Molecular Characteristics and Biological Properties of Bean Yellow Mosaic Virus Isolates from Slovakia
Abstract
1. Introduction
2. Materials and Methods
2.1. Determination of Complete BYMV Genomes Using HTS
2.2. Biological Experiments
2.3. Estimation of Virus Accumulation in Plants
3. Results and Discussion
3.1. Genome Characterization of Slovak BYMV Isolates
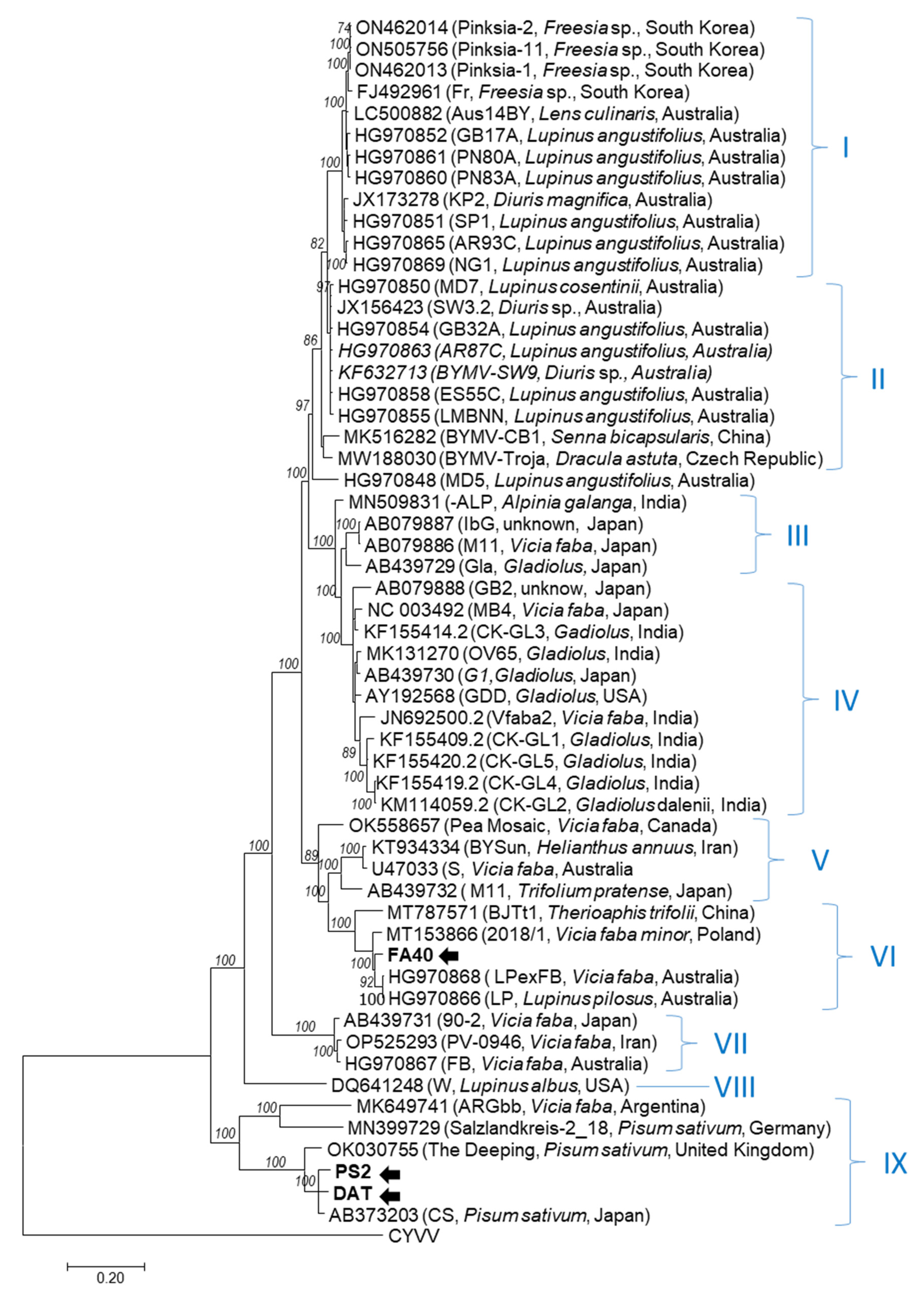
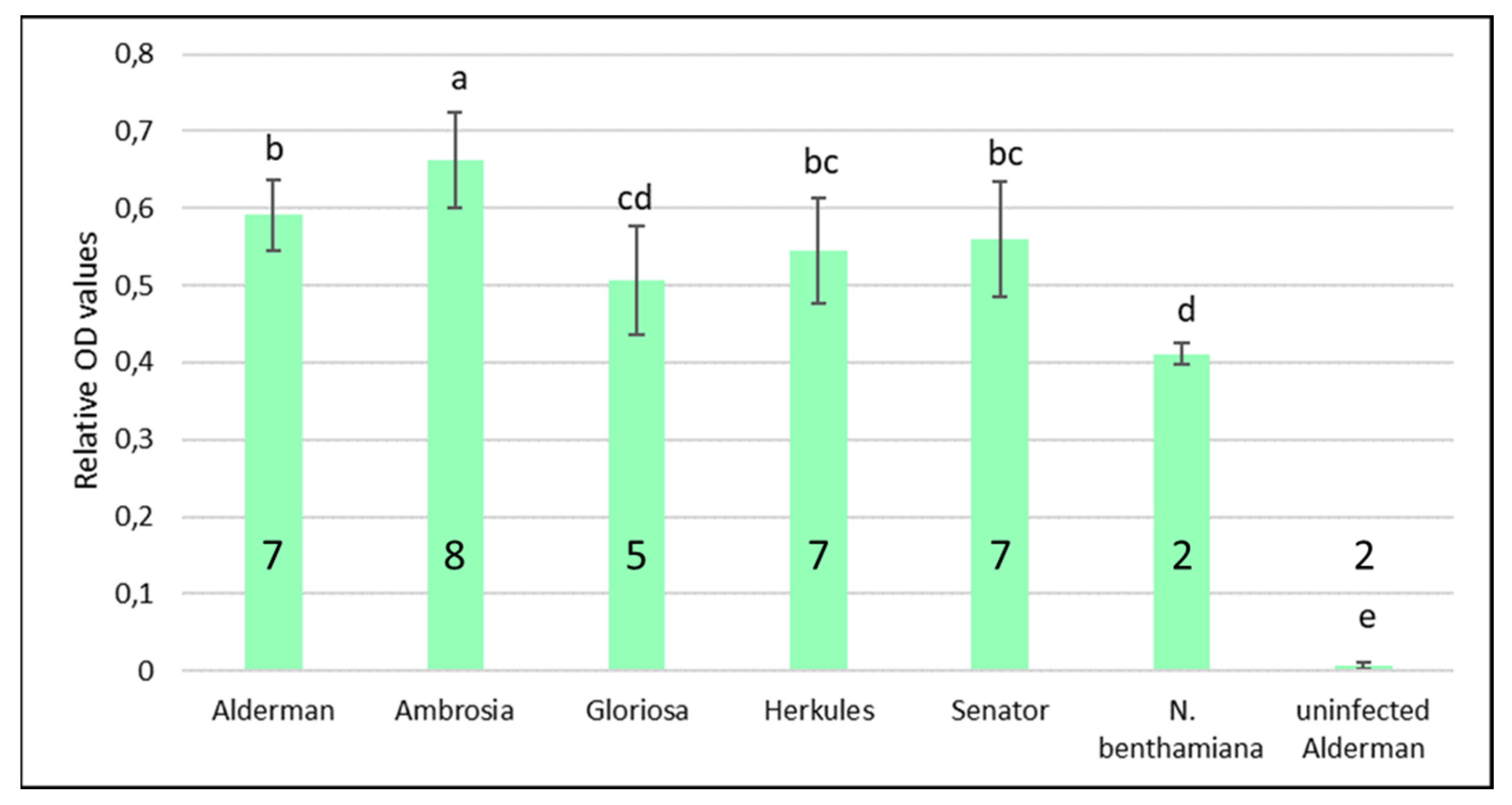
3.2. Experimental Infection of Pea Genotypes and Analysis of Virus Accumulation in Plants
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rashed, A.; Feng, X.; Prager, S.M.; Porter, L.D.; Knodel, J.J.; Karasev, A.; Eigenbrode, S.D. Vector-Borne Viruses of Pulse Crops, With a Particular Emphasis on North American Cropping System. Ann. Entomol. Soc. Am. 2018, 111, 205–227. [Google Scholar] [CrossRef]
- Chatzivassiliou, E.K. An Annotated List of Legume-Infecting Viruses in the Light of Metagenomics. Plants 2021, 10, 1413. [Google Scholar] [CrossRef] [PubMed]
- Jha, U.C.; Nayyar, H.; Chattopadhyay, A.; Beena, R.; Lone, A.A.; Naik, Y.D.; Thudi, M.; Prasad, P.V.V.; Gupta, S.; Dixit, G.P.; et al. Major viral diseases in grain legumes: Designing disease resistant legumes from plant breeding and OMICS integration. Front. Plant Sci. 2023, 14, 1183505. [Google Scholar] [CrossRef]
- Affrifah, N.S.; Uebersax, M.A.; Amin, S. Nutritional significance, value-added applications, and consumer perceptions of food legumes: A review. Legume Sci. 2023, 5, e192. [Google Scholar] [CrossRef]
- Wylie, S.J.; Adams, M.; Chalam, C.; Kreuze, J.; López-Moya, J.J.; Ohshima, K.; Praveen, S.; Rabenstein, F.; Stenger, D.; Wang, A.; et al. ICTV Virus Taxonomy Profile: Potyviridae. J. Gen. Virol. 2017, 98, 352–354. [Google Scholar] [CrossRef]
- Yang, X.; Li, Y.; Wang, A. Research Advances in Potyviruses: From the Laboratory Bench to the Field. Annu. Rev. Phytopathol. 2021, 59, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Wylie, S.J.; Coutts, B.A.; Jones, M.G.K.; Jones, R.A.C. Phylogenetic analysis of Bean yellow mosaic virus isolates from four continents: Relationship between the seven groups found and their hosts and origins. Plant Dis. 2008, 92, 1596–1603. [Google Scholar] [CrossRef]
- Kehoe, M.A.; Coutts, B.A.; Buirchell, B.J.; Jones, R.A. Split personality of a Potyvirus: To specialize or not to specialize? PLoS ONE 2014, 9, e105770. [Google Scholar] [CrossRef]
- Kehoe, M.A.; Coutts, B.A.; Buirchell, B.J.; Jones, R.A. Plant virology and next generation sequencing: Experiences with a Potyvirus. PLoS ONE 2014, 9, e104580. [Google Scholar] [CrossRef][Green Version]
- Maina, S.; Zheng, L.; King, S.; Aftab, M.; Nancarrow, N.; Trębicki, P.; Rodoni, B. Genome Sequence and Phylogeny of a Bean Yellow Mosaic Virus Isolate Obtained from a 14-Year-Old Australian Lentil Sample. Microbiol. Resour. Announc. 2020, 9, e01437-19. [Google Scholar] [CrossRef]
- Baradar, A.; Hosseini, A.; Ratti, C.; Hosseini, S. Phylogenetic analysis of a Bean yellow mosaic virus isolate from Iran and selecting the phylogenetic marker by comparing the individual genes and complete genome trees of BYMV isolates. Physiol. Mol. Plant Pathol. 2021, 114, 101632. [Google Scholar] [CrossRef]
- Villamor, D.E.V.; Ho, T.; Al Rwahnih, M.; Martin, R.R.; Tzanetakis, I.E. High Throughput Sequencing For Plant Virus Detection and Discovery. Phytopathology 2019, 109, 716–725. [Google Scholar] [CrossRef]
- Maclot, F.; Candresse, T.; Filloux, D.; Malmstrom, C.M.; Roumagnac, P.; van der Vlugt, R.; Massart, S. Illuminating an Ecological Blackbox: Using High Throughput Sequencing to Characterize the Plant Virome Across Scales. Front. Microbiol. 2020, 11, 578064. [Google Scholar] [CrossRef]
- Bejerman, N.; Giolitti, F.; Trucco, V.; de Breuil, S.; Dietzgen, R.G.; Lenardon, S. Complete genome sequence of a new enamovirus from Argentina infecting alfalfa plants showing dwarfism symptoms. Arch. Virol. 2016, 161, 2029–2032. [Google Scholar] [CrossRef]
- Filardo, F.F.; Thomas, J.E.; Webb, M.; Sharman, M. Faba bean polerovirus 1 (FBPV-1); a new polerovirus infecting legume crops in Australia. Arch. Virol. 2019, 164, 1915–1921. [Google Scholar] [CrossRef]
- Naito, F.Y.; Melo, F.L.; Fonseca, M.E.; Santos, C.A.; Chanes, C.R.; Ribeiro, B.M.; Gilbertson, R.L.; Boiteux, L.S.; de Cássia Pereira-Carvalho, R. Nanopore sequencing of a novel bipartite new world begomovirus infecting cowpea. Arch. Virol. 2019, 164, 1907–1910. [Google Scholar] [CrossRef] [PubMed]
- Glasa, M.; Šoltys, K.; Predajňa, L.; Sihelská, N.; Budiš, J.; Mrkvová, M.; Kraic, J.; Mihálik, D.; Ruiz- Garcia, A.B. High-throughput sequencing of Potato virus M from tomato in Slovakia reveals a divergent variant of the virus. Plant Protect. Sci. 2019, 55, 159–166. [Google Scholar] [CrossRef]
- Elmore, M.G.; Groves, C.L.; Hajimorad, M.R.; Stewart, T.P.; Gaskill, M.A.; Wise, K.A.; Sikora, E.; Kleczewski, N.M.; Smith, D.L.; Mueller, D.S.; et al. Detection and discovery of plant viruses in soybean by metagenomic sequencing. Virol. J. 2022, 19, 149. [Google Scholar] [CrossRef]
- Moury, B.; Simon, V. dN/dS-Based methods detect positive selection linked to trade-offs between different fitness traits in the coat protein of potato virus Y. Mol. Biol. Evol. 2011, 28, 2707–2717. [Google Scholar] [CrossRef]
- Nigam, D.; LaTourrette, K.; Souza, P.F.N.; Garcia-Ruiz, H. Genome-Wide Variation in Potyviruses. Front. Plant Sci. 2019, 10, 1439. [Google Scholar] [CrossRef]
- Stobbe, A.; Roossinck, M.J. Plant Virus Diversity and Evolution. Curr. Res. Top. Plant Virol. 2016, 22, 197–215. [Google Scholar] [CrossRef]
- Rubio, L.; Galipienso, L.; Ferriol, I. Detection of Plant Viruses and Disease Management: Relevance of Genetic Diversity and Evolution. Front Plant Sci. 2020, 11, 1092. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, A.; Sharma, A.; Belludi, R. Natural and Engineered Resistance Mechanisms in Plants against Phytoviruses. Pathogens 2023, 12, 619. [Google Scholar] [CrossRef] [PubMed]
- Mrkvová, M.; Hančinský, R.; Predajňa, L.; Alaxin, P.; Achs, A.; Tomašechová, J.; Šoltys, K.; Mihálik, D.; Olmos, A.; Ruiz-García, A.B.; et al. High-Throughput Sequencing Discloses the Cucumber Mosaic Virus (CMV) Diversity in Slovakia and Reveals New Hosts of CMV from the Papaveraceae Family. Plants 2022, 11, 1665. [Google Scholar] [CrossRef]
- Clark, M.F.; Adams, A.N. Characteristics of the microplate method of enzyme-linked immunosorbent assay for the detection of plant viruses. J. Gen. Virol. 1977, 34, 475–483. [Google Scholar] [CrossRef]
- Šubr, Z.; Matisová, J. Preparation of diagnostic monoclonal antibodies against two potyviruses. Acta Virol. 1999, 43, 255–257. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org/ (accessed on 18 February 2023).
- Nováková, S.; Klaudiny, J.; Kollerová, E.; Šubr, Z.W. Expression of a part of the Potato virus A non-structural protein P3 in Escherichia coli for the purpose of antibody preparation and P3 immunodetection in plant material. J. Virol. Methods 2006, 137, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.B.; López-Moya, J.J. When Viruses Play Team Sports: Mixed Infections in Plants. Phytopathology 2020, 110, 29–48. [Google Scholar] [CrossRef]
- Tomašechová, J.; Hančinský, R.; Predajňa, L.; Kraic, J.; Mihálik, D.; Šoltys, K.; Vávrová, S.; Böhmer, M.; Sabanadzovic, S.; Glasa, M. High-Throughput Sequencing Reveals Bell Pepper Endornavirus Infection in Pepper (Capsicum annum) in Slovakia and Enables Its Further Molecular Characterization. Plants 2020, 9, 41. [Google Scholar] [CrossRef]
- Maliogka, V.I.; Salvador, B.; Carbonell, A.; Sáenz, P.; León, D.S.; Oliveros, J.C.; Delgadillo, M.O.; García, J.A.; Simón-Mateo, C. Virus variants with differences in the P1 protein coexist in a Plum pox virus population and display particular host-dependent pathogenicity features. Mol. Plant Pathol. 2012, 13, 877–886. [Google Scholar] [CrossRef]
- Della Bartola, M.; Byrne, S.; Mullins, E. Characterization of Potato virus Y Isolates and Assessment of Nanopore Sequencing to Detect and Genotype Potato Viruses. Viruses 2020, 12, 478. [Google Scholar] [CrossRef] [PubMed]
- Glasa, M.; Hančinský, R.; Šoltys, K.; Predajňa, L.; Tomašechová, J.; Hauptvogel, P.; Mrkvová, M.; Mihálik, D.; Candresse, T. Molecular Characterization of Potato Virus Y (PVY) Using High-Throughput Sequencing: Constraints on Full Genome Reconstructions Imposed by Mixed Infection Involving Recombinant PVY Strains. Plants 2021, 10, 753. [Google Scholar] [CrossRef] [PubMed]
- Sasaya, T.; Iwasaki, M.; Yamamoto, T. Seed Transmission of Bean Yellow Mosaic Virus in Broad Bean (Vicia faba). Ann. Phytopath. Soc. Jpn. 1993, 59, 559–562. [Google Scholar] [CrossRef]
- Mali, V.R.; Šubr, Z.; Kúdela, O. Seed Transmission of Como and Potyviruses in Fababean and Vetch Genotypes Introduced into Slovakia. Acta Phytopathol. Entomol. Hung. 2003, 38, 87–97. [Google Scholar] [CrossRef]
- Atreya, C.D.; Pirone, T.P. Mutational analysis of the helper component-proteinase gene of a potyvirus: Effects of amino acid substitutions, deletions, and gene replacement on virulence and aphid transmissibility. Proc. Natl. Acad. Sci. USA 1993, 90, 11919–11923. [Google Scholar] [CrossRef] [PubMed]
- Shiboleth, Y.M.; Haronsky, E.; Leibman, D.; Arazi, T.; Wassenegger, M.; Whitham, S.A.; Gaba, V.; Gal-On, A. The conserved FRNK box in HC-Pro, a plant viral suppressor of gene silencing, is required for small RNA binding and mediates symptom development. J. Virol. 2007, 81, 13135–13148. [Google Scholar] [CrossRef]
- Kadaré, G.; Haenni, A. Virus-encoded RNA helicases. J. Virol. 1997, 71, 2583–2590. [Google Scholar] [CrossRef]
- Deng, P.; Wu, Z.; Wang, A. The multifunctional protein CI of potyviruses plays interlinked and distinct roles in viral genome replication and intercellular movement. Virol. J. 2015, 12, 141. [Google Scholar] [CrossRef]
- Li, Y.; Xia, F.; Wang, Y.; Yan, C.; Jia, A.; Zhang, Y. Characterization of a highly divergent Sugarcane mosaic virus from Canna indica L. by deep sequencing. BMC Microbiol. 2019, 19, 260. [Google Scholar] [CrossRef]
- Hong, Y.; Hunt, A.G. RNA polymerase activity catalyzed by a potyvirus-encoded RNA-dependent RNA polymerase. Virology 1996, 226, 146–151. [Google Scholar] [CrossRef]
- Shen, W.; Shi, Y.; Dai, Z.; Wang, A. The RNA-Dependent RNA Polymerase NIb of Potyviruses Plays Multifunctional, Contrasting Roles during Viral Infection. Viruses 2020, 12, 77. [Google Scholar] [CrossRef]
- Wylie, S.J.; Kueh, J.; Welsh, B.; Smith, L.J.; Jones, M.G.; Jones, R.A. A non-aphid-transmissible isolate of bean yellow mosaic potyvirus has an altered NAG motif in its coat protein. Arch. Virol. 2002, 147, 1813–1820. [Google Scholar] [CrossRef]
- Worrall, E.A.; Hayward, A.C.; Fletcher, S.J.; Mitter, N. Molecular characterization and analysis of conserved potyviral motifs in bean common mosaic virus (BCMV) for RNAi-mediated protection. Arch. Virol. 2019, 164, 181–194. [Google Scholar] [CrossRef]
- Zheng, L.; Wayper, P.J.; Gibbs, A.J.; Fourment, M.; Rodoni, B.C.; Gibbs, M.J. Accumulating variation at conserved sites in potyvirus genomes is driven by species discovery and affects degenerate primer design. PLoS ONE 2008, 3, e1586. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Fowkes, A.R.; McGreig, S.; Pufal, H.; Duffy, S.; Howard, B.; Adams, I.P.; Macarthur, R.; Weekes, R.; Fox, A. Integrating High throughput Sequencing into Survey Design Reveals Turnip Yellows Virus and Soybean Dwarf Virus in Pea (Pisum sativum) in the United Kingdom. Viruses 2021, 13, 2530. [Google Scholar] [CrossRef] [PubMed]
- Hasiów-Jaroszewska, B.; Boezen, D.; Zwart, M.P. Metagenomic Studies of Viruses in Weeds and Wild Plants: A Powerful Approach to Characterise Variable Virus Communities. Viruses 2021, 13, 1939. [Google Scholar] [CrossRef] [PubMed]
- Meziadi, C.; Blanchet, S.; Geffroy, V.; Pflieger, S. Genetic resistance against viruses in phaseolus vulgaris l.: State of the art and future prospects. Plant Sci. 2017, 265, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, W.T.; Provvidenti, R. A Common Gene for Resistance to Bean Yellow Mosaic Virus and Watermelon Mosaic Virus 2 in Pisum sativum. Phytopathology 1971, 61, 846–848. [Google Scholar] [CrossRef]
- Jurík, M.; Lebeda, A.; Gallo, J. Resistance of green peas to legume viruses. Acta Virol. 1994, 38, 97–99. [Google Scholar] [PubMed]
- van Leur, J.A.G.; Kumari, S.G.; Aftab, M.; Leonforte, A.; Moore, S. Virus resistance of Australian pea (Pisum sativum) varieties. N. Z. J. Crop Hortic. Sci. 2013, 41, 86–101. [Google Scholar] [CrossRef]
- Provvidenti, R.; Hampton, R.O. Inheritance of resistance to White lupin mosaic virus in common pea. HortScience 1993, 28, 836–837. [Google Scholar] [CrossRef]
- Green, S.K.; Kuo, Y.J.; Lee, D.R. Uneven distribution of two potyviruses (feathery mottle virus and sweet potato latent virus) in sweet potato plants and its implication on virus indexing of meristem derived plants. Trop. Pest Manag. 1988, 34, 298–302. [Google Scholar] [CrossRef]
- Dovas, C.I.; Mamolos, A.P.; Katis, N.I. Fluctuations in concentration of two potyviruses in garlic during the growing period and sampling conditions for reliable detection by ELISA. Ann. Appl. Biol. 2002, 140, 21–28. [Google Scholar] [CrossRef]
- Kogovšek, P.; Kladnik, A.; Mlakar, J.; Znidarič, M.T.; Dermastia, M.; Ravnikar, M.; Pompe-Novak, M. Distribution of Potato virus Y in potato plant organs, tissues, and cells. Phytopathology 2011, 101, 1292–1300. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rajamäki, M.L.; Valkonen, J.P. Viral genome-linked protein (VPg) controls accumulation and phloem-loading of a potyvirus in inoculated potato leaves. Mol. Plant Microbe Interact. 2002, 15, 138–149. [Google Scholar] [CrossRef]
- Glasa, M.; Šoltys, K.; Predajňa, L.; Sihelská, N.; Nováková, S.; Šubr, Z.; Kraic, J.; Mihálik, D. Molecular and Biological Characterisation of Turnip mosaic virus Isolates Infecting Poppy (Papaver somniferum and P. rhoeas) in Slovakia. Viruses 2018, 10, 430. [Google Scholar] [CrossRef] [PubMed]
- Mehle, N.; Kovač, M.; Petrovič, N.; Pompe Novak, M.; Baebler, Š.; Krečič Stres, H.; Gruden, K.; Ravnikar, M. Spread of potato virus Y NTN in potato cultivars (Solanum tuberosum L.) with different levels of sensitivity. Physiol. Mol. Plant Pathol. 2004, 64, 293–300. [Google Scholar] [CrossRef]



| Sample | Natural Host | Location | Year of Sampling | Leaf Symptoms | Viruses Identified in the Sample 1 |
|---|---|---|---|---|---|
| DAT | Trifolium pratense | Bratislava | 2020 | Mosaic | BYMV, SbDV, BGCV-2 |
| PS2 | Pisum sativum | Pezinok | 2021 | Mottling, mild leaf distortions | BYMV |
| FA40 | Phaseolus vulgaris | Vrbová nad Váhom | 2022 | Severe mosaic, yellowing | BYMV, CMV |
| Sample | Total Number of Reads/Average Length (bp) | Reads Mapped against BYMV Reference NC_003492 | Percentage of the Full-Length Genome Covered | Coverage Depth | The Closest BLAST Relative |
|---|---|---|---|---|---|
| DAT | 4,449,806/137.1 | 662,105 | 99.9% | 9764.5× | AB373203, pea, Japan |
| PS2 | 4,025,394/103.7 | 414,925 | 99.9% | 4794.5× | AB373203, pea, Japan |
| FA40 | 16,948,908/117.5 | 1,075,143 | 99.9% | 14,139.7× | HG970866, lupine, Australia |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mrkvová, M.; Kemenczeiová, J.; Achs, A.; Alaxin, P.; Predajňa, L.; Šoltys, K.; Šubr, Z.; Glasa, M. Molecular Characteristics and Biological Properties of Bean Yellow Mosaic Virus Isolates from Slovakia. Horticulturae 2024, 10, 262. https://doi.org/10.3390/horticulturae10030262
Mrkvová M, Kemenczeiová J, Achs A, Alaxin P, Predajňa L, Šoltys K, Šubr Z, Glasa M. Molecular Characteristics and Biological Properties of Bean Yellow Mosaic Virus Isolates from Slovakia. Horticulturae. 2024; 10(3):262. https://doi.org/10.3390/horticulturae10030262
Chicago/Turabian StyleMrkvová, Michaela, Jana Kemenczeiová, Adam Achs, Peter Alaxin, Lukáš Predajňa, Katarína Šoltys, Zdeno Šubr, and Miroslav Glasa. 2024. "Molecular Characteristics and Biological Properties of Bean Yellow Mosaic Virus Isolates from Slovakia" Horticulturae 10, no. 3: 262. https://doi.org/10.3390/horticulturae10030262
APA StyleMrkvová, M., Kemenczeiová, J., Achs, A., Alaxin, P., Predajňa, L., Šoltys, K., Šubr, Z., & Glasa, M. (2024). Molecular Characteristics and Biological Properties of Bean Yellow Mosaic Virus Isolates from Slovakia. Horticulturae, 10(3), 262. https://doi.org/10.3390/horticulturae10030262





