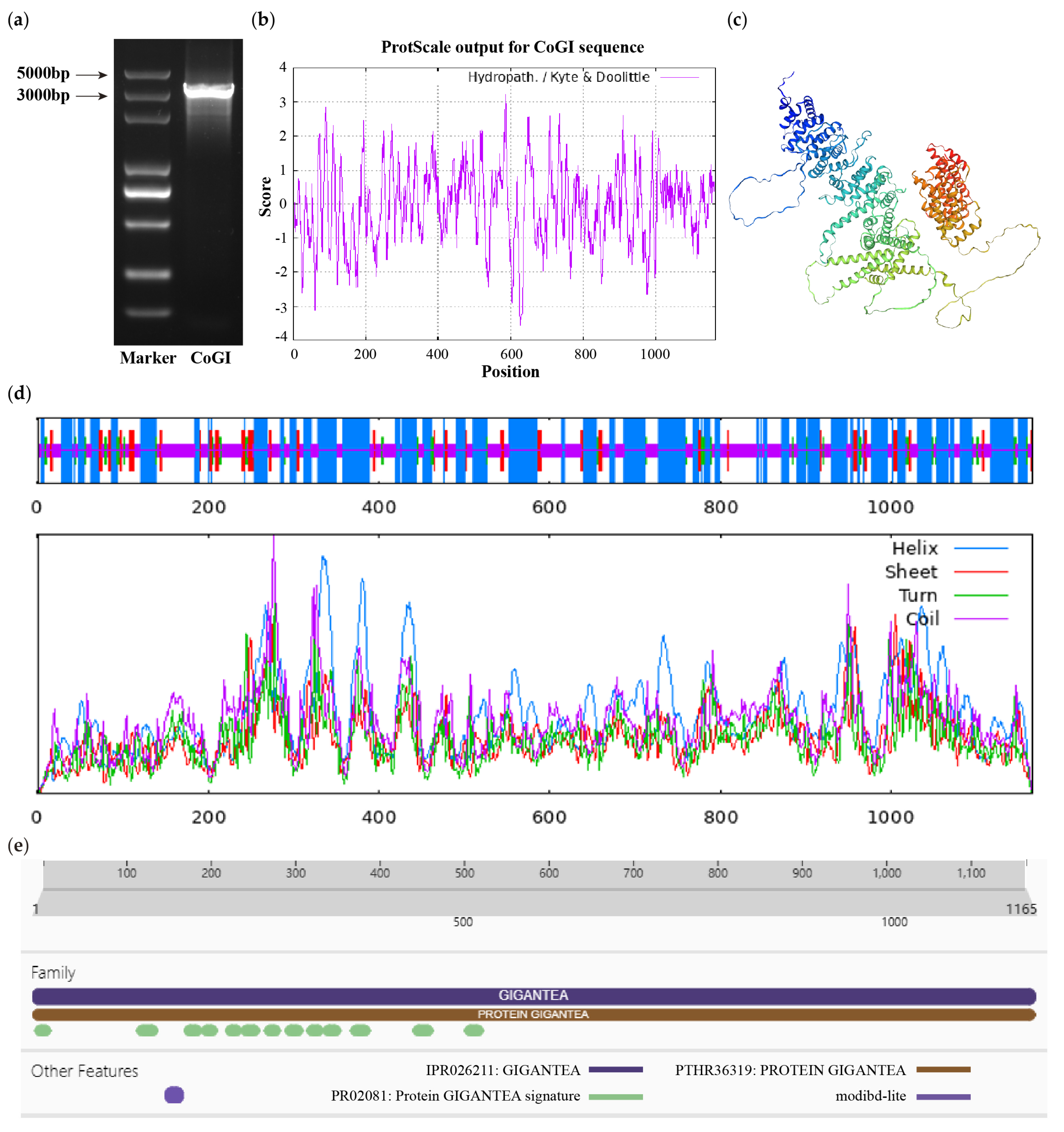
2.3. Clone and Vector Construction of CoGI
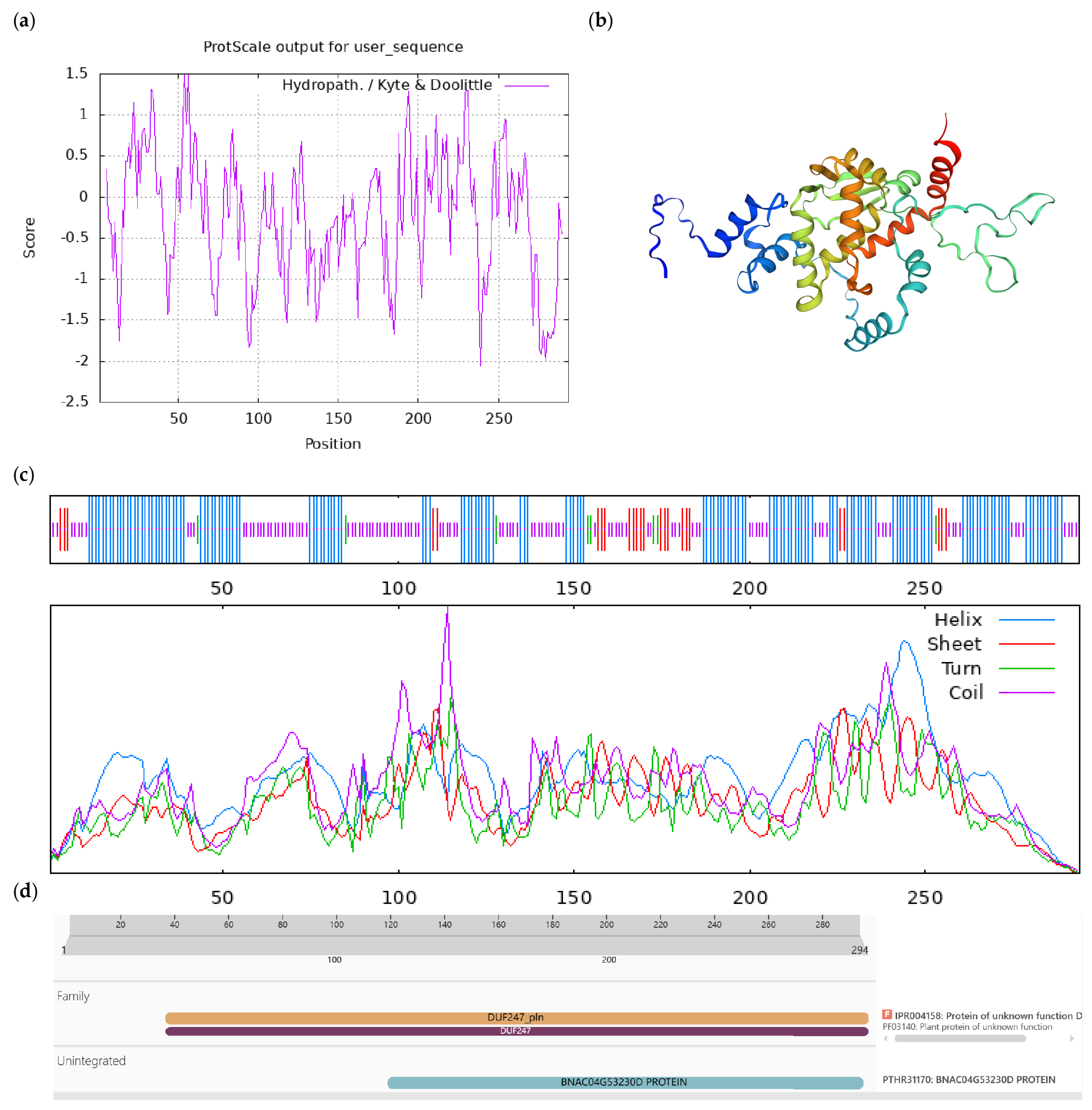
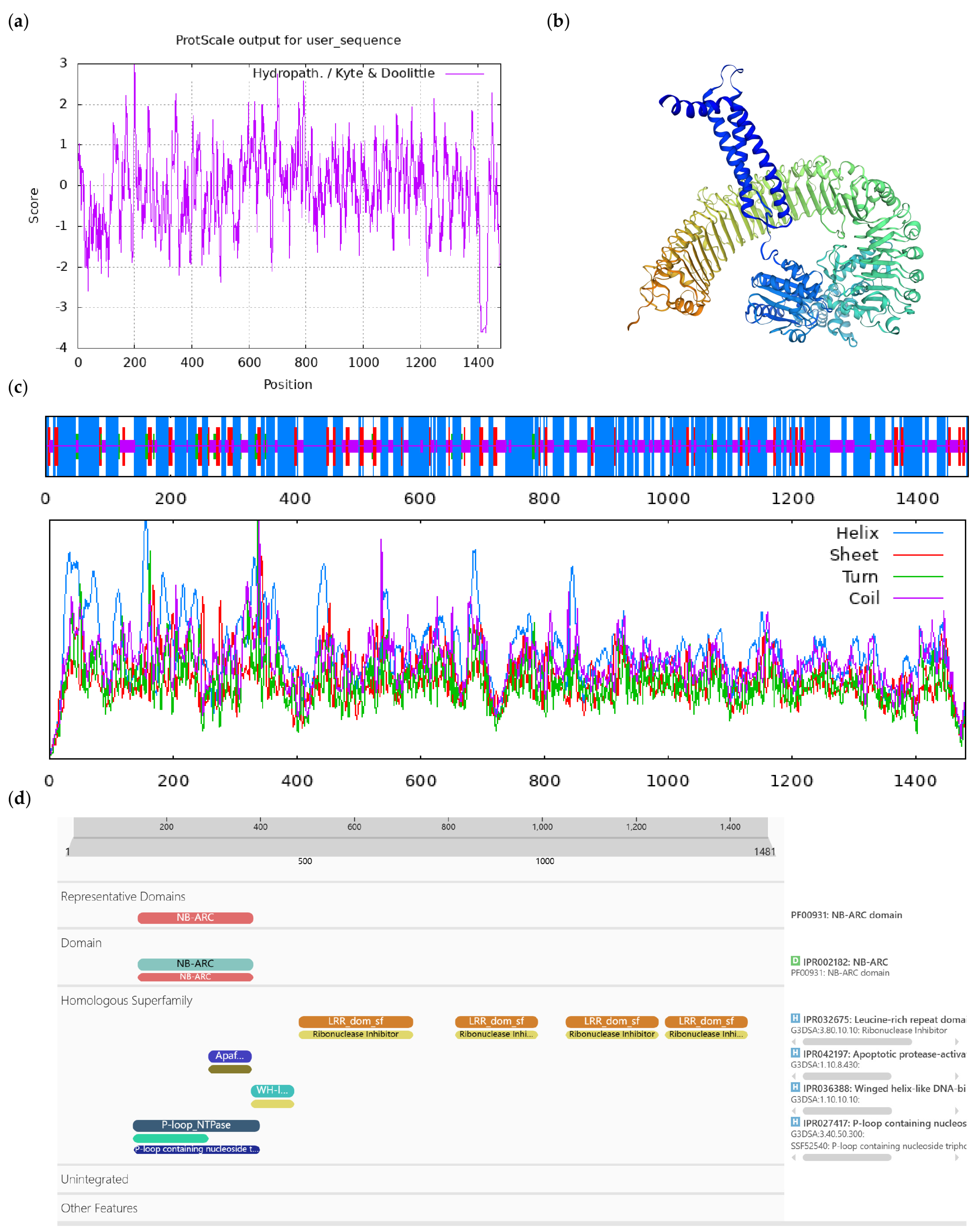
2.4. Bioinformatics Analysis of CoGI
2.5. Expression Analysis of CoGI
2.6. Transformation and Screening of Arabidopsis
2.7. Flowering Phenotypic Observation of Overexpression of CoGI in Transgenic Arabidopsis
2.8. The Autoactivation Activity Test of CoGI
2.9. Screen of Yeast Two-Hybrid Library
3. Results
3.1. Cloning of the CoGI Coding Sequence and Analysis of the Protein
3.2. Homology Analysis and Phylogenetic Tree Construction of CoGI Protein from C. oleifera
3.3. Expression Analysis of CoGI in C. oleifera
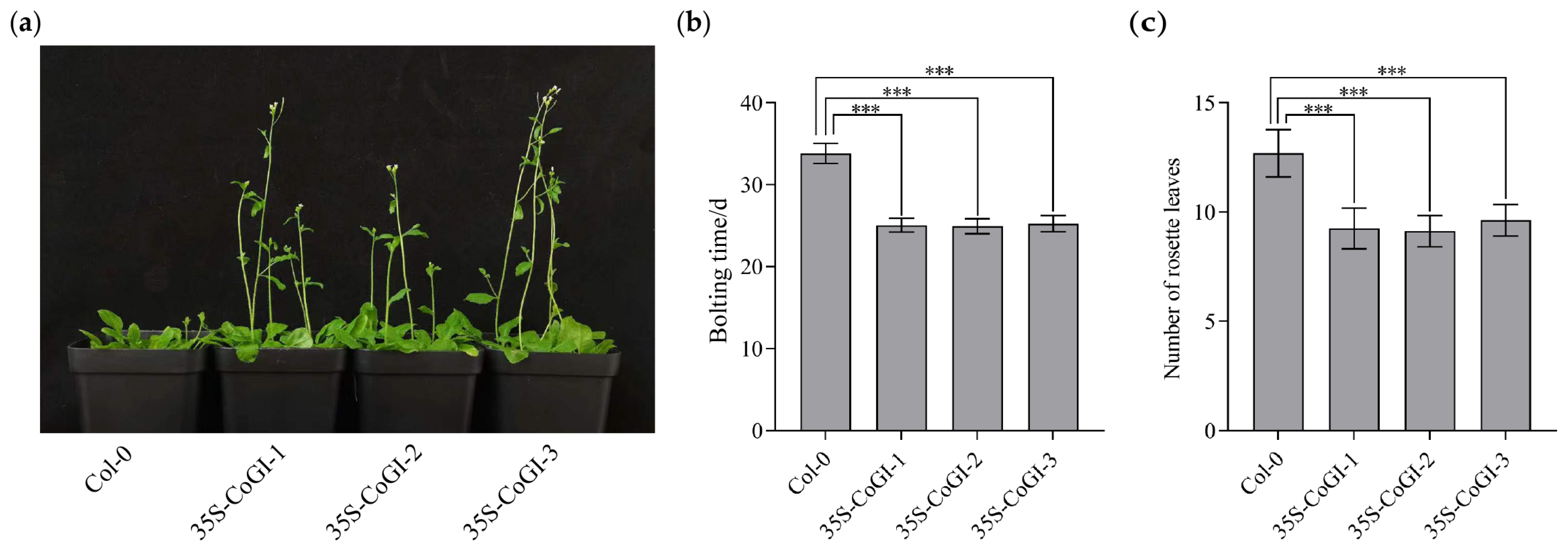
3.4. Heterologous Overexpression of CoGI in Arabidopsis
3.5. Autoactivation Activity Test and Yeast Two-Hybrid Screening of CoGI
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Primer Name | Primer Sequence (5′ to 3′) |
|---|---|
| CoGI-2300-F | GGTACCCGGGGATCCATGGCTACTTCATGCGAAAGG |
| CoGI-2300-R | TCCTCTAGAGGATCCAATTGATATGGTACATCCTAATTCC |
| CoGI-qPCR-F | TCAACAAAGCCGAACCA |
| CoGI-qPCR-R | CAGCAGGGAAACAACAGAG |
| GAPDH-F | CTACTGGAGTTTTCACCGA |
| GAPDH-R | TAAGACCCTCAACAATGCC |
| CoGI-BD-F | AATTCCCGGGGATCCATGGCTACTTCATGCGAAAGG |
| CoGI-BD-R | CAGGTCGACGGATCCAATTGATATGGTACATCCTAATTCC |
| Step | Temperature/°C | Time | Cycles |
|---|---|---|---|
| Predenaturation | 95 | 3 min | 35 |
| Denaturation | 95 | 15 s | |
| Annealing | 58 | 15 s | |
| Extension | 72 | 2 min | |
| Complete extension | 72 | 5 min | |
| Finish | 22 | Forever |
| Step | Temperature/°C | Time | Cycles |
|---|---|---|---|
| Predenaturation | 95 | 5 min | 45 |
| Denaturation | 95 | 30 s | |
| Annealing | 58 | 30 s | |
| Extension | 72 | 30 s |
| Protein | Accession | Species |
|---|---|---|
| GmGI | NP_001341719.1 | Glycine max |
| DlGI | AII99806.1 | Dimocarpus longan |
| SlGI | XP_004237832.1 | Solanum lycopersicum |
| VvGI | XP_010665062.1 | Vitis vinifera |
| ZmGI | NP_001288586.1 | Zea mays |
| OsGI | NP_001396355.1 | Oryza sativa |
| Protein | Accession | Species | Protein | Accession | Species |
|---|---|---|---|---|---|
| CsGI | XP_028106519.1 | Camellia sinensis | QoGI-like | XP_030952170.1 | Quercus lobata |
| ClGI | KAI8004298.1 | Camellia lanceoleosa | QlGI-like | XP_030952183.1 | |
| RcGI | XP_024166287.1 | Rosa chinensis | CiGI-X1 | XP_042962978.1 | Carya illinoinensis |
| VvGI | XP_059590911.1 | Vitisvinifera | CiGI-X2 | XP_042962979.1 | |
| DlGI | XP_052190440.1 | Diospyros lotus | PdGI | AJC01622.1 | Prunus dulcis |
| AcGI | PSS17443.1 | Actinidia chinensis | PdGI-X1 | XP_034196939.1 | |
| QrGI | XP_050269337.1 | Quercus robur | PdGI-X2 | XP_034196940.1 | |
| DzGI-like | XP_022767964.1 | Durio zibethinus | PmGI-X1 | XP_008237481.1 | Prunus mume |
| TcGI | XP_017981038.1 | Theobroma cacao | PmGI-X2 | XP_016650762.1 | |
| JrGI-like | XP_018846578.1 | Juglans regia | PaGI-X1 | XP_021818210.1 | Prunus avium |
| QsGI | XP_023907641.2 | Quercus suber | PaGI-X2 | XP_021818214.1 | |
| VrGI-like | XP_034674793.1 | Vitis riparia | PpGI-X1 | XP_007199688.2 | Prunus persica |
| HuGI | XP_021285158.1 | Herrania umbratica | PpGI-X2 | XP_020426395.1 | |
| BpGI | ALL25874.1 | Betula platyphylla | ApGI-X2 | XP_027367357.1 | Abrus precatorius |

References
- Wang, R.; He, Z.; Zhang, Y.; Zhang, Z.; Wang, X.; Chen, Y. Physiological and Molecular Responses of Camellia oleifera Seedlings to Varied Nitrogen Sources. Horticulturae 2023, 9, 1243. [Google Scholar] [CrossRef]
- Luo, J.; Wei, S.; Zhou, X.; Tian, Y.; Chen, Y.; Song, Q.; Chen, L. Nutrient contents in the organs and soil of young and mature Camellia oleifera C. Abel forests in China. Bangladesh J. Bot. 2022, 51, 359–369. [Google Scholar]
- Wu, L.L.; Li, J.A.; Gu, Y.Y.; Zhang, F.H.; Gu, L.; Tan, X.F.; Shi, M.W. Effect of Chilling Temperature on Chlorophyll Florescence, Leaf Anatomical Structure, and Physiological and Biochemical Characteristics of Two Camellia oleifera Cultivars. Int. J. Agric. Biol. 2020, 23, 777–785. [Google Scholar]
- Zhang, Y.Q.; Guo, Q.Q.; Luo, S.Q.; Pan, J.W.; Yao, S.; Gao, C.; Guo, Y.Y.; Wang, G. Light Regimes Regulate Leaf and Twigs Traits of Camellia oleifera (Abel) in Pinus massoniana Plantation Understory. Forests 2022, 13, 918. [Google Scholar] [CrossRef]
- Hoong-Yeet, Y. Solar rhythm in the regulation of photoperiodic flowering of long-day and short-day plants. J. Exp. Bot. 2013, 64, 2643–2652. [Google Scholar] [CrossRef]
- Chow, B.Y.; Kay, S.A. Global approaches for telling time: Omics and the Arabidopsis circadian clock. Semin. Cell Dev. Biol. 2013, 24, 383–392. [Google Scholar] [CrossRef]
- Suarez-Lopez, P.; Wheatley, K.; Robson, F.; Onouchi, H.; Valverde, F.; Coupland, G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 2001, 410, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Andrés, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nature reviews. Genetics 2012, 13, 627–639. [Google Scholar] [CrossRef]
- Shuai, M.M. Evolution Mechanism of Photoperiodic Floral Genes GIGANTEA and CONSTANS. Master’s Thesis, Zhejiang A&F University, Hangzhou, China, 2018. [Google Scholar]
- Oliverio, K.A.; Crepy, M.; Martin-Tryon, E.L.; Milich, R.; Harmer, S.L.; Putterill, J.; Yanovsky, M.J.; Casal, J.J. GIGANTEA regulates phytochrome A-mediated photomorphogenesis independently of its role in the circadian clock. Plant Physiol. 2007, 144, 495–502. [Google Scholar] [CrossRef][Green Version]
- Qi, X.H.; Wu, D.T.; Li, G.Z.; Zhao, J.L.; Li, M.L. Research progress of flower formation regulation pathway of Arabidopsis. J. Shanxi Agric. Univ. (Nat. Sci. Ed.) 2018, 38, 36. [Google Scholar] [CrossRef]
- Nohales, M.A.; Kay, S.A. Molecular mechanisms at the core of the plant circadian oscillator. Nat. Struct. Mol. Biol. 2016, 23, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Usman, M.A.; Amanda, D.; Jon, S.D.; Marcel, Q. Photoperiod sensing of the circadian clock is controlled by EARLY FLOWERING 3 and GIGANTEA. Plant J. Cell Mol. Biol. 2020, 101, 1397–1410. [Google Scholar] [CrossRef]
- Kim, H.; Park, S.J.; Kim, Y.; Nam, H.G. Subcellular Localization of GIGANTEA Regulates the Timing of Leaf Senescence and Flowering in Arabidopsis. Front. Plant Sci. 2020, 11, 589707. [Google Scholar] [CrossRef]
- Liang, Q.Z.; Song, K.H.; Lu, M.S.; Dai, T.; Yang, J.; Wan, J.X.; Li, L.; Chen, J.J.; Zhan, R.L.; Wang, S.B. Transcriptome and Metabolome Analyses Reveal the Involvement of Multiple Pathways in Flowering Intensity in Mango. Front. Plant Sci. 2022, 13, 933923. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Geng, R.; Gallenstein, R.A.; Somers, D.E. The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development 2013, 140, 4060–4069. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wang, X.F.; Zheng, C.S.; Xing, S.Y.; Shu, H.R. Full-length cDNA cloning, sequence information and quantitative expression analysis of CmGI (GIGANTEA) output gene from Chrysanthemum circadian clock. Chin. J. Agric. Sci. 2012, 45, 2690–2703. [Google Scholar]
- Izawa, T.; Mihara, M.; Suzuki, Y.; Gupta, M.; Itoh, H.; Nagano, A.J.; Motoyama, R.; Sawada, Y.; Yano, M.; Hirai, M.Y.; et al. Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell 2011, 23, 1741–1755. [Google Scholar] [CrossRef]
- Karlgren, A.; Gyllenstrand, N.; Källman, T.; Lagercrantz, U. Conserved function of core clock proteins in the gymnosperm Norway spruce (Picea abies L. Karst). PLoS ONE 2013, 8, e60110. [Google Scholar] [CrossRef]
- Yan, J.D. Molecular Genetics and Biochemical Analysis of Arabidopsis FKF1 Controlling Flowering by Regulating Gibberellin Signaling. Ph.D. Thesis, Hunan University, Changsha, China, 2020. [Google Scholar] [CrossRef]
- Ren, S.S. Cloning and Functional Study of Blue Light Receptor Gene CoZTL in Camellia oleifolia. Master’s Thesis, Central South University of Forestry and Technology, Changsha, China, 2023. [Google Scholar] [CrossRef]
- You, L.N. Screening and Functional Analysis of VfbZIP and VfWRKY Genes Involved in Oil Synthesis of Alnia oleifera. Master’s Thesis, Central South University of Forestry and Technology, Changsha, China, 2023. [Google Scholar] [CrossRef]
- Wang, X.N. Research on Phenology and Blossom Biology of Camellia oleifera Abel. Master’s Thesis, Central South University of Forestry and Technology, Changsha, China, 2011. [Google Scholar]
- Tai, Y.; Wei, C.; Yang, H.; Zhang, L.; Chen, Q.; Deng, W.; Wei, S.; Zhang, J.; Fang, C.; Ho, C.; et al. Transcriptomic and phytochemical analysis of the biosynthesis of characteristic constituents in tea (Camellia sinensis) compared with oil tea (Camellia oleifera). BMC Plant Biol. 2015, 15, 190. [Google Scholar] [CrossRef]
- Wiktorek-Smagur, A.; Hnatuszko-Konka, K.; Kononowicz, A.K. Flower bud dipping or vacuum infiltration-two methods of Arabidopsis thaliana transformation. Russ. J. Plant Physiol. 2009, 56, 560–568. [Google Scholar] [CrossRef]
- Ren, S.S.; Juan, L.M.; He, J.C.; Liu, Q.; Yan, J.D.; Li, J.A. Expression Analysis and Interaction Protein Screening of CoZTL in Camellia oleifera Abel. Horticulturae 2023, 9, 833. [Google Scholar] [CrossRef]
- Makuch, L. Yeast Two-Hybrid Screen. Methods Enzymol. 2014, 539, 31–51. [Google Scholar] [CrossRef] [PubMed]
- Soellick, T.R.; Uhrig, J.F. Development of an optimized interaction-mating protocol for large-scale yeast two-hybrid analyses. Genome Biol. 2001, 2, research0052.0051. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Q.; Ling, H.E.; Jun, W.; Cheng, J. Screening of hepatocyte proteins binding with C-terminally truncated surface antigen middle protein of hepatitis B virus (MHBst167) by a yeast two-hybrid system. Mol. Med. Rep. 2014, 10, 1259–1263. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Wang, T.; Che, J.J.; Yu, F.; Liu, H.L.; Zuo, Z.Y.; Yang, Z.H.; Fan, H.D. Screening and identification of proteins interacting with IL-24 by the yeast two-hybrid screen, Co-IP, and FRET assays. Anti-Cancer Drugs 2016, 27, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Kundu, P.; Sahu, R. GIGANTEA confers susceptibility to plants during spot blotch attack by regulating salicylic acid signalling pathway. Plant Physiol. Biochem. 2021, 167, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Seo, P.J. Dependence and independence of the root clock on the shoot clock in Arabidopsis. Genes Genom. 2018, 40, 1063–1068. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhang, X.M.; Hu, R.B.; Wu, F.Q.; Ma, J.H.; Meng, Y.; Fu, Y.F. Identification and molecular characterization of FKF1 and GI homologous genes in soybean. PLoS ONE 2013, 8, e79036. [Google Scholar] [CrossRef]
- Zhou, X.C.; Han, H.Q.; Chen, H.Y.; Liu, Y. Cloning and expression analysis of biorhythm clock output gene BnGI in Brassica napus. J. Shanghai Jiao Tong Univ. (Agric. Sci. Ed.) 2014, 32, 5–11. [Google Scholar]
- Tang, W.; Yan, H.; Su, Z.X.; Park, S.C.; Liu, Y.J.; Zhang, Y.J.; Wang, X.; Kou, M.; Ma, D.F.; Kwak, S.S.; et al. Cloning and characterization of a novel GIGANTEA gene in sweet potato. Plant Physiol. Biochem. 2017, 116, 27–35. [Google Scholar] [CrossRef]
- Karsai, R.F.; Odgerel, K.; Jose, J.; Bánfalvi, Z. In Silico Characterization and Expression Analysis of GIGANTEA Genes in Potato. Biochem. Genet. 2022, 60, 2137–2154. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.F. Expression Analysis and Functional Verification of ClGI Gene in Citrus limon. Master’s Thesis, Huazhong Agricultural University, Wuhan, China, 2019. [Google Scholar] [CrossRef]
- Yuan, X.; Liu, J.M.; Guo, C.H.; Kang, C.; Zhang, Z.G.; Quan, S.W.; Niu, J.X. Cloning and expression analysis of JrGI gene in walnut. Chin. J. Bioeng. 2023, 39, 640–652. [Google Scholar] [CrossRef]
- Song, Y.; Jiang, Y.; Kuai, B.; Li, L. CIRCADIAN CLOCK-ASSOCIATED 1 Inhibits Leaf Senescence in Arabidopsis. Front. Plant Sci. 2018, 9, 280. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, H.J.; Vu, Q.T.; Jung, S.; McClung, C.R.; Hong, S.; Nam, H.G. Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 8448–8453. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Wang, Y.; Wei, H.; Li, N.; Tian, W.W.; Chong, K.; Wang, L. Circadian Evening Complex Represses Jasmonate-Induced Leaf Senescence in Arabidopsis. Mol. Plant 2018, 11, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Takato, I.; Thomas, F.S.; Frank, G.H.; Lindsey, A.H.; Steve, A.K. FKF1 F-Box Protein Mediates Cyclic Degradation of a Repressor of CONSTANS in Arabidopsis. Science 2005, 309, 293–297. [Google Scholar] [CrossRef]
- Kim, W.Y.; Fujiwara, S.; Suh, S.S.; Kim, J.; Kim, Y.; Han, L.Q.; David, K.; Putterill, J.; Nam, H.G.; Somers, D.E. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 2007, 449, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Mariko, S.; Dmitri, A.N.; Steve, A.K.; Takato, I. FKF1 and GIGANTEA Complex Formation is Required for Day-Length Measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef]
- Fabio, F.; Kishore, C.S.P.; Lionel, G.; Nicolas, S.; Mark, R.; José, A.J.; George, C. Arabidopsis DOF Transcription Factors Act Redundantly to Reduce CONSTANS Expression and Are Essential for a Photoperiodic Flowering Response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef]
- Tang, W.; Liu, Y.J.; Zhang, Y.G.; Wang, X.; Hou, M.; Yan, H.; Ma, D.F.; Li, Q. Research progress on physiological functions of plant GI genes. Mol. Plant Breed. 2014, 12, 1044–1049. [Google Scholar] [CrossRef]
- Sawa, M.; Kay, S.A. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 11698–11703. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.Y.; Lee, S.M.; Seo, P.J.; Yang, M.S.; Park, C.M. Identification and molecular characterization of a Brachypodium distachyon GIGANTEA gene: Functional conservation in monocot and dicot plants. Plant Mol. Biol. 2010, 72, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Kudo, T.; Kawai, A.; Yamaya, T.; Hayakawa, T. Cellular distribution of ACT domain repeat protein 9, a nuclear localizing protein, in rice (Oryza sativa). Physiol. Plant. 2008, 133, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.S.; Chen, Y.J.; Hsieh, W.Y.; Li, Y.C.; Hsieh, M.H. Arabidopsis ACT DOMAIN REPEAT9 represses glucose signaling pathways. Plant Physiol. 2023, 192, 1532–1547. [Google Scholar] [CrossRef] [PubMed]
- Cortaga, C.Q.; Latina, R.A.; Habunal, R.R.; Lantican, D.V. Identification and characterization of genome-wide resistance gene analogs (RGAs) of durian (Durio zibethinus L.). J. Genet. Eng. Biotechnol. 2022, 20, 29. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.J.; Chen, H.B.; Hu, Z.Q.; Lu, X.Y.; Wang, H.Y.; Liu, H.; Zhou, B.Y. Comparative proteomics of phloem exudates reveals long-distance signals potentially involved in Litchi chinensis flowering. Biol. Plant. 2020, 64, 220–224. [Google Scholar] [CrossRef]
- Liu, R.X. Molecular Mechanism of Differential Expression of Transcriptome and Cell Wall Invertase Genes in Different Cupr-resistant Populations of Elsholtzia Haizhou. Ph.D. Thesis, Wuhan University, Wuhan, China, 2023. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.J.; Li, C.G.; Chen, S.K.; Tang, Q.L.; Xiao, Y.F.; Zhong, L.X.; Chen, Y.Y.; Chen, B. Gene expression programs during callus development in tissue culture of two Eucalyptus species. BMC Plant Biol. 2022, 22, 1. [Google Scholar] [CrossRef]
- Miao, W.; Dai, J.; Wang, Y.T.; Wang, Q.Q.; Lu, C.; La, Y.M.; Niu, J.Y.; Tan, F.; Zhou, S.X.; Wu, Y.F.; et al. Roles of IDM3 and SDJ1/2/3 in Establishment and/or Maintenance of DNA Methylation in Arabidopsis. Plant Cell Physiol. 2021, 62, 1409–1422. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.G.; Wang, X.G.; Xie, S.J.; Pan, L.; Miki, D.; Tang, K.; Hsu, C.C.; Lei, M.G.; Zhong, Y.L.; Hou, Y.; et al. A pair of transposon-derived proteins function in a histone acetyltransferase complex for active DNA demethylation. Cell Res. 2017, 27, 226–240. [Google Scholar] [CrossRef]
- Lang, Z.B.; Lei, M.G.; Wang, X.G.; Tang, K.; Miki, D.; Zhang, H.M.; Mangrauthia, S.K.; Liu, W.S.; Nie, W.F.; Ma, G.J.; et al. The Methyl-CpG-Binding Protein MBD7 Facilitates Active DNA Deme-thylation to Limit DNA Hyper-Methylation and Transcriptional Gene Silencing. Mol. Cell 2015, 57, 971–983. [Google Scholar] [CrossRef]
- Guo, Y.; Zhu, J.; Xu, Z.C.; Zhang, S.C. Research progress of plant ascorbate oxidase. Chin. Agric. Sci. Bull. 2008, 3, 196–199. [Google Scholar]
- Dowdle, J.; Ishikawa, T.; Gatzek, S.; Rolinski, S.; Smirnoff, N. Two Genes in Arabidopsis Tha-liana Encoding GDP-L-Galactose Phosphorylase Are Required for Ascor-bate Biosynthesis and Seedling Viability. Plant J. Cell Mol. Biol. 2007, 52, 673–689. [Google Scholar] [CrossRef] [PubMed]
- Aniqa, A.; Nudrat, A.A.; Muhammad, A. Influence of natural and synthetic vitamin C (ascorbic acid) on primary and secondary metabolites and associated metabolism in quinoa (Chenopodium quinoa Willd.) plants under water deficit regimes. Plant Physiol. Biochem. 2018, 123, 192–203. [Google Scholar] [CrossRef]
- Li, C. Cloning and Low Temperature Response Mechanism of Tobacco Ascorbate Oxidase Promoter. Master’s Thesis, Henan Agricultural University, Zhengzhou, China, 2021. [Google Scholar] [CrossRef]
- Kim, M.; Ohr, H.; Lee, J.W.; Hyun, Y.; Fischer, R.L.; Choi, Y. Temporal and spatial downregulation of Arabidopsis MET1 activity results in global DNA hypomethylation and developmental defects. Mol. Cells 2008, 26, 611–615. [Google Scholar] [CrossRef] [PubMed]
 = 100%,
= 100%,  ≥ 75%,
≥ 75%,  > 50%; (b) phylogenetic tree of GI protein family from C. oleifera and other species.
> 50%; (b) phylogenetic tree of GI protein family from C. oleifera and other species.
 = 100%,
= 100%,  ≥ 75%,
≥ 75%,  > 50%; (b) phylogenetic tree of GI protein family from C. oleifera and other species.
> 50%; (b) phylogenetic tree of GI protein family from C. oleifera and other species. 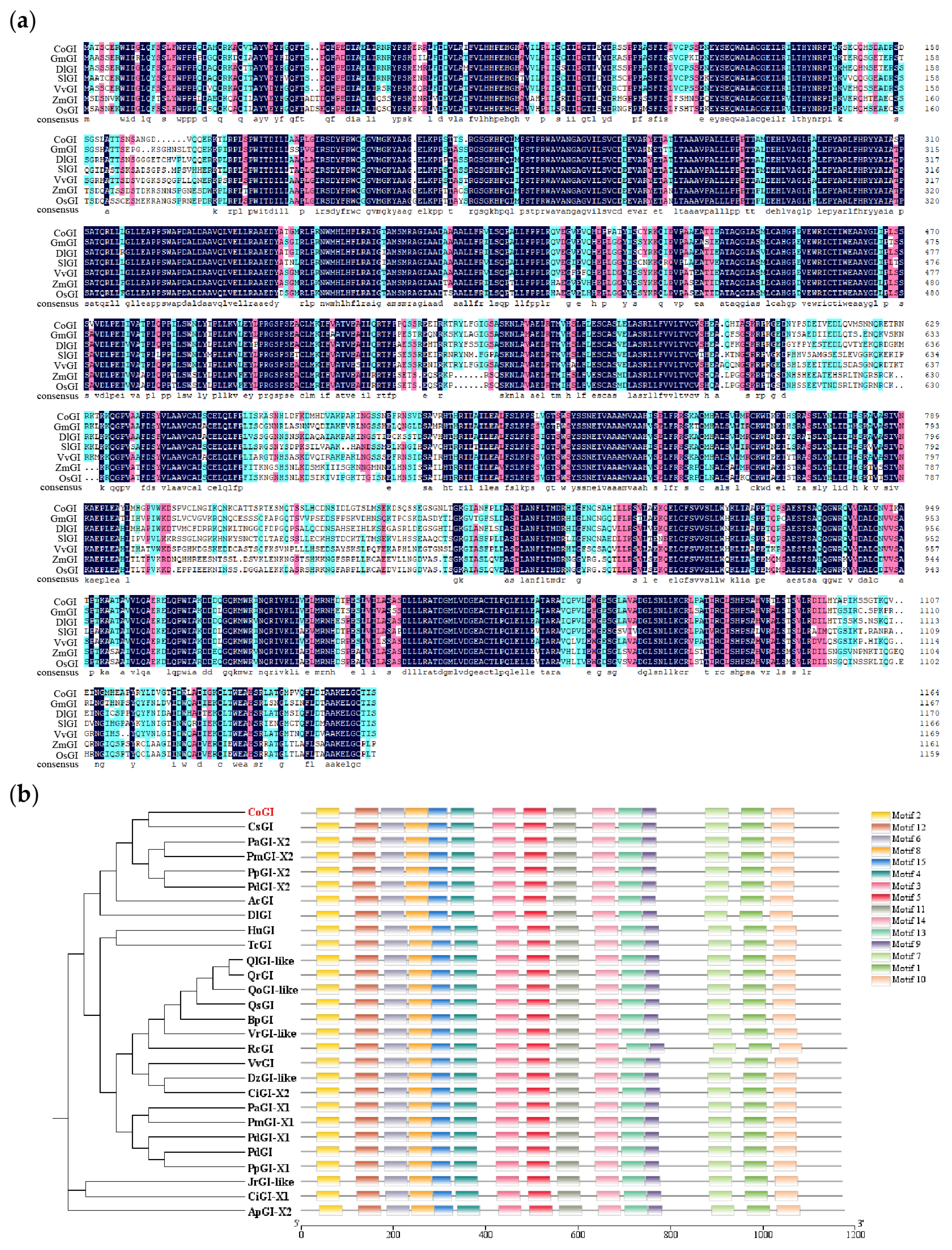

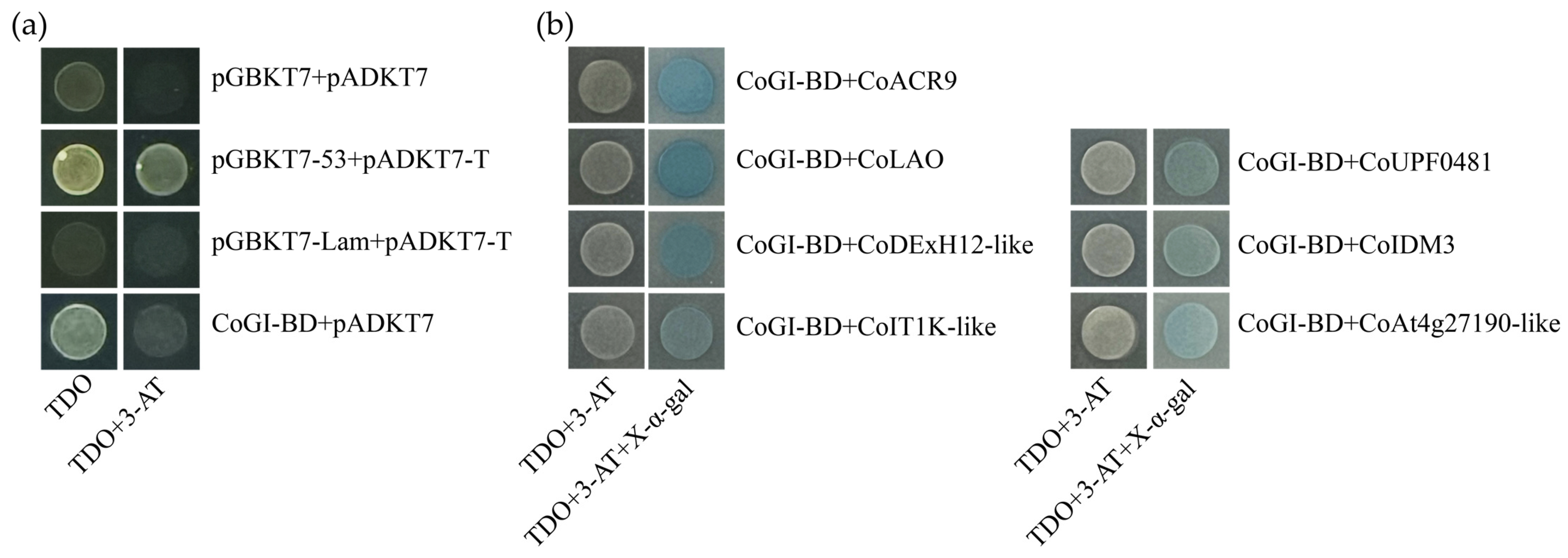
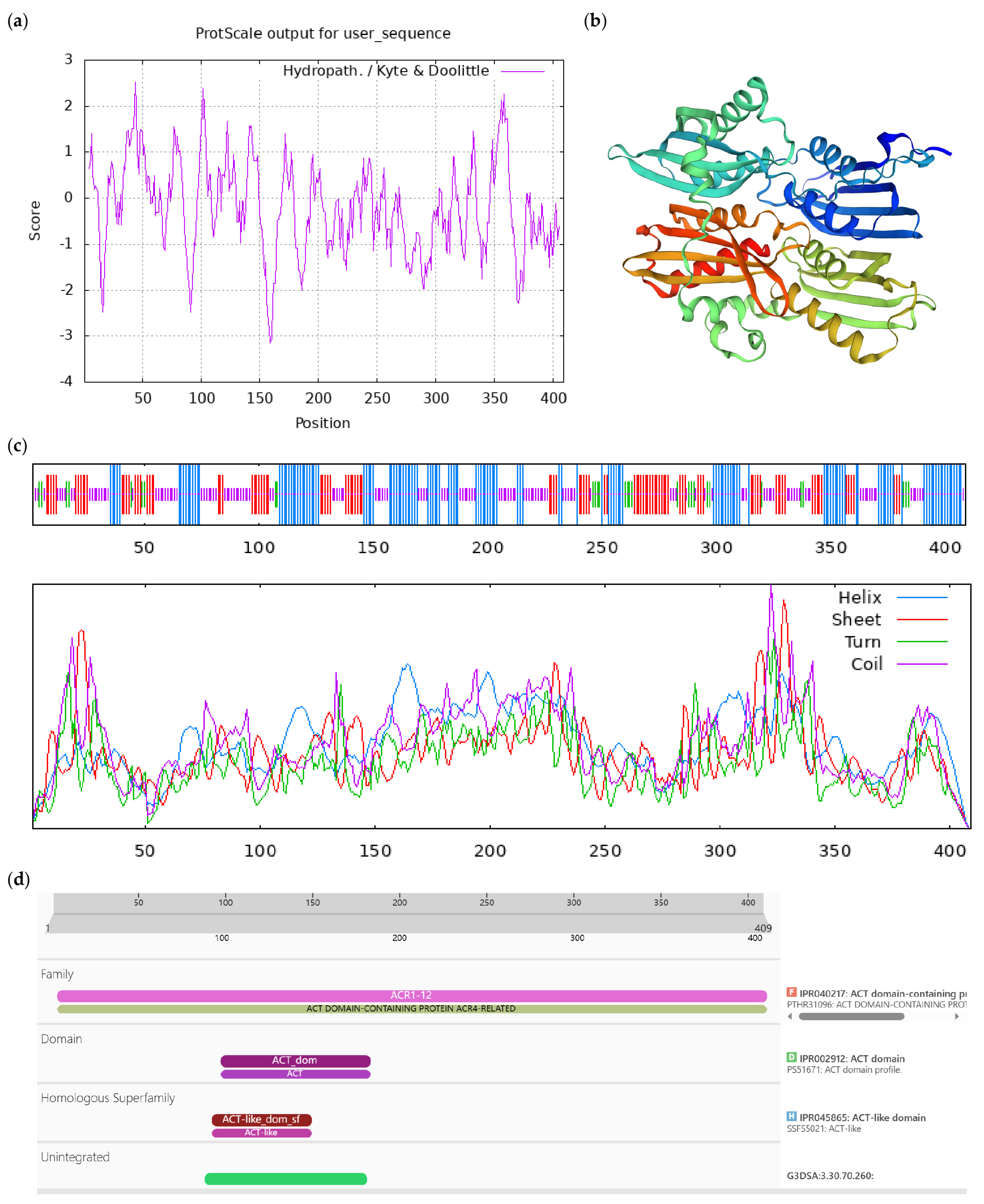
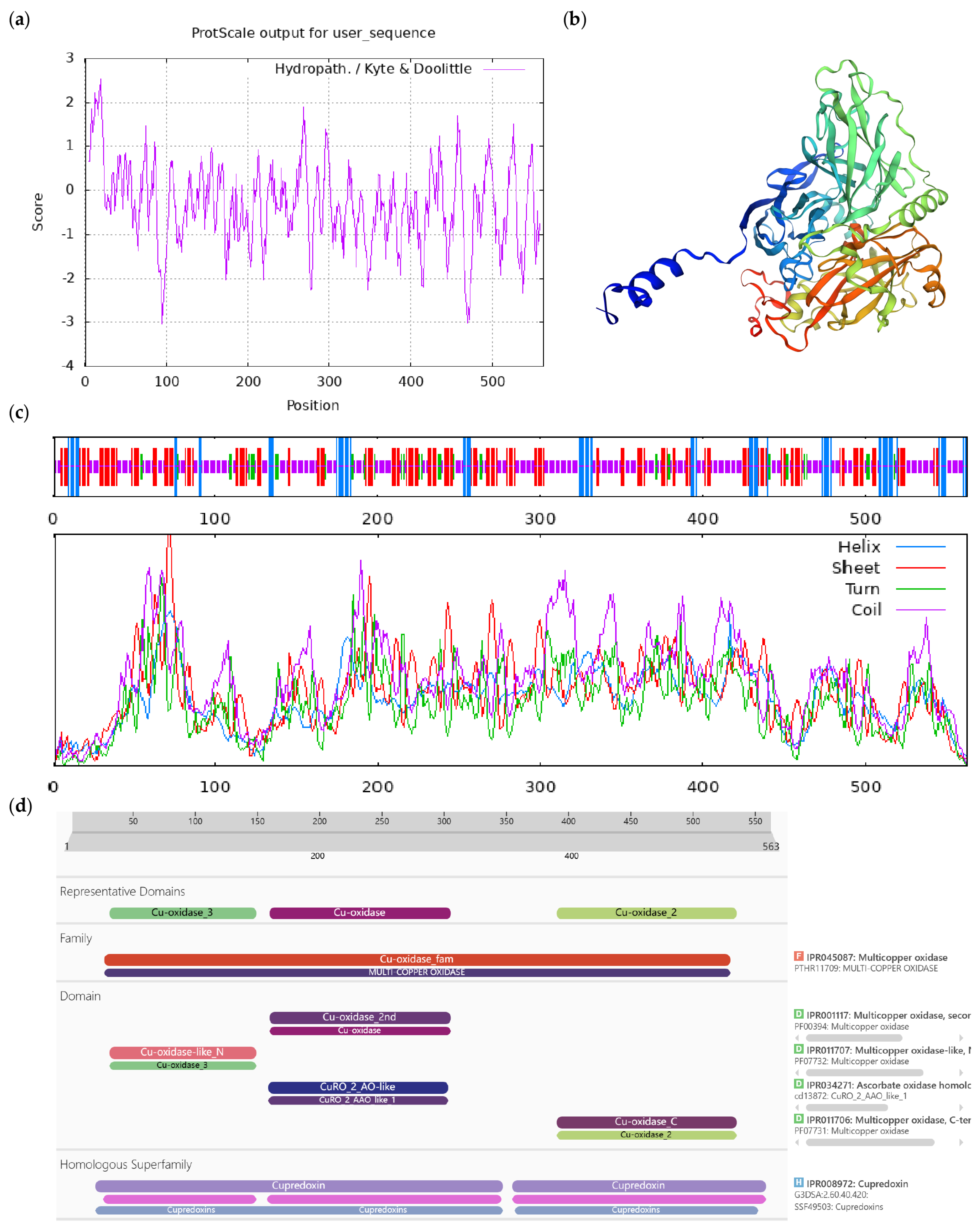
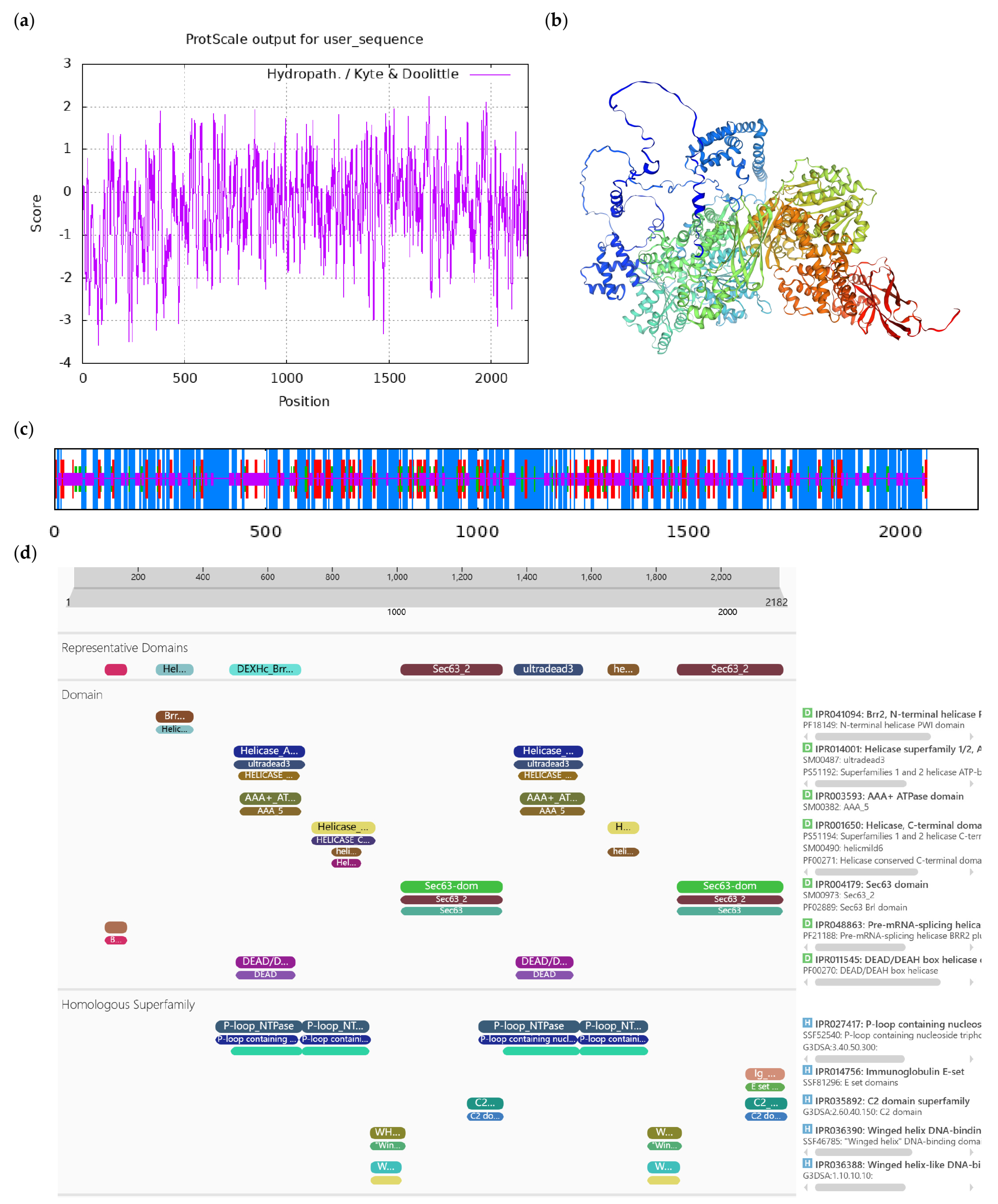
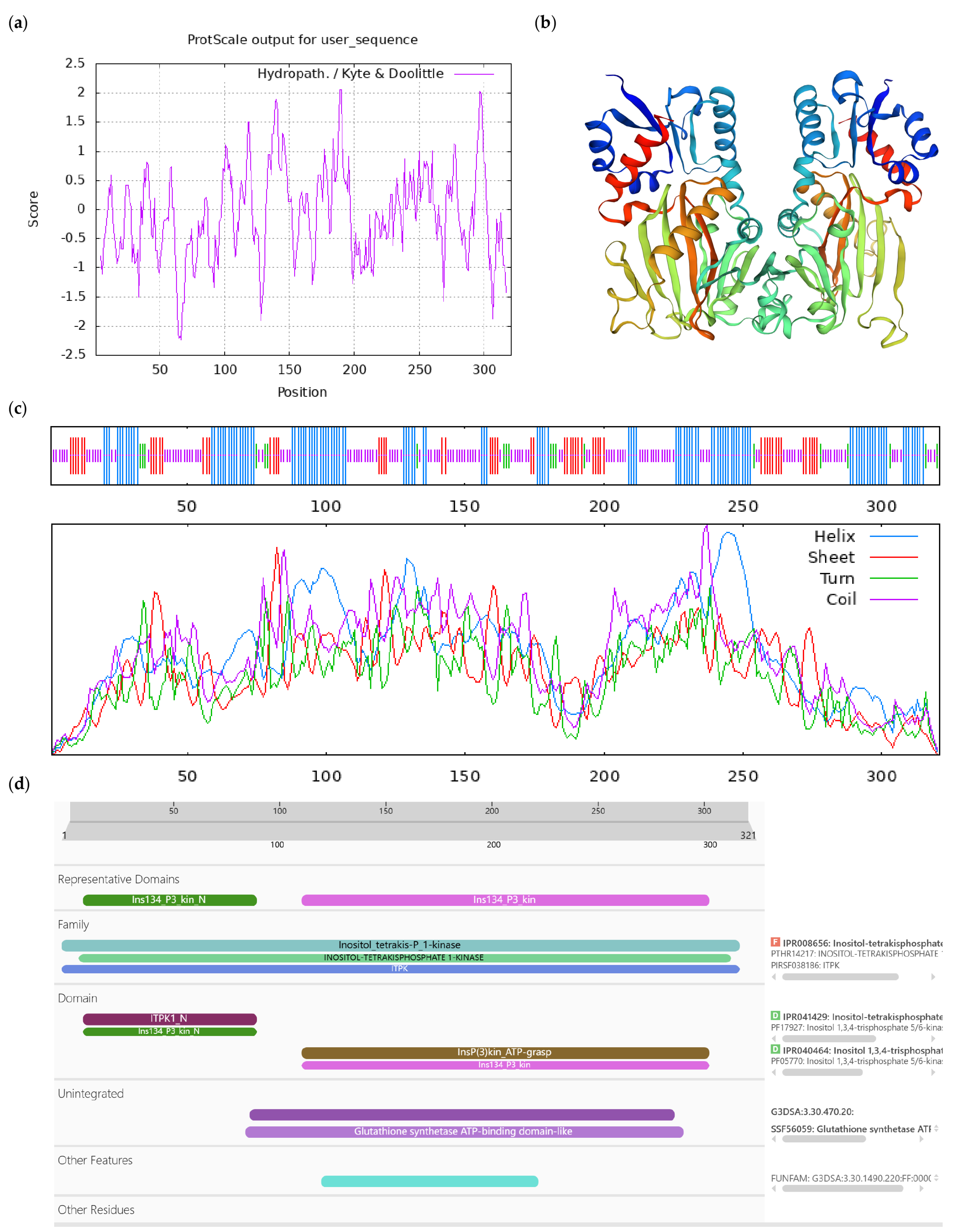
| Number | Accession | Protein Name | Short Name |
|---|---|---|---|
| 1 | XM_028241103.1 | ACT domain-containing protein ACR9 | ACR9 |
| 2 | XM 028270551.1 | L-ascorbate oxidase homolog | LAO |
| 3 | XM_028263935.1 | DExH-box ATP- dependent RNA helicase DExH12-like | DExH12-like |
| 4 | XM 028256937.1 | Inositol-tetrakisphosphate 1-kinase 1-like | IT1K-like |
| 5 | JANUSC010000011.1 | UPF0481 protein | UPF0481 |
| 6 | JANUSC010000015.1 | Increased DNA methylation 3 | IDM3 |
| 7 | XM 028245028.1 | Disease resistance protein At4g27190-like | At4g27190-like |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juan, L.; Ren, S.; Liu, Q.; Zhang, L.; Yan, J.; Li, J. Expression Analysis and Interaction Protein Screening of CoGI, the Key Factor in Photoperiod Regulation of Flowering in Camellia oleifera Abel. Horticulturae 2024, 10, 715. https://doi.org/10.3390/horticulturae10070715
Juan L, Ren S, Liu Q, Zhang L, Yan J, Li J. Expression Analysis and Interaction Protein Screening of CoGI, the Key Factor in Photoperiod Regulation of Flowering in Camellia oleifera Abel. Horticulturae. 2024; 10(7):715. https://doi.org/10.3390/horticulturae10070715
Chicago/Turabian StyleJuan, Lemei, Shuangshuang Ren, Qian Liu, Liling Zhang, Jindong Yan, and Jian’an Li. 2024. "Expression Analysis and Interaction Protein Screening of CoGI, the Key Factor in Photoperiod Regulation of Flowering in Camellia oleifera Abel" Horticulturae 10, no. 7: 715. https://doi.org/10.3390/horticulturae10070715
APA StyleJuan, L., Ren, S., Liu, Q., Zhang, L., Yan, J., & Li, J. (2024). Expression Analysis and Interaction Protein Screening of CoGI, the Key Factor in Photoperiod Regulation of Flowering in Camellia oleifera Abel. Horticulturae, 10(7), 715. https://doi.org/10.3390/horticulturae10070715






