Abstract
Fruit locule number is an important agronomic trait that affects fruit appearance, quality, and yield. CLAVATA3 (SlCLV3) is a candidate gene of the fasciated (fas) locus that plays a role in controlling the number of flower organs and fruit locules in tomato. The SlCLV3 encoding signal peptide mainly acts by inhibiting the expression of WUSCHEL (WUS) but there is little research about how the receptor transmits the CLV3 signal to WUS and inhibits its expression. The CRISPR/Cas9 method was employed to edit the first exon of tomato SlCLV3 in this study, leading to the functional deletion of SlCLV3. As a result, flowers with a high number of organs and fruits with a high number of locules were produced. We screened six candidate genes using the transcriptome of clv3 mutants, analyzed expression variations in these genes between the cultivated allele and wild-type allele of fas, and showed that only SlLET6 and SlGIF1 (GRF1-interacting factor 1) were influenced by the fas locus. SlLET6 overexpression resulted in an increase in flower carpels and fruit locules. These results suggest that SlLET6 may be the downstream gene of SlCLV3 regulating the number of carpels and fruit locules in tomato.
1. Introduction
Tomato (Solanum lycopersicum) is an extensively grown vegetable crop that serves as a model plant for investigating fruit development. Fruit size is an important modern breeding target since it directly impacts fruit appearance, quality, and yield. The eventual size of the fruit is determined by the division of carpel cells and the number of carpels [1]. The fruit locule is derived from the carpel of the flower, and the locule number has a significant influence on the size of fruit. All wild tomato and small-fruit tomato cultivars have only 2–4 locules, while most other cultivars have 6 or more locules [1]. Increased locule number is also thought to have played an important role in the evolution of extremely large fruits [2]. Increased locule number is strongly correlated with the number of floral organs, and it is controlled by multiple quantitative trait loci (QTL) [3]. Two major loci controlling this trait have been previously identified: the fasciated (fas) and locule number (lc) locus [4].
The lc locus regulates the locule number, and the cultivated allele of lc has 2–4 additional locules than the wild type of lc in tomato [5,6]. The lc locus includes a 1600-base-pair non-coding region that is located 1080 bp downstream of WUSCHEL (WUS), which encodes a transcription factor that maintains the homeostasis of stem cells in the shoot apical meristem [6]. The increase in locule number caused by the cultivated allele of lc may be attributed to two single nucleotide polymorphisms (SNPs) in a putative repressor CArG motif, which is bound by AGAMOUS to suppress the expression of WUS in Arabidopsis [6,7]. Research has demonstrated that the CArG motif mutation leads to an increase in locule number in tomato [8]. The cultivated allele of lc exhibited a higher expression of WUS compared to the wild type of lc. Decreased expression of WUS has been shown to reduce the number of floral organs and fruit locules in tomato [9]. Therefore, tomato WUS may be the candidate gene for the lc locus.
The fas locus exerts a more significant influence and yields a higher number of locules compared to the lc locus in tomato [5,6]. YABBY2b had been proposed as the candidate gene of the fas locus, and reduced YABBY2b expression was shown to increase the number of fruit locules [10]. Evidence has shown that the inversion of a 294 kb fragment on chromosome 11 produces the cultivated allele of fas [11], located between intron 1 of the YABBY2b gene and 1 kb upstream of CLAVATA3 (SlCLV3). Introgression of fasciated locus into S. pimpinellifolium (Sp-fas) produces half of all fruits producing three locules when the full SlCLV3 gene is introduced into Sp-fas plants, resulting in fruit rescued to two locules, confirming that SlCLV3 is the candidate gene of the fas locus. Additionally, the study revealed that CLE9 and SlCLV3 genes belong to the EMBRYO-SURROUNDING REGION (CLE) genes family and exhibit functional redundancy [12]. An analysis was conducted to examine the epistatic relationship between SlCLV3 (fas locus), WUS (lc locus), and CLE9 in tomato. The results revealed idiosyncratic and dose-dependent epistasis [13].
The shoot meristem size and floral organ number in Arabidopsis thaliana are determined by the feedback system between WUS and CLV (CLAVATA) signaling proteins [14]. First, a signal cascade of CLV3 suppresses WUS expression to prevent excessive proliferation of stem cells, and, in turn, WUS promotes SlCLV3 expression to limit WUS activity [15]. The CLV3 signal is perceived by three groups of receptors: CLV1 (CLAVATA1), CLV2 (CLAVATA2)/CORYNE (CRN), and receptor-like PROTEIN KINASE2 (RPK2), which are all located on the cell membrane. These receptors transmit the CLV3 signal to inhibit WUS expression [16,17,18].
In Arabidopsis, several studies have focused on understanding how the receptors of CLV3 perceive signals and transmit them to WUS. The phenotype of KAPP (Kinase-associated protein phosphatase) overexpression results in a phenotype that closely resembles that of CLV1-deficient mutants. In vitro experiments have shown that CLV1 phosphorylates KAPP, while KAPP attenuates CLV1 autophosphorylation, confirming that KAPP is a negative regulator in the CLV signaling pathway [19]. The characteristics of the clv3 mutant were greatly reduced when double deletion mutants of the phos-phatases POL (POLTERGEIST) and PLL (POL-like) were crossed with it, indicating their involvement in the CLV signaling pathway [20]. The phenotypes after POL and PLL mutation were similar to that of the WUS mutant and affected the expression level of WUS and the overexpression of WUS-rescued pol pll1 mutants, which has indicated that POL and PLL act downstream of CLV3 and upstream of WUS [20]. GTP binding proteins transmit signals from RPK2 to maintain stem cell homeostasis in the stem terminal meristem [21], while mitogen-activated protein kinases (MAPK) in the cytoplasm may play a role in the maintenance of stem meristem [22]. Calcium signaling is crucial for maintaining the shoot apex meristem, and treating Arabidopsis seedlings with calcium channel inhibitors causes the stem apex meristem to grow larger. Furthermore, the suppression of CLV3 on the shoot apex meristem can be reduced by calcium channel inhibitors, implying that calcium signaling may be downstream of CLV3 signaling pathways [23]. SHOOTMERISTEMLESS (STM) also has a crucial function in controlling the development of shoot and floral meristems, while clv1 and clv3 mutations partially complement the phenotype of the stm mutant. Additionally, stm mutations are dominant over clv phenotypes, suggesting that the phenotype of stm is influenced by CLV activity, while the phenotype of clv is also influenced by STM activity [24].
In tomato, the feedback system between the WUS transcription factor and CLV signaling protein is also highly conserved. The function of homologous protein CLV3 is consistent with CLV3 of Arabidopsis, whereas the application of CLV3 externally leads to a decrease in the size of the shoot apex meristem [12,25]. The receptors CLV1, CRN, and RPK2 in tomato are involved in transmitting the CLV3 signal, which leads to the inhibition of WUS expression [12]. Additionally, there have been investigations conducted on WUS interaction factors in tomato, such as BRI1-EMS-SUPPRESSOR 1 (BES1) protein [26,27]. However, there is little research about how the receptor transmits the CLV3 signal to WUS and inhibits its expression.
In this study, tomato clv3 mutant was obtained by the CRISPR/cas9 system, resulting in an increase in the number of flower organs and fruit locules. To identify the downstream genes of SlCLV3, we performed a transcriptome analysis and screened six candidate genes based on prior studies. To further screen candidate genes, we investigated the alterations in expression changes in candidate genes between the wild type and cultivated alleles of fas. The results showed that only SlLET6 and SlGIF1 (GRF1-interacting factor 1) were regulated by the fas locus, and the expression pattern analysis suggested that they may play an important role in flower development. Finally, transgenic experiments provided evidence that the SlLET6 function was related to the number of carpels and fruit locules. These results suggest that SlLET6 may act downstream of SlCLV3 and regulate the number of carpels and fruit locules in tomato.
2. Materials and Methods
2.1. Plant Materials
To verify the function of the SlCLV3, we constructed the clv3 mutant on the ‘Ailsa Craig’ background. The guide sequence of CLV3 was designed as GTCTCATGGTCTGTCTCTAA using the CRISPR Guide Design tool (http://crispr.mit.edu, accessed on 13 January 2021). A vector construction kit (View Biotech, Beijing, China) was used to construct the gRNA fragments into the CRISPR/Cas9 vector.
Tomato seeds of Solanum lycopersicum ‘Ailsa Craig’ were subjected to a 55 °C water bath for 20 min, followed by a 20-min treatment with a 33% sodium hypochlorite. The seeds were cultured in seed medium (1/2 MS) for 7 days under 16-h light/8-h dark at 28 °C. The middle of cotyledon was cut and cultured in pre-medium (KCMS culture medium) for a duration of 48 h under dark conditions at a temperature of 28°C. The explants and Agrobacterium were inserted into the CRISPR/Cas9 vector were immersed in suspension media (200 mM acetosyringone, 10 mM MgCl2, and 10 mM MES [pH 5.6]) and subjected to gentle agitation for a duration of 4 min. The dried explants were cultured in pre-medium (KCMS culture medium) for 48 h in darkness at 28 °C. The explants were transferred to the screening medium (MS containing 400 mg/L cefalexin and 30 mg/L hygromycin) and cultured under a 16-h light/8-h dark at 28 °C, with medium replaced every 15 days. The new shoots were cultivated in the rooting medium (MS containing 1 mg/L naphthalene acetic acid) for 15 days, and the transformed seedlings were transplanted to the breeding substrate in a greenhouse under routine management.
We generated overexpressed plants of SlLET6 and SlGIF1 on the ‘Ailsa Craig’ background to analyze the gene function. Specific primers were used to amplify the target gene using cDNA as a template (Table 1), and then the target gene fragment was inserted into pCAMBIA1300-35S-E9 vector using Uniclone One Step Seamless Cloning Kit (Genesand Biotech, Beijing, China). The genetic transformation approach employed was identical to that utilized for the creation of clv3 mutants.

Table 1.
Primers for amplifying the target gene.
Three tomato lines ‘MLK1’, ‘FL1’, and ‘Zhongshu6’ were also used in this study [9]. ‘MLK1’ is large fruit tomato with more than 10 locules, ‘FL1’ is small fruit tomato with 2–4 locules, and ‘Zhongshu6’ is medium fruit tomato with 6–8 locules. The cultivated allele of fas is present in ‘MLK1’ and the wild-type allele of fas is present in ‘Zhongshu 6’ and ‘FL1’.
2.2. Identification of Transgenic Plants
The identification of clv3 mutants was mainly based on sequence alignment. The leaves of transformed seedlings were collected and rapidly frozen in liquid nitrogen before DNA extraction using a plant DNA extraction kit (CWBiotech, Beijing, China). PCR was performed using primers: CLV3-F (CTGTTCTTTTGTTACTTTCC) and CLV3-R (CCATACTTACATTTTCGTTT), taq enzymes, and buffers to amplify SlCLV3 gene fragments on the DNA template of each transformed seedling. PCR products were sequenced by ThermoFisher (Beijing, China), gene and amino acid sequence alignments were conducted using DNAMAN software (Version 10).
The identification of overexpressed plants was mostly determined through RT-PCR analysis conducted on the early vegetative meristem of transformed seedlings.
2.3. Phenotypic Analysis of Flowers and Fruits
Five flowers at the anthesis stage were selected from transgenic and wild-type plants, and the number of petals, sepals, stamens, and carpels were calculated. Five mature fruits of transgenic and wild-type plants were collected to count locule number. All data values are included with standard error. The IBM SPSS Statistics 23.0 software package was utilized to analyze significant differences with * indicating significant differences at p < 0.05 and ** indicating significant differences at p < 0.01 according to the Independent t-test.
2.4. Transcriptome Sequencing and Data Analysis
Early vegetative meristem from clv3 and WT plants was collected and frozen in liquid nitrogen. There were three biological replicates with 50 early vegetative meristems per replicate. Total RNA (5 μg) was extracted using a plant RNA extraction kit (CWBiotech, Beijing, China). The concentration and purity of each RNA sample were determined using NanoDrop 2000 (Thermo Fisher Scientific Inc., Waltham, MA, USA), while RNA integrity was assessed by 1% agarose gel electrophoresis. The sequencing library was prepared according to the manufacturer’s protocol (NEBNext® Ultra RNA Library Prep Kit for Illumina®, Ipswich, MA, USA), and the library was then sequenced using an Illumina HiSeq instrument (Illumina, San Diego, CA, USA). A cutoff of p < 0.05 and logFC > 1 was used to identify differentially expressed genes. GO functional classification of the differentially expressed genes was carried out using the gene ontology database (http://www.geneontology.org/, accessed on 20 December 2021) and KEGG pathway analysis of the differentially expressed genes was performed based on the KEGG pathway database (http://www.genome.jp/kegg/, accessed on 22 December 2021).
2.5. Total RNA Extraction and Quantitative RT-PCR
Early vegetative meristems were collected from clv3 mutant and WT plants to verify the transcriptome data. Early vegetative meristems were collected from three tomato lines, including ‘MLK1’, ‘FL1’, and ‘Zhongshu6’ for further screening of candidate genes. In addition, several parts of the tomato ‘Ailsa Craig’ plant, including leaves, apical meristem, floral/inflorescence meristem, young flower buds, flowers, fruits, and roots, were used to analyze the expression pattern of SlLET6 and SlGIF1. Three biologically replicated samples were collected and frozen in liquid nitrogen. Total RNA (5 μg) was extracted using a plant RNA extraction kit (CWBiotech, Beijing, China). Quantitative RT-PCR was performed using gene-specific primers (Table 2) and TB Green Premix (CWBiotech, Beijing, China) on an ABI7500 system with ABI7500 software v2.3 used for analysis. The IBM SPSS Statistics 23.0 software package was utilized to analyze significant differences with * indicating significant differences at p < 0.05, ** indicating significant differences at p < 0.01 according to Independent t-test, and small letters indicating significant differences at p < 0.05 according to Duncan’s multiple range tests.

Table 2.
Primers for quantitative RT-PCR.
3. Results
3.1. CRISPR/Cas9-Engineered Mutations in SlCLV3 Cause an Increase in the Number of Carpels and Fruit Locules
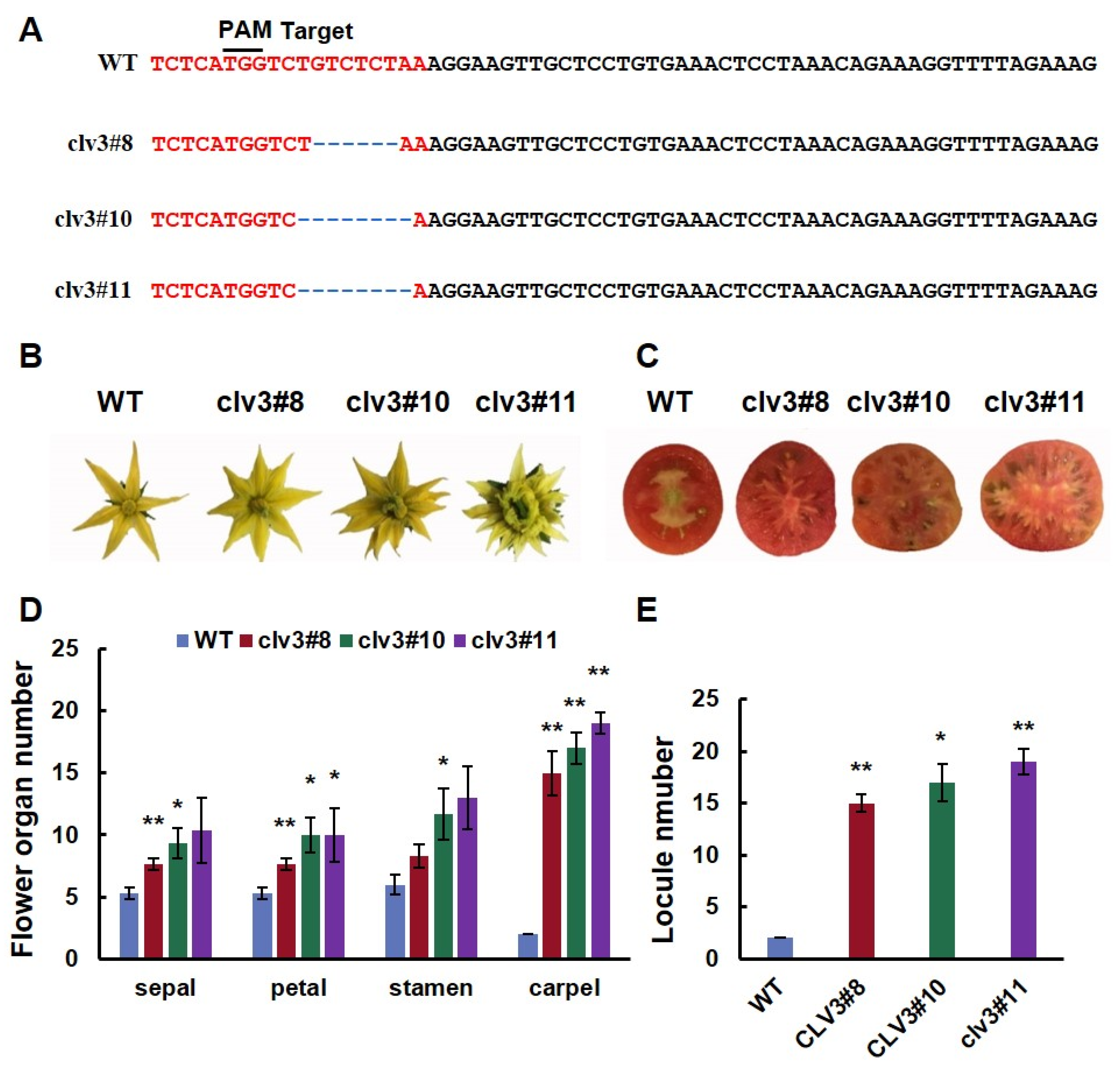
To investigate the locule-related function of SlCLV3 in fruit, a CRISPR/Cas9 construct with one SlCLV3 target was designed and transformed into ‘Ailsa Craig’. Sequence analysis verified that three T0 plants were successfully edited. Three mutant lines were homozygotes, Line 8 contained a six-base deletion at the target site, while lines 10 and 11 contained eight-base deletions at the target site (Figure 1A). The deletion of six bases in line 8 resulted in the absence of two amino acids in the CLV3 protein. Deleting eight nucleotides in lines 10 and 11 caused frameshift mutations in the CLV3 protein (Figure 2). Three mutant lines exhibited increases in the number of floral organs and fruit locules (Figure 1B,C). The number of sepals, petals, stamens, and carpels of three mutant lines increased significantly compared to the WT (Figure 1D). The number of fruit locules in the three mutant lines was significantly higher than that of the WT (Figure 1E).
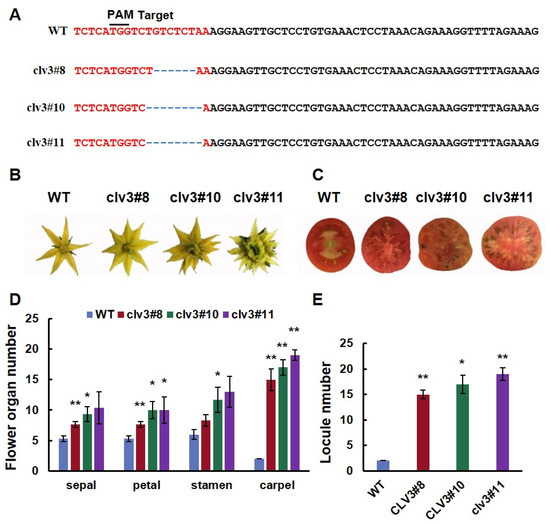
Figure 1.
Gene sequence and phenotype analysis of WT and clv3: (A) Sequence alterations observed in clv3 mutants; (B) flowers of WT and clv3 at anthesis; (C) mature fruits of WT and clv3; (D) comparative analysis of the organ number in the four floral whorls at the anthesis stage; (E) comparison of locule number of WT and clv3. PAM: protospacer adjacent motif; ‘-’ refers to a base deletion. * indicates significant differences at p < 0.05, ** indicates significant differences at p < 0.01, according to Independent t-test.

Figure 2.
Amino acid sequence analysis of clv3 and WT.
The amino acid sequence was deduced from the DNA sequence and compared using DNAMAN software (Version 10).
3.2. Transcriptome Analysis of WT and clv3 Mutants
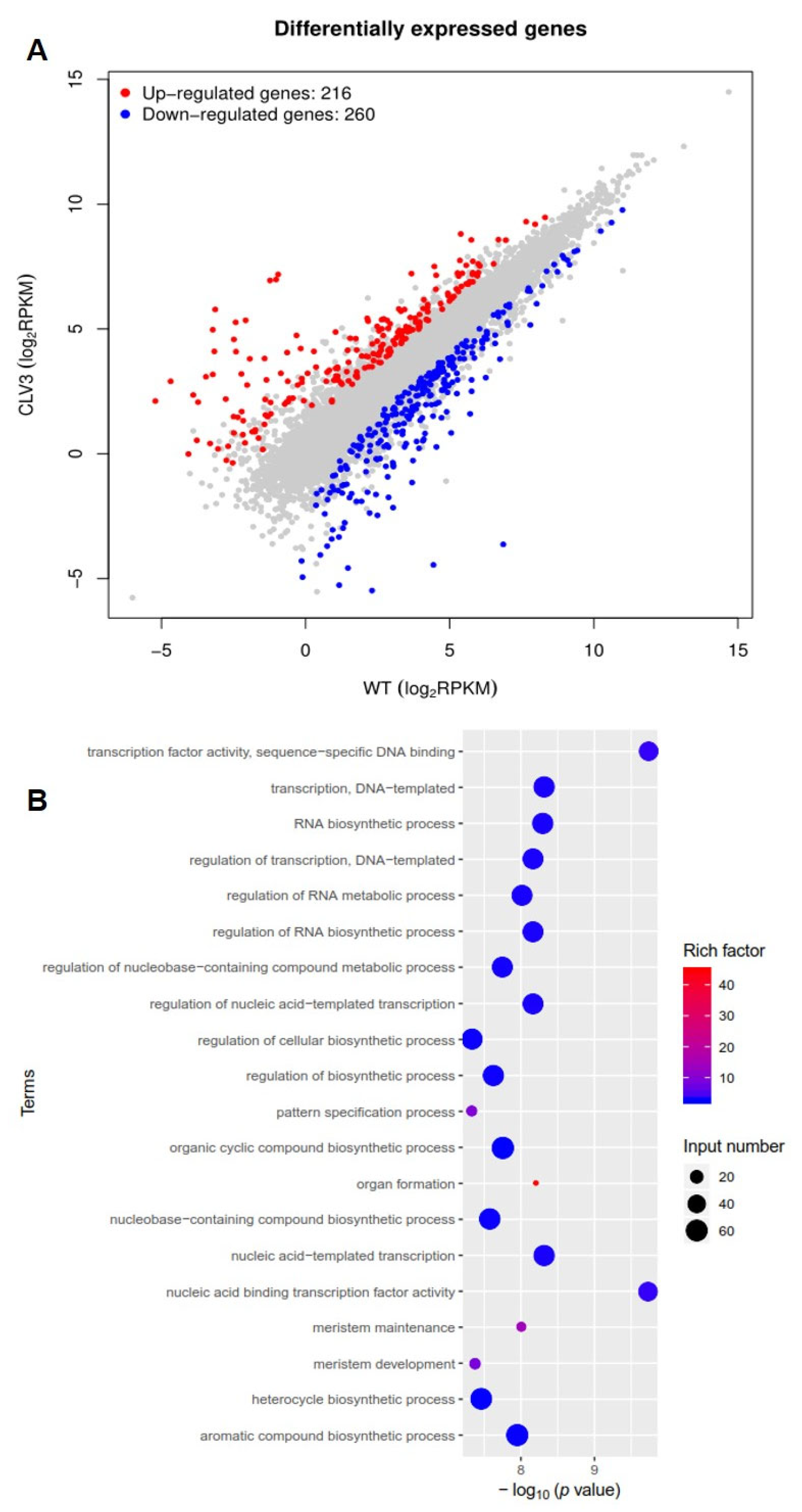
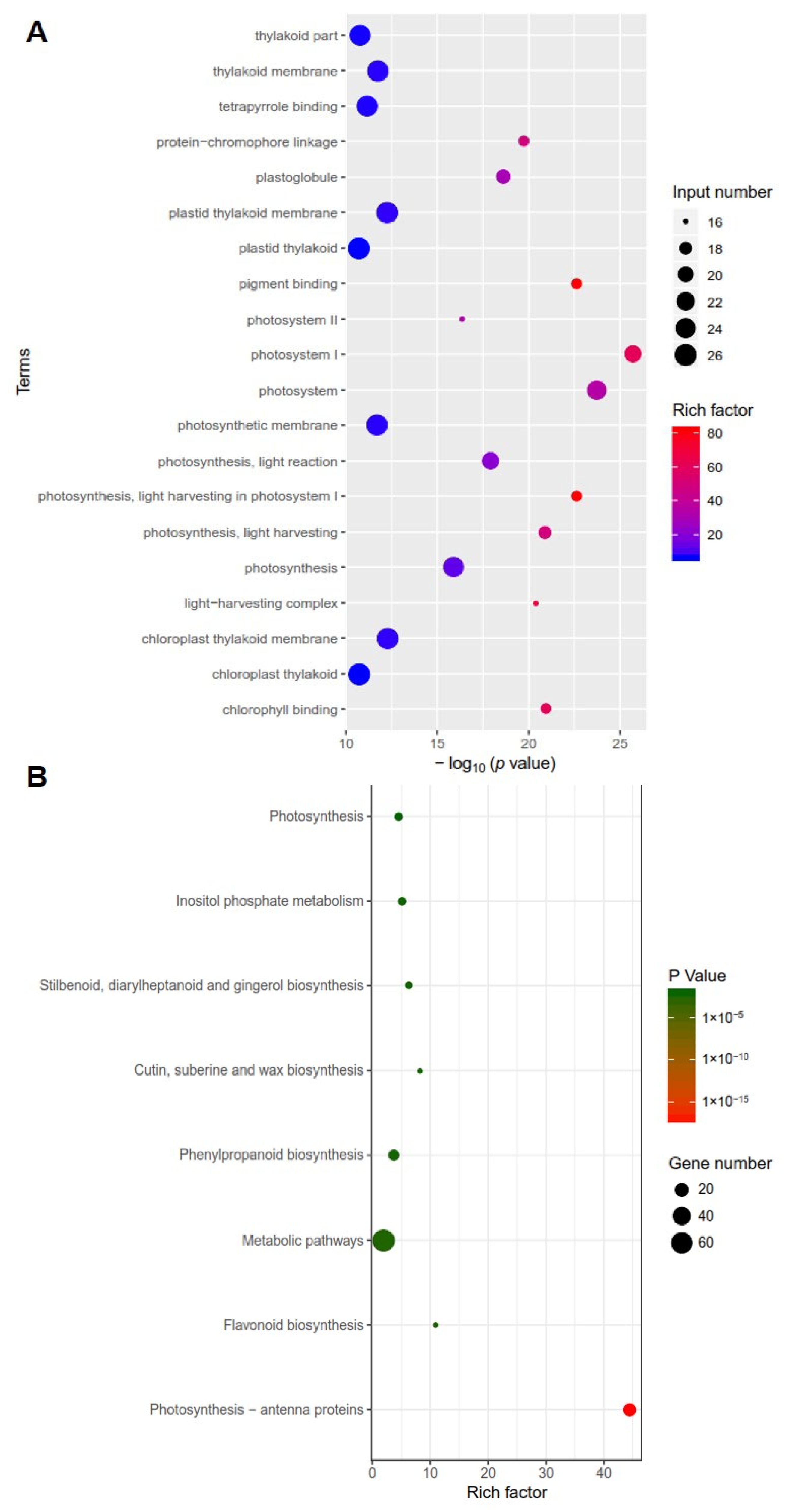
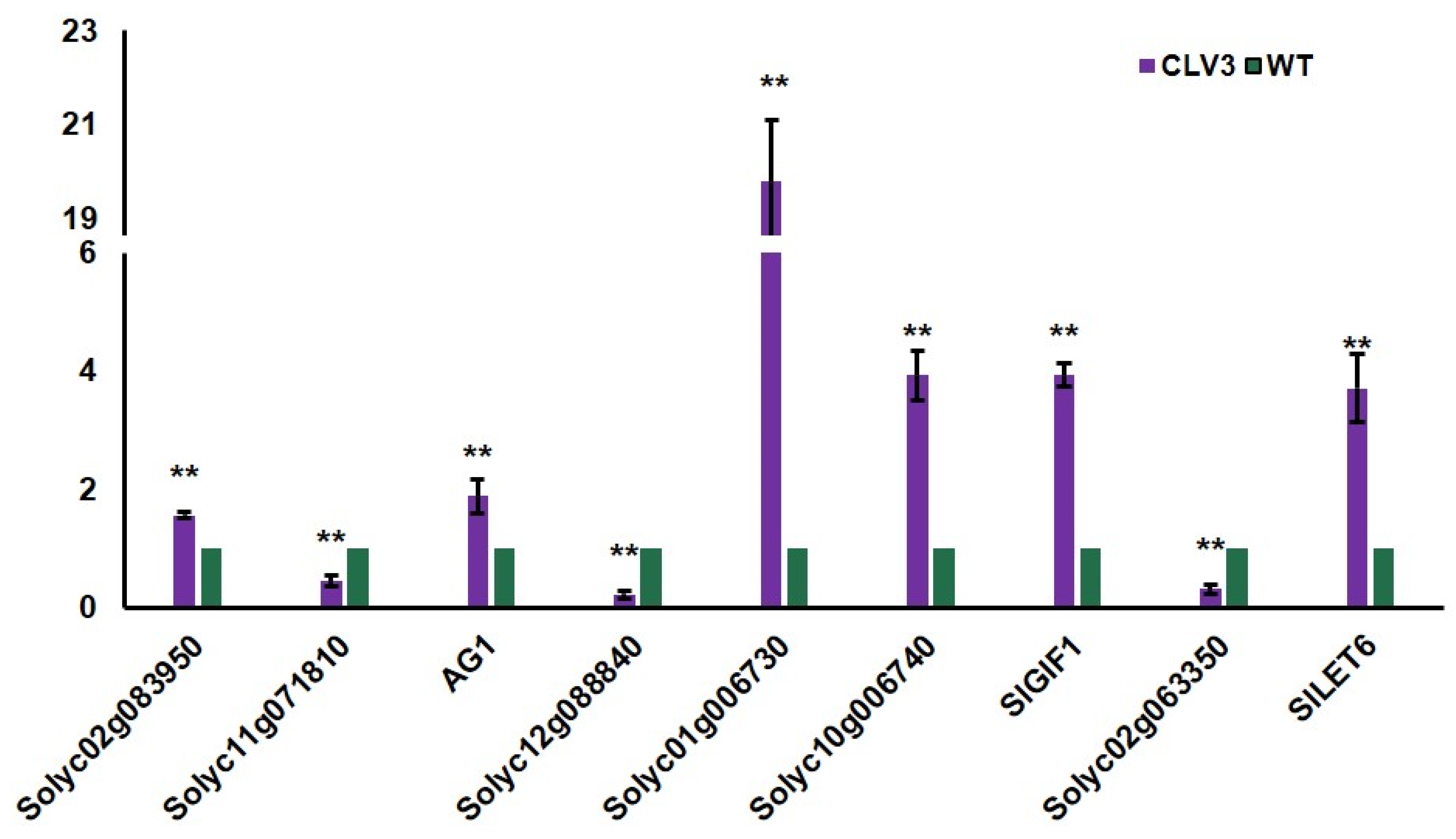
To identify differentially expressed genes in clv3 mutant compared to the wild type, RNA-seq libraries were created and sequenced from the early vegetative meristems of the WT and clv3 mutant plants. A total of 476 differentially expressed genes (DEGs) were found, with 216 up-regulated and 260 down-regulated (Figure 3A). GO functional annotation was performed on up-regulated DEGs, and GO terms were organ formation, meristem maintenance, organic cyclic compound biosynthetic process, meristem development, regulation of cellular biosynthetic process, and so on (Figure 3B). GO functional annotation was also performed on down-regulated DEGs, and GO terms were photosystem I, photosystem, photosynthesis, light harvesting in photosystem I, pigment binding, chlorophyll binding, photosynthesis, and so on (Figure 4A). The pathway enrichment analysis revealed that the DEGs were associated with many biological processes, including photosynthesis-antenna proteins, flavonoid biosynthesis, metabolic pathways, phenylpropanoid biosynthesis, cutin, suberine, and wax biosynthesis (Figure 4B). According to previous studies, KAPP (Kinase-associated protein phosphatase), the phosphatases POL (POLTERGEIST) and PLL (POL-like), GTP binding proteins, mitogen-activated protein kinases (MAPK), calcium signaling genes, and STM (SHOTMERISTEMLESS) may be potentially involved downstream of SlCLV3. WUSCHEL, YABBY2b, TAG1, and six candidate genes of SlCLV3 downstream, including five calcium signaling-related genes as well as SlLET6 (the STM ortholog), were discovered in the DEGs (Table 3). RT-PCR was employed to validate the chosen differentially expressed genes (DEGs) and confirm the reliability of the RNA-seq results (Figure 5).
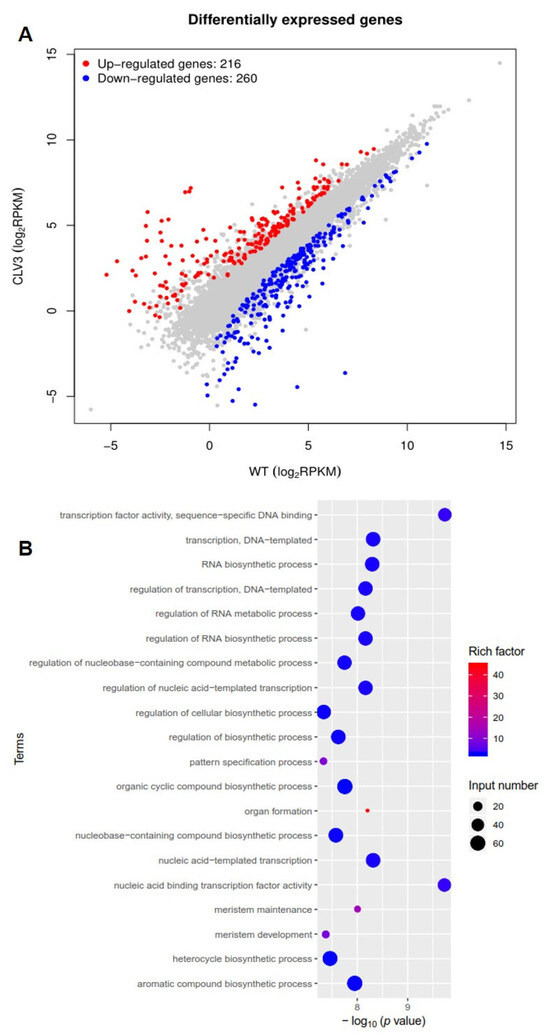
Figure 3.
Transcriptome analysis of clv3 and WT plants: (A) Differentially expressed genes (DEGs); (B) GO functional annotation of up-regulated DEGs. DEGs were selected based on their absolute fold change value of logFC > 1 and a p-value < 0.05. A significance level of <0.05 was employed as a standard for screening enriched GO data.
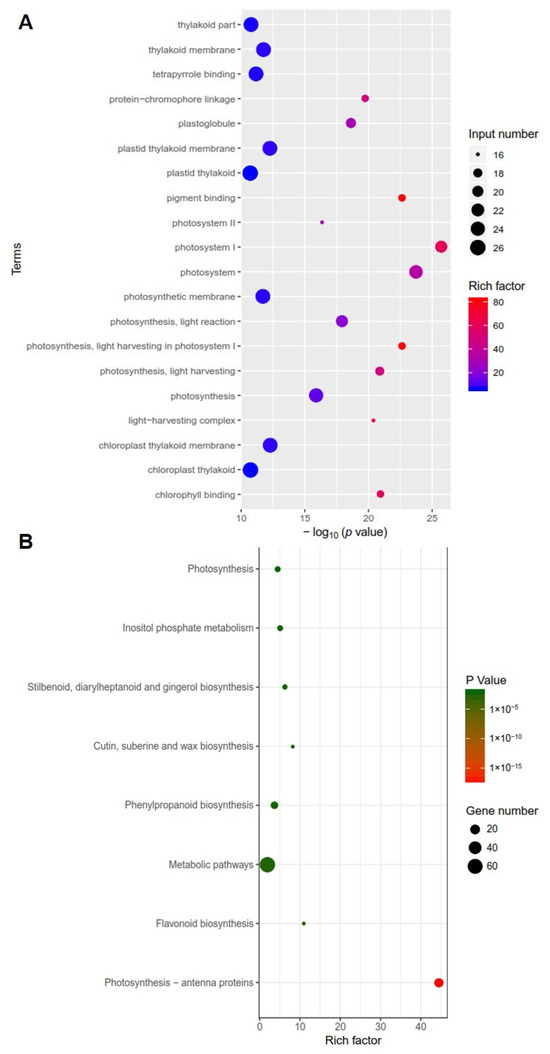
Figure 4.
Transcriptome analysis of clv3 and WT plants: (A) GO functional annotation of down-regulated DEGs; (B) pathway enrichment analysis of the DEGs. p-value < 0.05 was used as the standard for screening enriched GO data and KEGG pathway databases.

Table 3.
DEGs involved in the CLV-WUS pathway.
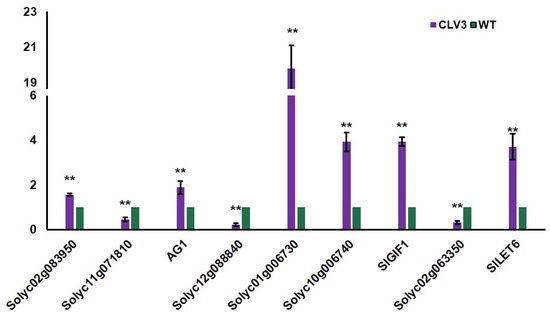
Figure 5.
Quantitative RT-PCR analysis on DEGs in the early vegetative meristem of WT and clv3. ** indicates significant differences at p < 0.01, according to Independent t-test.
3.3. Higher Expression of SlLET6 and SlGIF1 Is Found on a Fas Mutation Background
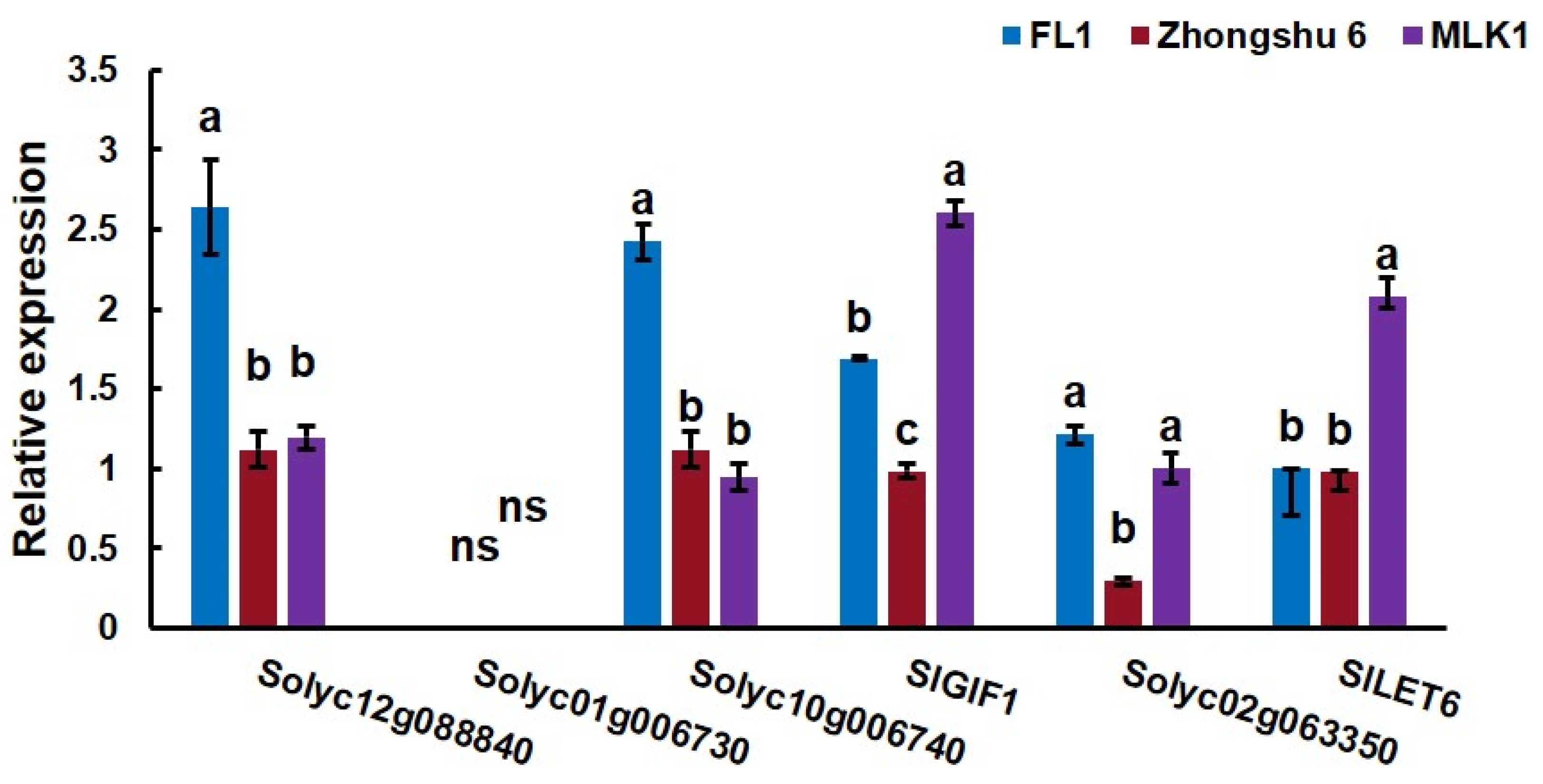
To identify genes that act downstream of SlCLV3, expression levels of six candidate genes were analyzed in the early vegetative meristems of three tomato lines, including ‘MLK1’, ‘FL1’, and ‘Zhongshu6’. ‘MLK1’ is large fruit tomato with more than 10 locules, ‘FL1’ is small fruit tomato with 2–4 locules, and ‘Zhongshu6’ is medium fruit tomato with 6–8 locules. The cultivated allele of fas is present in ‘MLK1’ and the wild-type allele of fas is present in ‘Zhongshu 6’ and ‘FL1’. The results showed that the expression levels of SlLET6 and SlGIF1 were higher in ‘MLK1’ compared to ‘FL1’ and ‘Zhongshu6’ (Figure 6), which indicated that the expression of SlLET6 and SlGIF1 was the difference between the wild-type and cultivated allele of fas. The expression levels of other candidate genes in ‘MLK1’ were not significantly different from those in ‘FL1’ and ‘Zhongshu6’.
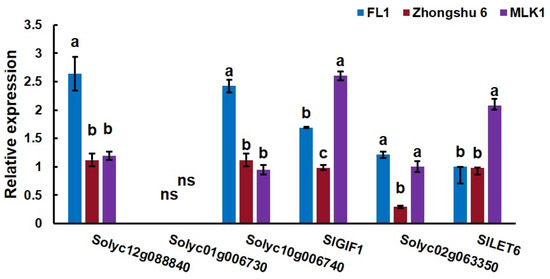
Figure 6.
Expression levels analysis of candidate genes in the early vegetative meristem of three tomato lines, including ‘MLK1’, ‘FL1’, and ‘Zhongshu6’. The small letters indicate significant differences at p < 0.05 according to Duncan’s multiple range tests.
3.4. SlLET6 and SlGIF1 Were Highly Expressed during Flower Development
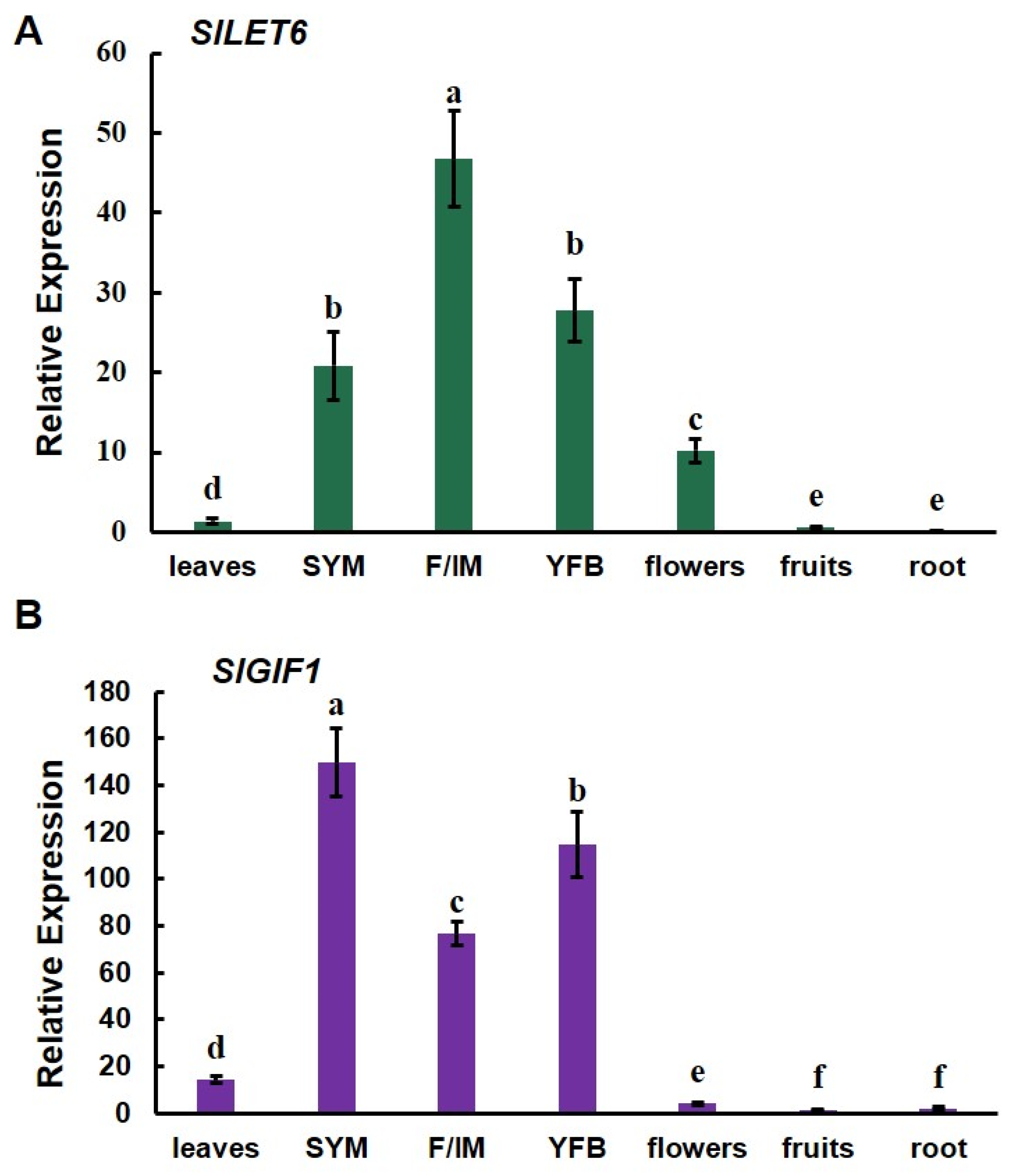
Gene expression patterns were analyzed by detecting the expression levels of the SlLET6 and SlGIF1 genes in all tissues. SlLET6 was highly expressed in the vegetative meristems, floral/inflorescence meristems, young flower buds, and flowers, while low expression was found in the leaves, fruits, and roots (Figure 7A). SlGIF1 exhibited highly expressed in the vegetative meristems, floral/inflorescence meristems, and young flower buds, while showing minimal expression in the leaves, flowers, fruits, and roots (Figure 7B). Importantly, SlLET6 and SlGIF1 were highly expressed during tomato flower development, indicating that they may play a role in this process.
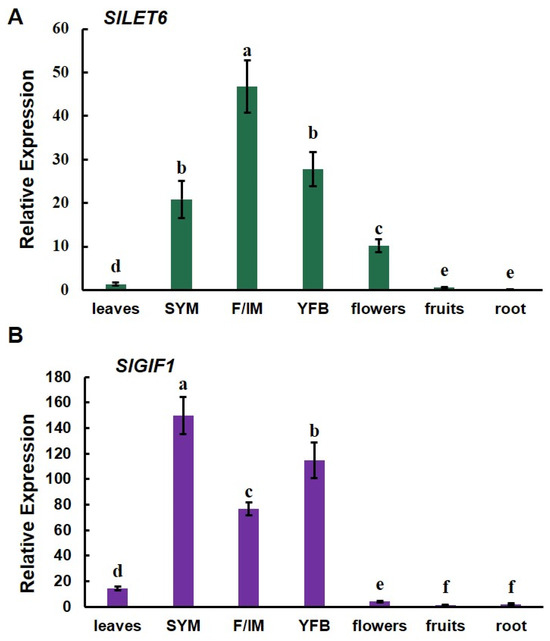
Figure 7.
Expression pattern analysis of SlLET6 and SlGIF1: (A) Expression pattern analysis of SlLET6; (B) expression pattern analysis of SlGIF1. SYM, apical meristem; F/IM, floral/inflorescence meristems; YFB, young flower buds. The small letters indicate significant differences at p < 0.05 according to Duncan’s multiple range test.
3.5. The Overexpression of SlLET6 Increased Tomato Fruit Locules
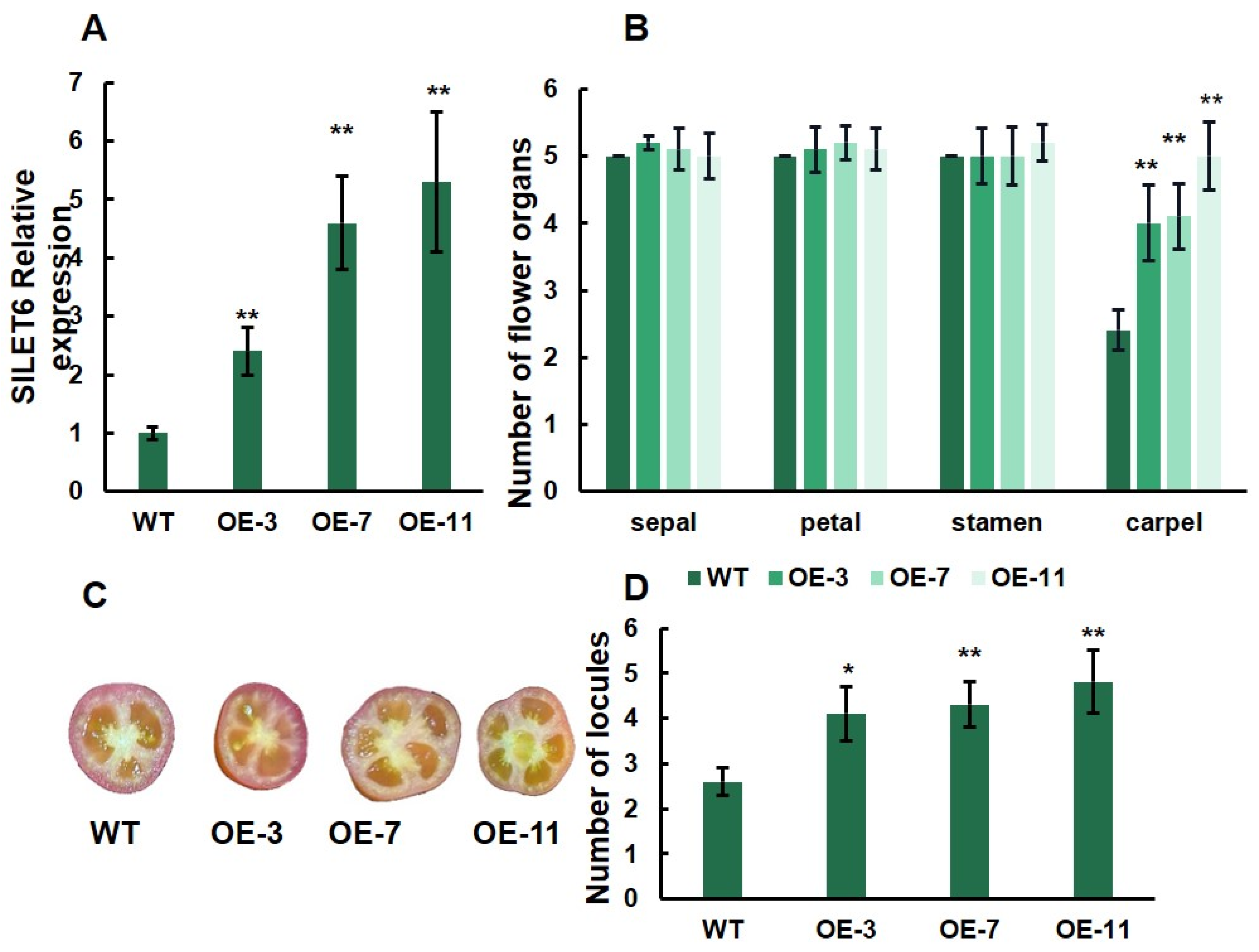
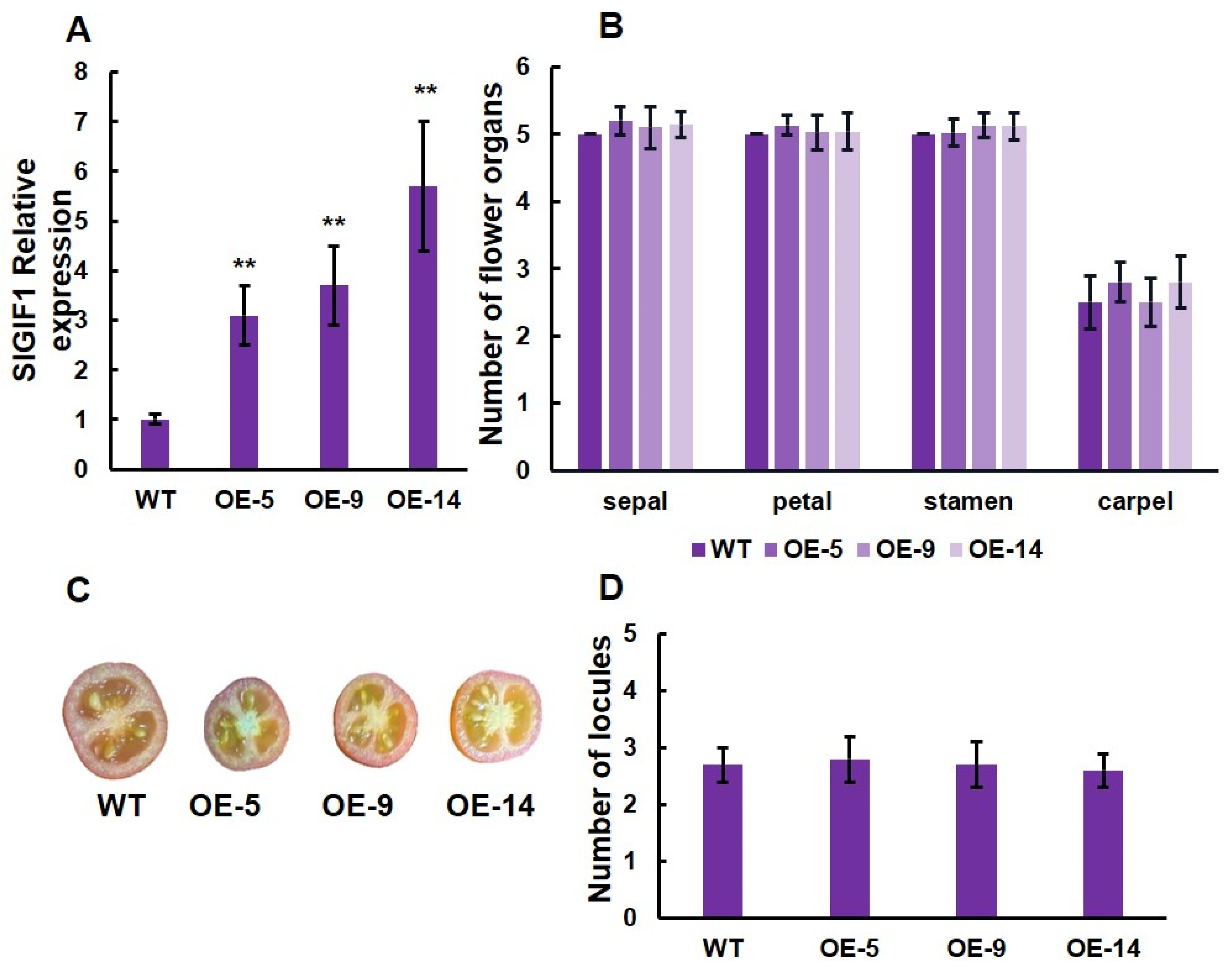
To further understand the function of SlLET6 and SlGIF1, we generated three overexpression transgenic lines (OE-3, OE-7, and OE-11) of SlLET6 and three overexpression transgenic lines (OE-5, OE-9, and OE-14) of SlGIF1. RT-PCR analysis revealed that the expression of SlLET6 in OE-3, OE-7, and OE-11 was more than twice that of the WT (Figure 8A). The carpel number of three transgenic lines increased significantly compared to the WT, while there were no significant alterations in the number of sepals, petals, and stamens (Figure 8B). The number of fruit locules of the three transgenic lines were consistently higher than that of the WT (Figure 8C,D). The above indicates that the function of SlLET6 is to control the carpel number in flowers and ultimately affects the locule number in fruit. The expression of SlGIF1 in OE-5, OE-9, and OE-14 were more than three times that of the WT (Figure 9A). The number of sepals, petals, stamens, and carpels of the three transgenic lines did not exhibit any significant alteration when compared to those of the WT (Figure 9B). There was also no significant difference in the locule number of fruits between the transgenic lines and WT (Figure 9C, D). All of the above suggests that the SlGIF1 function is unrelated to regulating the number of flower organs and fruit locules.
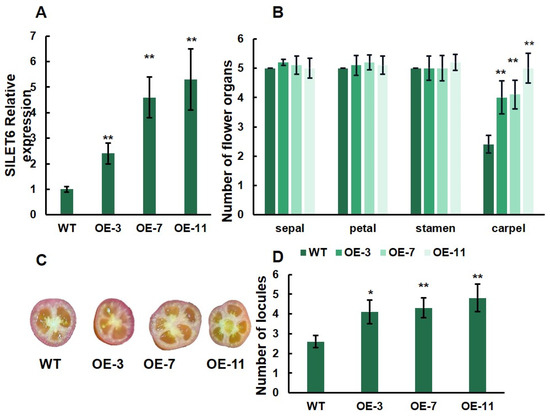
Figure 8.
Gene expression level and phenotypic analysis of overexpression transgenic lines of SlLET6 and WT: (A) SlLET6 expression analysis of overexpression transgenic lines and WT; (B) mean comparison of the organ number in the four floral whorls at the anthesis stage; (C) mature fruits of overexpression transgenic lines and WT; (D) mean comparison of locule number of overexpression transgenic lines and WT. * indicates significant differences at p < 0.05, ** indicates significant differences at p < 0.01, according to Independent t-test.
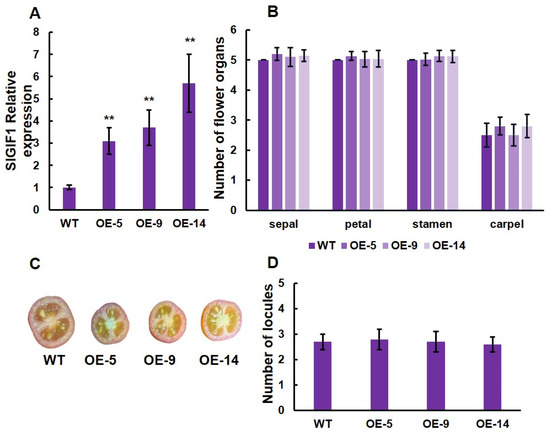
Figure 9.
Gene expression level and phenotypic analysis of SlGIF1 in overexpression transgenic lines and WT: (A) SlGIF1 expression analysis in overexpression transgenic lines and WT; (B) mean comparison of the organ number in the four floral whorls at the anthesis stage; (C) mature fruits of overexpression transgenic lines and WT; (D) Mean comparison of locule number of overexpression transgenic lines and WT. ** indicates significant differences at p < 0.01, according to Independent t-test.
4. Discussion
4.1. The Role of CLV3 in Regulating the Number of Flower Organs and Fruit Locules in Tomato
Introgression of fas locus into S. pimpinellifolium (Sp-fas) produced half of all fruits developing three locules, while the full SlCLV3 gene was introduced into Sp-fas plants, resulting in fruit rescued to two locules, those confirmed that SlCLV3 is the candidate gene of fasciated locus [12]. Additionally, modifying the SlCLV3 promoter resulted in a significant increase in the number of flower organs and locules in tomato [8]. In this study, the first exon of the tomato SlCLV3 gene was edited by the CRISPR/cas9 system, resulting in the functional deletion of SlCLV3, producing flowers with a large number of organs and fruits with a large number of locules. The results are consistent with previous views, and provide additional confirmation of the role of SlCLV3 in regulating the number of flower organs and fruit locules in tomato.
4.2. SlLET6 Plays an Important Role in Regulating the Flower Development of Tomato
SlLET6 encodes class 1 knotted-like homeodomain protein and is an ortholog of AtSTM, which regulates floral fate and coordinates the meristem and boundary [28,29]. In situ hybridization has previously demonstrated that this gene has significant expression in young flower buds and ovules of tomato [30]. The overexpression of SlLET6 in tomato has been shown to cause fasciated flowers with a large number of petals and bifurcated fasciated fruit developing from an abnormal flower [31]. In this study, expression pattern analysis revealed that SlLET6 exhibited a prominent expression pattern in the apical meristems, floral/inflorescence meristems, young flower buds, and flowers. The overexpression of SlLET6 increased flower carpels and fruit locules. In conclusion, it is indicated that SlLET6 has a crucial function in regulating flower development, especially in controlling the number of carpels, hence influencing the number of fruit locules in tomato.
4.3. SlLET6 May Play an Important Role Downstream of CLV3
Previous studies have indicated that several factors, including KAPP (Kinase-associated protein phosphatase) [18], the phosphatases POL (POLTERGEIST) and PLL (POL-like) [20,32], GTP binding proteins [21], mitogen-activated protein kinases (MAPK) [22], calcium signaling genes [23], and STM (SHOTMERISTEMLESS) [33], may all play roles downstream of CLV3.
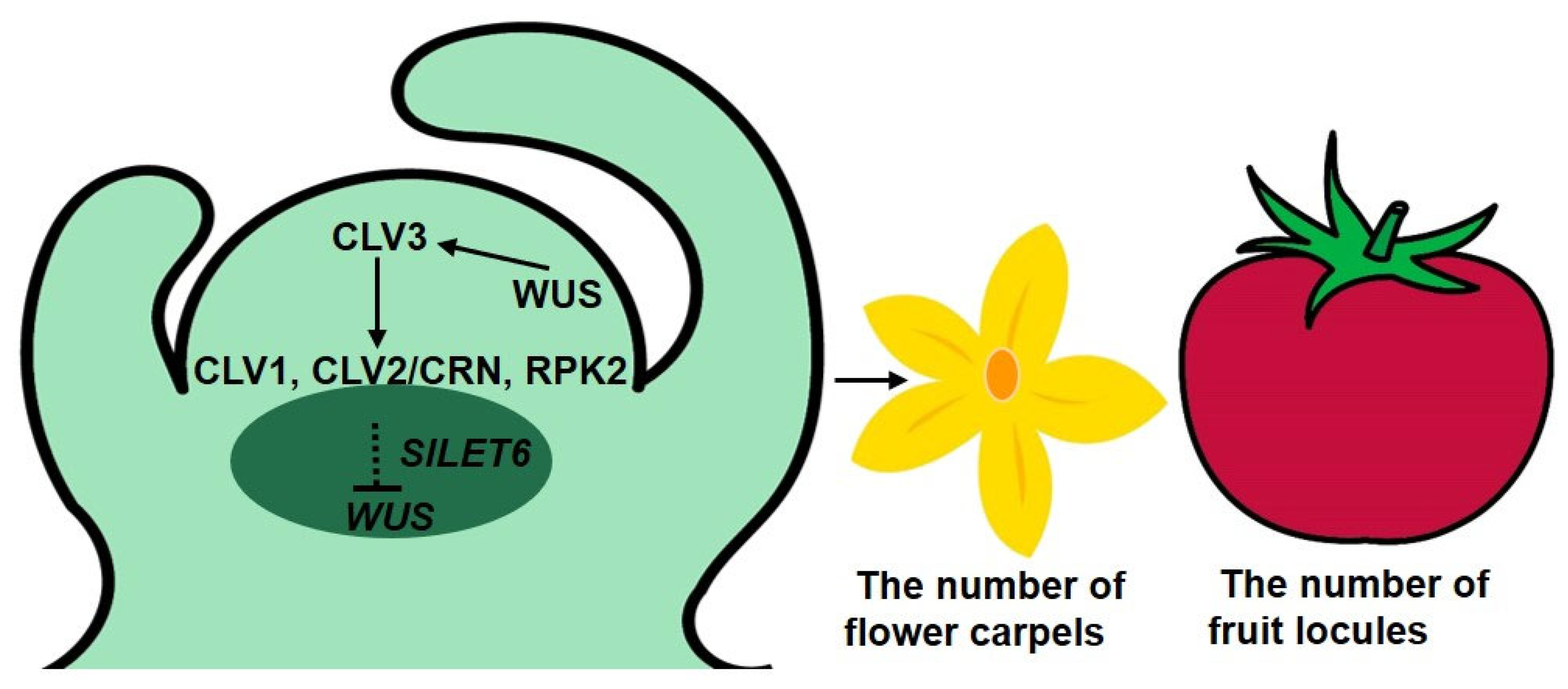
We screened six candidate genes, including five calcium-related genes and SlLET6 (the STM ortholog) using a transcriptome analysis of clv3 mutants and the WT. Analysis of expression alterations of candidate genes between the cultivated and wild-type alleles of fas revealed that only SlLET6 and SlGIF1 were regulated by the fas locus. SlCLV3 is identified as the candidate gene of the fas locus, SlLET6 and SlGIF1 are regulated by the fas locus, suggesting that SlLET6 and SlGIF1 may be the downstream genes of SlCLV3. Transgenic experiments demonstrated that SlLET6 and SlCLV3 exhibited similar functions in regulating the number of flower carpels and fruit locules in tomato. This provides more evidence supporting the hypothesis that SlLET6 may be the downstream gene of SlCLV3 regulating the number of carpels and fruit locules in tomato. In this study, we proposed a working model of SlCLV3 regulating the number of flower carpels and fruit locules mediated by SlLET6 in tomato (Figure 10). In tomato apical meristem, WUS protein promotes SlCLV3 expression, in turn, the signal peptide SlCLV3 is recognized by cell membrane receptors CLV1, CLV2/CRN, and RPK2, this recognition triggers a signal that is transmitted to SlLET6, resulting in the inhibition of WUS expression. This pathway controls the apical meristem size and ultimately affects the number of flower carpels and fruit locules. In the future, we will conduct a series of experiments to confirm the relationship between SlCLV3 and SlLET6 in tomato.
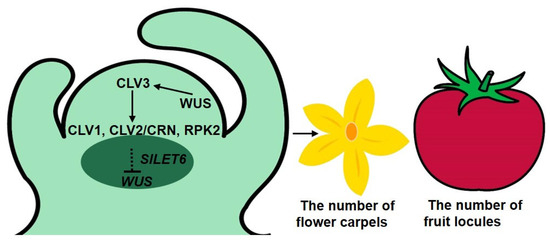
Figure 10.
SlCLV3 regulates the number of flower carpels and fruit locules mediated by SlLET6 in tomato. Previous studies have confirmed that in tomato apical meristem, a feedback system composed of CLV-WUS regulates the size of apical meristem. Firstly, WUS protein promotes the expression of SlCLV3, and SlCLV3 signal peptide is recognized by cell membrane receptors CLV1, CLV2/CRN, and RPK2, ultimately leading to the inhibition of WUS expression [12]. Our study verifies that SlLET6 may act downstream of CLV3, inhibiting WUS expression, controlling apical meristem size, and ultimately impacting the number of flower carpels and fruit locules.
5. Conclusions
The number of flower carpels and fruit locules can affect the yield and quality of tomato fruit. Tomato SlCLV3 plays a crucial role in controlling the number of flower carpels and fruit locules, mainly by inhibiting WUS expression. However, the factors mediating CLV3 protein to inhibit WUS expression have yet to be identified. Therefore, we analyzed the transcriptome of the clv3 mutant and the expression changes in differentially expressed genes between the wild-type and cultivated alleles of fas and screened two candidate genes of SlCLV3 downstream. The expression pattern analysis and transgenic experiments confirmed that SlLET6 may be the downstream gene of SlCLV3 to regulate the number of flower carpels and fruit locules. The study results offer a fresh outlook on comprehending the mechanism of the CLV-WUS feedback system regulating the number of flower carpels and fruit locules.
Author Contributions
Conceptualization, T.L.; methodology, M.Q.; software, M.S.; validation, Y.Z.; formal analysis, Y.M.; investigation, H.S.; resources, L.H.; data curation, L.T.; writing—review and editing, Y.Z.; visualization, H.S.; supervision, M.S.; project administration, M.Q.; funding acquisition, T.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Shanxi Basic Research Project (202103021223171, 20210302124242), Shanxi Agricultural University Doctoral Research Project (2021BQ31), and Shanxi Province Key R&D Plan (202102140601013).
Data Availability Statement
The sequence data in this work can be searched in the NCBI database (SRA data: PRJNA1053672).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tanksley, S.D. The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 2004, 16 (Suppl. 1), S181–S189. [Google Scholar] [CrossRef] [PubMed]
- Lippman, Z.; Tanksley, S.D. Dissecting the Genetic Pathway to Extreme Fruit Size in Tomato Using a Cross Between the Small-Fruited Wild Species Lycopersicon pimpinellifolium and L. esculentum var. Giant Heirloom. Genetics 2001, 158, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Barrero, L.S.; Tanksley, S.D. Evaluating the genetic basis of multiple-locule fruit in a broad cross section of tomato cultivars. Theor. Appl. Genet. 2004, 109, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, G.R.; Munos, S.; Anderson, C.; Sim, S.C.; Michel, A.; Causse, M.; Gardener, B.B.; Francis, D.; van der Knaap, E. Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 2011, 156, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Barrero, L.S.; Cong, B.; Wu, F.; Tanksley, S.D. Developmental characterization of the fasciated locus and mapping of Arabidopsis candidate genes involved in the control of floral meristem size and carpel number in tomato. Genome 2006, 49, 991–1006. [Google Scholar] [CrossRef] [PubMed]
- Munos, S.; Ranc, N.; Botton, E.; Berard, A.; Rolland, S.; Duffe, P.; Carretero, Y.; Le Paslier, M.C.; Delalande, C.; Bouzayen, M.; et al. Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol. 2011, 156, 2244–2254. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Kim, Y.J.; Muller, R.; Yumul, R.E.; Liu, C.; Pan, Y.; Cao, X.; Goodrich, J.; Chen, X. AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 2011, 23, 3654–3670. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Leal, D.; Lemmon, Z.H.; Man, J.; Bartlett, M.E.; Lippman, Z.B. Engineering Quantitative Trait Variation for Crop Improvement by Genome Editing. Cell 2017, 171, 470–480. [Google Scholar] [CrossRef]
- Li, H.; Qi, M.; Sun, M.; Liu, Y.; Liu, Y.; Xu, T.; Li, Y.; Li, T. Tomato Transcription Factor SlWUS Plays an Important Role in Tomato Flower and Locule Development. Front. Plant Sci. 2017, 8, 457. [Google Scholar] [CrossRef]
- Cong, B.; Barrero, L.S.; Tanksley, S.D. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 2008, 40, 800–804. [Google Scholar] [CrossRef]
- Huang, Z.; Knaap, E.V.D. Tomato fruit weight 11.3 maps close to fasciated on the bottom of chromosome 11. Theor. Appl. Genet. 2011, 123, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Liberatore, K.L.; MacAlister, C.A.; Huang, Z.; Chu, Y.H.; Jiang, K.; Brooks, C.; Ogawa-Ohnishi, M.; Xiong, G.; Pauly, M.; et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 2015, 47, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, L.; Hendelman, A.; Hutton, S.F.; McCandlish, D.M.; Lippman, Z.B. Idiosyncratic and dose-dependent epistasis drives variation in tomato fruit size. Science 2023, 328, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Stahl, Y.; Simon, R. Plant primary meristems: Shared functions and regulatory mechanisms. Curr. Opin. Plant Biol. 2010, 13, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.; Jürgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Gou, X.; He, K.; Yang, H.; Yuan, T.; Lin, H.; Clouse, S.D.; Li, J. Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana. BMC Genom. 2010, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Muller, R.; Bleckmann, A.R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 2008, 20, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Atsuko, K.; Shigeyuki, B.; Yuriko, O.; Shinji, M.; Shingo, N.; Yvonne, S.; Rüdiger, S.; Kazuko, Y.S.; Hiroo, F.; Shinichiro, S. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 2010, 137, 3911–3920. [Google Scholar]
- Williams, R.W.; Wilson, J.M.; Meyerowitz, E.M. A possible role for kinase-associated protein phosphatase in the Arabidopsis CLAVATA1 signaling pathway. Proc. Natl. Acad. Sci. USA 1997, 94, 10467–10472. [Google Scholar] [CrossRef]
- Song, S.K.; Clark, S.E. POL and related phosphatases are dosage-sensitive regulators of meristem and organ development in Arabidopsis. Dev. Biol. 2005, 285, 272–284. [Google Scholar] [CrossRef]
- Takashi, I.; Ryo, T.; Masashi, Y.; Mitsuhiro, A.; Kanako, M.; Masayuki, F.; Katsushi, Y.; Shuji, S.; Masayuki, H.; Hiroyuki, T. Heterotrimeric G proteins control stem cell proliferation through CLAVATA signaling in Arabidopsis. EMBO Rep. 2015, 15, 1202–1209. [Google Scholar]
- Shigeyuki, B.; Fuminori, T.; Atsuko, K.; Hiroki, M.; Kazuo, S.; Hiroo, F.; Shinichiro, S. Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant Cell Physiol. 2011, 52, 14–29. [Google Scholar]
- Chou, H.; Zhu, Y.; Ma, Y.; Berkowitz, G.A. The CLAVATA signaling pathway mediating stem cell fate in shoot meristems requires Ca2+ as a secondary cytosolic messenger. Plant J. 2016, 85, 494–506. [Google Scholar] [CrossRef]
- Clark, S.E.; Jacobsen, S.E.; Levin, J.Z.; Meyerowitz, E.M. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 1996, 122, 1567–1575. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, S.; Song, Y.; Wang, J. Genome-wide characterization, expression and functional analysis of CLV3/ESR gene family in tomato. BMC Genom. 2014, 15, 827. [Google Scholar] [CrossRef]
- Su, D.; Xiang, W.; Wen, L.; Lu, W.; Shi, Y.; Liu, Y.; Li, Z. Genome-wide identification, characterization and expression analysis of BES1 gene family in tomato. BMC Plant Biol. 2021, 21, 161. [Google Scholar] [CrossRef]
- Su, D.; Wen, L.; Xiang, W.; Shi, Y.; Lu, W.; Liu, Y.; Xian, Z.; Li, Z. Tomato transcriptional repressor SlBES1.8 influences shoot apical meristem development by inhibiting the DNA binding ability of SlWUS. Plant. J. 2022, 110, 482–498. [Google Scholar] [CrossRef]
- Roth, O.; Alvarez, J.P.; Levy, M.; Bowman, J.L.; Ori, N.; Shani, E. The KNOXI Transcription Factor SHOOT MERISTEMLESS Regulates Floral Fate in Arabidopsis. Plant Cell 2018, 30, 1309–1321. [Google Scholar] [CrossRef]
- Scofield, S.; Murison, A.; Jones, A.; Fozard, J.; Aida, M.; Band, L.R.; Bennett, M.; Murray, J.A.H. Coordination of meristem and boundary functions by transcription factors in the SHOOT MERISTEMLESS regulatory network. Development 2018, 145, 157081. [Google Scholar] [CrossRef]
- Janssen, B.J.; Williams, A.; Chen, J.J.; Mathern, J.; Hake, S.; Sinha, N. Isolation and characterization of two knotted-like homeobox genes from tomato. Plant Mol. Biol. 1998, 36, 417–425. [Google Scholar] [CrossRef]
- Janssen, B.J.; Lund, L.; Sinha, N. Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol. 1998, 117, 771–786. [Google Scholar] [CrossRef]
- Song, S.K.; Lee, M.M.; Clark, S.E. POL and PLL1 phosphatases are CLAVATA1 signaling intermediates required for Arabidopsis shoot and floral stem cells. Development 2006, 133, 4691–4698. [Google Scholar] [CrossRef]
- Lenhard, M.; Jürgens, G.; Laux, T. The WUSCHEL and SHOOTMERISTEMLESS genes fulfil complementary roles in Arabidopsis shoot meristem regulation. Development 2002, 129, 3195. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).