Establishment and Optimization of an Experiment System for Flow Cytometry in Oil-Seed Camellia
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Reagents and Instrumentation
2.3. Experimental Method
2.3.1. Establishment of the Flow Cytometry Detection Method
2.3.2. Identification of Oil-Seed Camellia Ploidy by Flow Cytometry
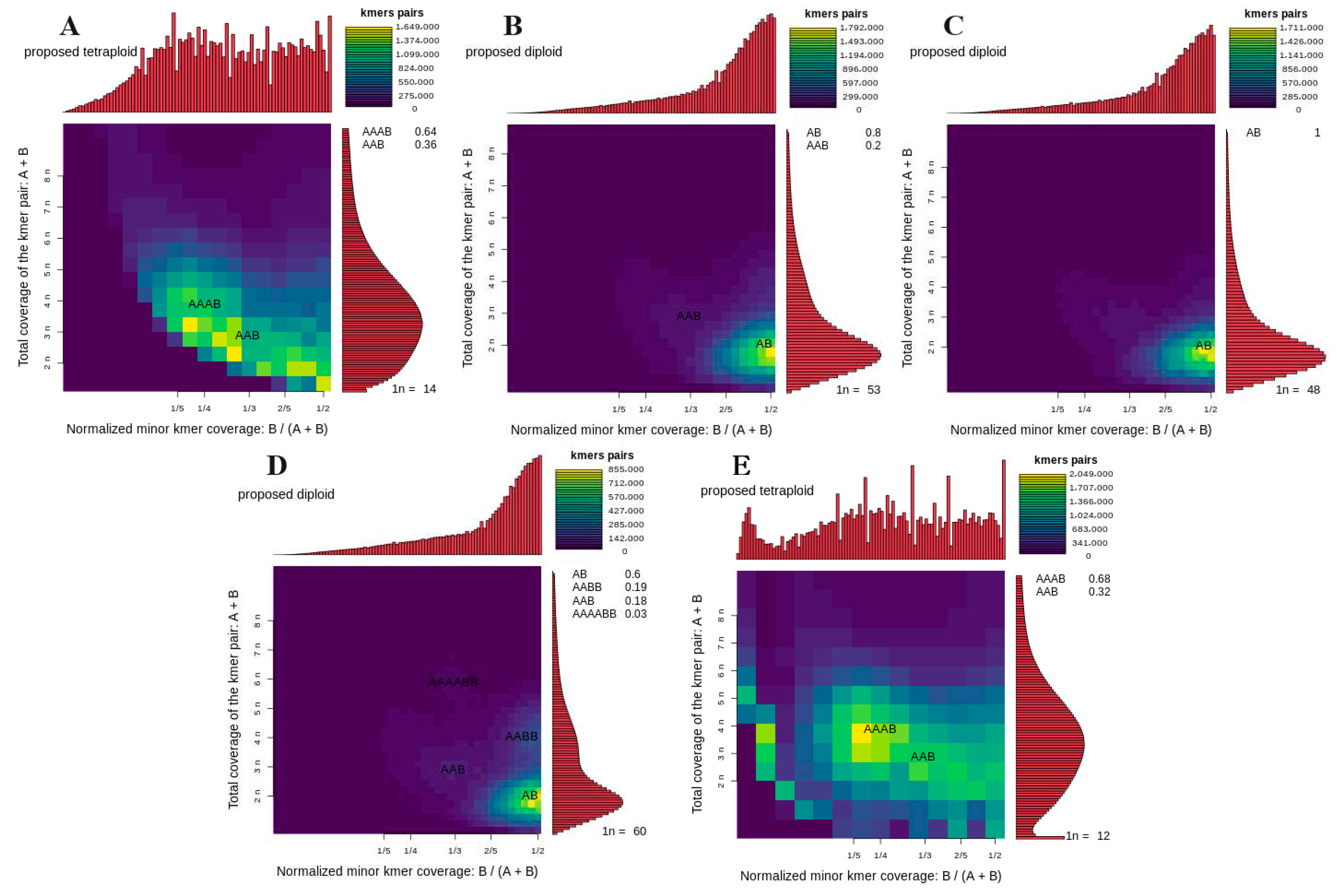
2.3.3. K-Mer Analysis to Identify the Ploidy of Oil-Seed Camellia
3. Results
3.1. Influence of Different Tissue Organs on Detection Effect
3.2. Influence of Different Lysis and Dyeing Times on Detection Effect
3.3. Ploidy Identification of Oil-Seed Camellia Using Flow Cytometry
3.4. K-Mer Analysis to Identify the Ploidy of Oil-Seed Camellia
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Y.N.; Xie, X.Y.; Wu, Y.; Zhu, Z.G.; Ye, S.C.; Huang, J.J.; Liu, W.X. CoBBX22 from Camellia oleifera negatively regulates drought stress tolerance in transgenic Arabidopsis thaliana. Plant Physiol. J. 2023, 59, 383–392. [Google Scholar]
- Chen, Y.Z.; Deng, S.H.; Chen, L.S.; Ma, L.; He, H.; Wang, X.N.; Peng, S.F.; Liu, C.X.; Wang, R.; Xu, Y.M.; et al. A new view on the development of oil tea camellia industry. J. Nanjing For. Univ. 2020, 44, 1–10. [Google Scholar]
- Zhuang, R.L. Camellia oleifera in China, 2nd ed.; China Forestry Publishing House: Beijing, China, 2008; pp. 65–108. [Google Scholar]
- Li, T.Z.; Tian, D.L.; Wuyun, T.N.; Tan, X.F.; Xiang, W.H.; Yan, W.D. Identification and variation of tetraploid Camellia oleifera. Sci. Silvae Sin. 2009, 45, 150–154. [Google Scholar]
- Ye, T.W.; Yuan, D.Y.; Li, Y.M.; Xiao, S.X.; Gong, S.F.; Zhang, J.; Li, S.F.; Luo, J. Ploidy Identification of Camellia hainanica. Sci. Silvae Sin. 2021, 57, 61–69. [Google Scholar]
- Jingade, P.; Huded, A.K.C.; Mishra, M.K. First report on genome size and ploidy determination of five indigenous coffee species using flow cytometry and stomatal analysis. Braz. J. Bot. 2021, 44, 381–389. [Google Scholar] [CrossRef]
- Kallamadi, P.R.; Mulpuri, S. Ploidy analysis of Helianthus species by flow cytometry and its use in hybridity confirmation. Nucleus 2016, 59, 123–130. [Google Scholar] [CrossRef]
- Feng, X.Q.; Chen, H.L.; Xiao, B.H.; Wu, Q.; Zhang, J.Y.; Zhang, N.; Li, P.P.; Wang, L.; Yin, J.R.; Sui, Z.H. Ploidy Identification by Flow Cytometry and Application of the Method to Characterize Seasonal Ploidy Variation of Wild Populations of the Red Alga Gracilariopsis lemaneiformis. Mar. Biotechnol. 2022, 24, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.W.; Ye, B.H.; Song, Q.Y.; Pu, W.T.; Shen, J.J.; Li, H.B. Estimation of ploidy levels by flow cytometry and identification of Torreya grandis cultivars and breeding strains by SSR markers. Trees 2022, 36, 1735–1745. [Google Scholar] [CrossRef]
- Tian, L.M.; Qi, D.; Cao, Y.F.; Dong, X.G.; Zhang, Y.; Huo, H.L. Studies on chromosome ploidy of Pyrus germplasm collection by flow cytometry. Acta Hortic. 2021, 1303, 79–84. [Google Scholar] [CrossRef]
- Ma, Y.P.; Wei, J.X.; Yu, Z.Y.; Qin, B.; Dai, S.L. Characterization of ploidy levels in Chrysanthemum L. by flow cytometry. J. For. Res. 2015, 26, 771–775. [Google Scholar] [CrossRef]
- Huang, M.G.; Shen, N.; Liu, X.Y.; Wu, X.S.; Chen, A.P.; Meng, Y.; Hu, X.G.; Tong, Z.K.; Huang, H.H.; Lou, X.Z. Identification of Chromosomal Ploidy and DNA Content in Betula by Flow Cytometry. J. Southwest For. Univ. 2023, 43, 49–57. [Google Scholar]
- Yan, R.; Ruan, C.J.; Wu, B.; Liu, L.Y.; Wang, H.M.; Zhang, W.C. SSR-PAGE and SSRseq identification of ploidy on Camellia oleifera germplasm. Mol. Plant Breed. 2018, 16, 6390–6397. [Google Scholar]
- Jia, W.Q. Fundamental Study on Reproductive Biology and Ploidy Breeding of Camellia Flower. Ph.D. Thesis, Chinese Academy Forestry, Beijing, China, 2015. [Google Scholar]
- Xu, X.D.; Shao, W.Z.; Zheng, W. Ploidy study on polyploidy cultivars of Camellia reticulata. Sci. Silvae Sin. 2018, 54, 44–48. [Google Scholar]
- Wu, F.Y.; Cai, Y.; Hao, B.Q.; Jia, Y.X.; Ye, H.; Zhang, Z.Y.; Ma, J.L. Establishment of a flow cytometry method for genome size determination of Camellia osmantha and Camellia vietnamensis and its application. Chin. J. Trop. Crops 2023, 44, 1542–1550. [Google Scholar]
- Gong, Q.Y. A Comparative Study on Resource Characteristics Andaffiliation of Hainan Oil-Tea. Master’s Thesis, Central South University of Forestry and Technology, Changsha, China, 2018. [Google Scholar]
- Yang, X.L.; Tong, Y.; Lei, X.L.; Xu, L.C.; Gao, L.Z. Estimation of Genome Sizes of 15 Camellia oleifera Varieties Using Flow Cytometry Analysis. J. Southwest For. Univ. (Nat. Sci.) 2018, 38, 46–51. [Google Scholar]
- Xiong, W.Y.; Ran, P.; Liu, L.Y.; Chen, J.W.; Zhang, J.L. Estimation Ploidy and Genome Size of 27 Dendrobium Species by Flow Cytometry. Chin. J. Trop. Crops 2022, 43, 2249–2257. [Google Scholar]
- Lü, S.; Ren, Y.; Wang, F.; Hu, G.B.; Huang, B.Z.; Liu, W.Q.; He, J.Q.; Liu, J.P.; Zeng, L.S.; Zhou, J.K.; et al. Ploidy identification of 169 Musa germplasms by flow cytometry. J. Fruit Sci. 2018, 35, 668–684. [Google Scholar]
- Han, S.; Li, S.J.; Xu, X.D.; Su, C.; Jiao, F. Quick identification of chromosomal ploidy of mulberry cells by flow cytometry. Acta Sericol. Sin. 2013, 39, 1042–1048. [Google Scholar]
- Wu, Y.Q.; Zhou, X.M.; Chen, L.; Cheng, H.H.; Li, Y.S. Nuclear DNA content and chromosome ploidy determination of sweet cherryrootstock by flow cytometry. J. Fruit Sci. 2014, 31, 48–52. [Google Scholar]
- Wang, T.T.; Zhang, G.H.; Cai, W.B.; Guo, H.C. Optimization of Potato Ploidy Identification Method Using Flow Cytometry. Chin. Potato J. 2019, 33, 1–7. [Google Scholar]
- Wu, Q.Q.; Lai, B.; Zhu, W.C.; Zhong, P.L. Method Optimization for Detection of Strawberry Ploidy by Flow Cytometry. Mod. Agric. Sci. Tech. 2015, 20, 48–50. [Google Scholar]
- Gu, H.N. Optimization of Flow Cytometry and Its Application in Determination of Ploidy in Wild Barley. Master’s Thesis, Anhui Agricultural University, Hefei, China, 2017. [Google Scholar]
- Wei, G.M.; Fang, Y.M. Establishment and optimization of an experiment system for flow cytometry in Ouercus. J. Nanjing For. Univ. 2015, 39, 167–172. [Google Scholar]
- Li, R.L. Ploidy Identification System of Camellia sinensis. Master’s Thesis, Anhui Agricultural University, Hefei, China, 2015. [Google Scholar]
- Ming, T.L. A Systematic Synopsis of the Genus Camellia. Acta Bot. Yunnan 1999, 21, 149–159. [Google Scholar]
- Li, G.T. New Advance in Karyotype Studies of Genus Camellia. Chin. Wild Plant Resour. 2001, 20, 9–14. [Google Scholar]
- Huang, H.T.; Yu, J.Z.; Ni, B.R. Research progress in tea polyploidy breeding. J. Tea 2006, 32, 14–17. [Google Scholar]
- Zhang, W.J.; Min, T.L. A Cytogeological Study of Genus Camellia. Acta Bot. Yunnan 1999, 2, 56–68. [Google Scholar]
- Huang, H.; Tong, Y.; Zhang, Q.J.; Gao, L.Z. Genome size variation among and within Camellia species by using flow cytometric analysis. PLoS ONE 2013, 8, e64981. [Google Scholar] [CrossRef]
- Zhou, L.P.; Wang, S.Z.; Ruan, S.L.; Ma, H.S.; Tong, J.X. Optimization of extraction method for flow cytometry in strawberry. Acta Agric. Zhejiangensis 2015, 27, 2024–2028. [Google Scholar]



| Germplasm ID | Species | Source | Germplasm ID | Species | Source |
|---|---|---|---|---|---|
| CK | C. azalea | NCOGRPC | C036 | C. fraterna | NCOGRPC |
| C001 | C. oleifera | Changde, Hunan | C037 | C. oleifera | NCOGRPC |
| C002 | C. oleifera | Changde, Hunan | C038 | C. vietnamensis | NCOGRPC |
| C003 | C. oleifera | Changde, Hunan | C039 | C. gauchowensis | NCOGRPC |
| C004 | C. oleifera | Changde, Hunan | C040 | C. chekiangoleosa | NCOGRPC |
| C005 | C. oleifera | Changde, Hunan | C041 | C. meiocarpa | NCOGRPC |
| C006 | C. oleifera | Changde, Hunan | C042 | C. meiocarpa | NCOGRPC |
| C007 | C. oleifera | Changde, Hunan | C043 | C. meiocarpa | NCOGRPC |
| C008 | C. oleifera | Changde, Hunan | C044 | C. meiocarpa | NCOGRPC |
| C009 | C. oleifera | Changde, Hunan | C045 | C. meiocarpa | NCOGRPC |
| C010 | C. oleifera | Changde, Hunan | C046 | C. meiocarpa | NCOGRPC |
| C011 | C. oleifera | Changde, Hunan | C047 | C. furfuracea | NCOGRPC |
| C012 | C. oleifera | Changde, Hunan | C048 | C. furfuracea | NCOGRPC |
| C013 | C. oleifera | Changde, Hunan | C049 | C. oleifera | NCOGRPC |
| C014 | C. oleifera | Changde, Hunan | C050 | C. oleifera | NCOGRPC |
| C015 | C. oleifera | Xiangxi, Hunan | C051 | C. gigantocarpa | NCOGRPC |
| C016 | C. oleifera | Xiangxi, Hunan | C052 | C. gigantocarpa | NCOGRPC |
| C017 | C. oleifera | Xiangxi, Hunan | C053 | C. sasanqua | NCOGRPC |
| C018 | C. oleifera | Xiangxi, Hunan | C054 | C. sasanqua | NCOGRPC |
| C019 | C. oleifera | Xiangxi, Hunan | C055 | C. brevistyla | NCOGRPC |
| C020 | C. oleifera | Xiangxi, Hunan | C056 | C. kissi var. megalantha | NCOGRPC |
| C021 | C. oleifera | Xiangxi, Hunan | C057 | C. brevistyla | NCOGRPC |
| C022 | C. oleifera | Xiangxi, Hunan | C058 | C. kissi var. megalantha | NCOGRPC |
| C023 | C. oleifera | Xiangxi, Hunan | C059 | C. brevistyla | NCOGRPC |
| C024 | C. oleifera | Xiangxi, Hunan | C060 | C. fraterna | NCOGRPC |
| C025 | C. oleifera | Xiangxi, Hunan | C061 | C. gauchowensis | NCOGRPC |
| C026 | C. oleifera | Xiangxi, Hunan | C062 | C. gauchowensis | NCOGRPC |
| C027 | C. oleifera | Xiangxi, Hunan | C063 | C. gauchowensis | NCOGRPC |
| C028 | C. oleifera | Xiangxi, Hunan | C064 | C. pitardii | NCOGRPC |
| C029 | C. pitardii | Xiangxi, Hunan | C065 | C. pitardii | NCOGRPC |
| C030 | C. pitardii | Xiangxi, Hunan | C066 | C. vietnamensis | NCOGRPC |
| C031 | C. yuhsienensis | Hengyang, Hunan | C067 | C. tunganica | NCOGRPC |
| C032 | C. handelii | Changsha, Hunan | C068 | C. tunganica | NCOGRPC |
| C033 | C. nitidissima | Jinhua, Zhejiang | C069 | C. pitardii | NCOGRPC |
| C034 | C. nitidissima | Jinhua, Zhejiang | C070 | C. semiserrata | NCOGRPC |
| C035 | C. oleifera | NCOGRPC |
| Germplasm ID | Peak Value | CV (%) | Ratio of DNA Content | Ploidy Estimation |
|---|---|---|---|---|
| CK | 25.82 | 4.37 | 1.00 | 2 |
| C001 | 69.14 | 4.75 | 2.68 | 6 |
| C002 | 50.02 | 4.19 | 1.94 | 4 |
| C003 | 56.93 | 2.15 | 2.20 | 4 |
| C004 | 45.76 | 5.85 | 1.77 | 4 |
| C005 | 52.28 | 6.90 | 2.02 | 4 |
| C006 | 74.95 | 3.54 | 2.90 | 6 |
| C007 | 68.91 | 6.36 | 2.67 | 6 |
| C008 | 71.14 | 3.74 | 2.76 | 6 |
| C009 | 69.21 | 4.99 | 2.68 | 6 |
| C010 | 55.76 | 3.45 | 2.16 | 4 |
| C011 | 54.86 | 3.63 | 2.12 | 4 |
| C012 | 54.27 | 4.92 | 2.10 | 4 |
| C013 | 54.44 | 4.04 | 2.11 | 4 |
| C014 | 54.57 | 6.43 | 2.11 | 4 |
| C015 | 107.03 | 2.45 | 4.15 | 8 |
| C016 | 101.83 | 4.51 | 3.94 | 8 |
| C017 | 101.92 | 2.64 | 3.95 | 8 |
| C018 | 93.77 | 3.68 | 3.63 | 8 |
| C019 | 69.05 | 4.77 | 2.67 | 6 |
| C020 | 108.92 | 2.88 | 4.22 | 8 |
| C021 | 101.80 | 4.29 | 3.94 | 8 |
| C022 | 82.19 | 3.03 | 3.18 | 6 |
| C023 | 76.62 | 4.66 | 2.97 | 6 |
| C024 | 93.54 | 3.00 | 3.62 | 8 |
| C025 | 100.93 | 3.34 | 3.91 | 8 |
| C026 | 103.13 | 3.54 | 3.99 | 8 |
| C027 | 82.12 | 5.95 | 3.18 | 6 |
| C028 | 55.80 | 7.51 | 2.16 | 4 |
| C029 | 100.17 | 3.29 | 3.88 | 8 |
| C030 | 98.45 | 3.73 | 3.81 | 8 |
| C031 | 47.56 | 7.18 | 1.84 | 4 |
| C032 | 30.84 | 7.59 | 1.19 | 2 |
| C033 | 25.11 | 7.57 | 0.97 | 2 |
| C034 | 20.91 | 6.90 | 0.81 | 2 |
| C035 | 70.79 | 3.71 | 2.74 | 6 |
| C036 | 27.21 | 5.29 | 1.05 | 2 |
| C037 | 44.96 | 4.41 | 1.74 | 4 |
| C038 | 95.46 | 2.90 | 3.70 | 8 |
| C039 | 53.53 | 3.85 | 2.07 | 4 |
| C040 | 22.45 | 4.57 | 0.87 | 2 |
| C041 | 44.85 | 4.53 | 1.74 | 4 |
| C042 | 49.30 | 4.88 | 1.91 | 4 |
| C043 | 50.42 | 4.29 | 1.95 | 4 |
| C044 | 44.85 | 5.29 | 1.74 | 4 |
| C045 | 47.10 | 4.61 | 1.82 | 4 |
| C046 | 48.95 | 4.20 | 1.90 | 4 |
| C047 | 20.84 | 4.76 | 0.81 | 2 |
| C048 | 22.42 | 3.75 | 0.87 | 2 |
| C049 | 70.34 | 5.18 | 2.72 | 6 |
| C050 | 70.51 | 4.58 | 2.73 | 6 |
| C051 | 24.22 | 4.27 | 0.94 | 2 |
| C052 | 30.97 | 4.63 | 1.20 | 2 |
| C053 | 55.51 | 7.35 | 2.15 | 4 |
| C054 | 68.69 | 4.28 | 2.66 | 6 |
| C055 | 22.86 | 6.10 | 0.89 | 2 |
| C056 | 24.91 | 6.41 | 0.96 | 2 |
| C057 | 28.28 | 6.35 | 1.10 | 2 |
| C058 | 45.26 | 6.02 | 1.75 | 4 |
| C059 | 48.02 | 3.01 | 1.86 | 4 |
| C060 | 72.99 | 6.51 | 2.83 | 6 |
| C061 | 71.36 | 3.49 | 2.76 | 6 |
| C062 | 83.53 | 3.74 | 3.24 | 6 |
| C063 | 80.84 | 7.58 | 3.13 | 6 |
| C064 | 81.53 | 5.04 | 3.16 | 6 |
| C065 | 104.49 | 4.96 | 4.05 | 8 |
| C066 | 55.66 | 7.75 | 2.16 | 4 |
| C067 | 47.79 | 4.95 | 1.85 | 4 |
| C068 | 29.71 | 6.17 | 1.15 | 2 |
| C069 | 20.90 | 5.28 | 0.81 | 2 |
| C070 | 22.38 | 4.69 | 0.87 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Zhang, Z.; Wang, X.; Wang, R.; He, Z.; Xiao, G.; Li, W.; Chen, Y. Establishment and Optimization of an Experiment System for Flow Cytometry in Oil-Seed Camellia. Horticulturae 2024, 10, 704. https://doi.org/10.3390/horticulturae10070704
Zhang Y, Zhang Z, Wang X, Wang R, He Z, Xiao G, Li W, Chen Y. Establishment and Optimization of an Experiment System for Flow Cytometry in Oil-Seed Camellia. Horticulturae. 2024; 10(7):704. https://doi.org/10.3390/horticulturae10070704
Chicago/Turabian StyleZhang, Ying, Zhen Zhang, Xiangnan Wang, Rui Wang, Zhilong He, Gaohong Xiao, Weiguo Li, and Yongzhong Chen. 2024. "Establishment and Optimization of an Experiment System for Flow Cytometry in Oil-Seed Camellia" Horticulturae 10, no. 7: 704. https://doi.org/10.3390/horticulturae10070704
APA StyleZhang, Y., Zhang, Z., Wang, X., Wang, R., He, Z., Xiao, G., Li, W., & Chen, Y. (2024). Establishment and Optimization of an Experiment System for Flow Cytometry in Oil-Seed Camellia. Horticulturae, 10(7), 704. https://doi.org/10.3390/horticulturae10070704







