Abstract
Every year, seafood waste produced globally contains about 10 million tons of wasted crab, shrimp and lobster shells, which are rich in chitin resources. The exploitation and utilization of chitin resources are of great significance to environmental protection, economic development and sustainable development. Lytic polysaccharide monooxygenases (LPMOs) can catalyze polysaccharides by oxidative breakage of glycosidic bonds and have catalytic activity for chitin and cellulose, so they play an important role in the transformation of refractory polysaccharides into biomass. Although there have been many studies related to LPMOs, the research related to lytic chitin monooxygenases (LCMs) is still very limited. The specific catalytic mechanism of LCMs has not been fully elucidated, which poses a challenge to their application in industrial biomass conversion. This review introduces the present situation of resource development and utilization in chitin, the origin and classification of different LCMs families, the structural characteristics of LCMs and the relationship between structure and function. The research results related to activity detection, screening, preparation and transformation of LCMs were summarized and discussed. Finally, the synergistic effect of LCMs and chitin enzyme on biomass degradation was reviewed, and the existing problems and future research directions were pointed out. This is the first review focusing on Chitin-Active LPMOs in recent years, intending to provide a reference for applying chitin degradation enzymes system in the industry.
1. Introduction
With the increasing depletion of petrochemical resources, the world is facing severe challenges of energy and resources. Solving the sustainable supply of energy and resources and realizing green sustainable development have become major challenges faced by all countries in the world [1,2]. The utilization of renewable biomass resources has been paid more and more attention. As an important renewable biomass, chitin is the second-largest natural polymer compound after cellulose and the second-largest nitrogen-containing organic compound after protein [3]. Chitin is a highly crystalline, water-insoluble nitrogen-containing polysaccharide composed of N-acetyl-D-glucosamine (GlcNAc) through β-1, 4 bonds. Chitin is ubiquitous in nature, being mainly found in the shell of crustaceans, the outer epidermis of insects and the cell wall of fungi. Shrimp crab shells derived from seafood waste are the most common and largest source of chitin. Although the annual output of shrimp and crab shells in the world exceeds 10 million tons, most of them are not fully utilized and are even discarded as waste, which not only causes a waste of resources but also brings environmental problems [4]. Compared with cellulose, the utilization rate of chitin is still low. Converting chitin’s biomass into valuable chemicals could have a positive impact on addressing the global energy crisis and reducing environmental pollution [5,6]. Therefore, realising the large-scale and high-value utilization of chitin resources is of great economic value and strategic significance. Under the extensive concern of scholars from all over the world, the development and utilization of chitin resources is becoming a hot topic in current research. Lytic polysaccharide monooxygenases (LPMOs) are newly discovered oxidases which can destroy the structure of chitin and cellulose, etc., by oxidative cleavage and have an important role in the efficient enzymatic hydrolysis of chitin and cellulose. In the past decade, many lytic chitin monooxygenases (LPMOs) with the potential for efficient treatment of biomass have been well studied and reviewed [2,7,8,9]. The chitin-active LPMOs (lytic chitin monooxygenases; LCMs; EC 1.14. 99.53) and glycoside hydrolases (GH) showed much higher chitin degradation activity than any single enzyme. At present, there are three kinds of LCMs: AA10 family LCMs, AA11 family LCMs and AA15 family LCMs. Compared with AA11 and AA15 family LCMs, AA10 family LCMs are more widely distributed and discovered earliest, but there is no literature to summarize their research progress. In this paper, the development status of chitin biomass resources, the research history and the classification of LPMO were systematically summarized, and the function and mechanism, activity determination and screening of LCMs with chitin activity were discussed. The discovery of AA10 chitin LPMs, the improvement of chitin enzymatic hydrolysis efficiency, catalytic mechanism, recombinant expression technology and its application were emphatically introduced, and the challenges faced by LCMs and the future research directions were prospected, hoping to provide a reference for the efficient utilization of LCMs in chitin resources, biofuel production and other biotechnology applications.
2. Present Situation of Biomass Resources Development in Chitin
The annual biomass of chitin in the world is 1010–1011 tons, and the main source of chitin resources is crustacean waste, especially seafood waste [4,10]. The shell waste produced by crustaceans is about 1.44 million metric tons of dry weight every year and contains about 7 wt% nitrogen, which can provide raw materials for green manufacturing of nitrogen-containing chemicals [11]. At present, the rapid development of the seafood processing industry and seafood consumption makes it more convenient to collect chitin resources. The shell waste ingredient includes proteins, minerals and chitin, producing a variety of products such as biofuels, biopolymers and health products. Chitin is formed by the polymerization of GlcNAc via β-1,4-glycosidic bonds, and adjacent polysaccharide chains are connected by hydrogen bonds, which have a highly ordered crystal structure. Chitin has strong intra- and inter-molecular hydrogen bonds, and crystalline chitin is insoluble in water, dilute acids, dilute bases and common organic solutions. Chitin has three types of crystal structures, α, β and γ; α-chitin is the most abundant chitin in nature [12] (Figure 1a). The main difference is that the sugar chains of α-chitin are arranged in the reverse parallel direction, the sugar chains of β-chitin are arranged in the forward parallel direction, and the sugar chains of γ-chitin have two mixed arrangements (Figure 1b). Among the three types of chitin, the α-chitin has the strongest hydrogen bonds, so α-chitin is the most stable and the most difficult chitin to degrade.
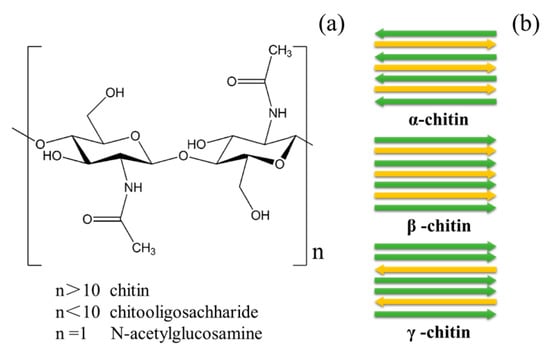
Figure 1.
(a) The chemical structures of chitin, chitooligosaccharides and N-Acetylglucosamine; (b) the crystalline forms of chitin.
Compared with cellulose, chitin hydrogen bonding is stronger, and acetylamino groups are highly hydrophobic [13,14]. Therefore, chitin is insoluble in water, dilute acids, dilute bases and most organic solvents. Conventional shell waste treatment techniques use a large amount of dilute acid/base. It leads to a large amount of wastewater in the process of demineralization and deproteinization. Wastewater not only contains a lot of calcium and protein but also has high salinity and corrosiveness, which leads to the inability to make full use of this wastewater. In addition, the loss of protein and minerals, long reaction time and low product yield are also problems in the current chitin processing. This is also a great challenge for the development and utilization of chitin resources [15,16]. At present, only a small part of chitin resources are used for the extraction and production of chitin-derived products such as chitooligosaccharides and N-acetylglucosamine, and the characteristics of environmental pollution and low profit are widespread in the process of extraction and utilization [16,17]. Chitin-derived products are soluble, renewable and non-toxic compounds which have many excellent biological properties such as antioxidant, antimicrobial and anti-cancer properties. Therefore, it has a wide range of applications (Figure 2). For example, oligosaccharides from chitin, in the field of medicine, have the functions of regulating immunity, regulating hormone secretion, lowering blood lipid, blood sugar, blood pressure, cholesterol, heavy metal ions, inhibiting the growth and metastasis of cancer cells and diminishing inflammation. In the field of food, it has the functions of seasoning and improving food characteristics. In the agricultural field, it is a new kind of natural, efficient, broad-spectrum and non-toxic plant growth regulator and has an activation effect on plant defence mechanisms. In addition, oligosaccharides from chitin are also widely used in environmental protection, chemical industry and other fields [7,17,18]. With the increasing global demand, chitin-derived products have shown unprecedented application prospects and economic value, and the production of chitin-derived products has become the main research direction of resource utilization in chitin.
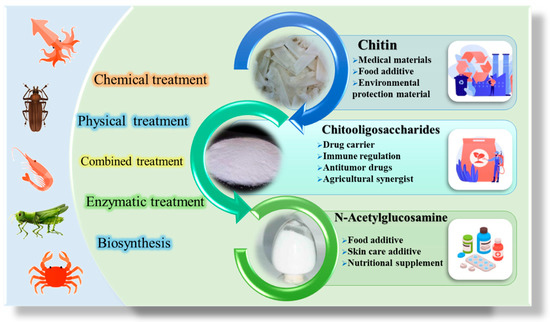
Figure 2.
Production of chitin, chitooligosaccharides and N-acetylglucosamine from various resources and their applications.
The preparation methods of chitin-derived products such as glucosamine and chitooligosaccharides mainly include chemical, physical, enzymatic hydrolysis, and biosynthesis [17,19,20,21,22]. Most of the raw materials used in chemical and physical production are chitin extracted from shrimp and crab shells. Compared with physical and chemical methods, enzymatic degradation of chitin has the advantages of relatively mild reaction conditions, strong control and environmental friendliness, which will be the development direction of the modern chitin industry. The bio-enzymatic method has been studied and applied to degrade chitin to produce high-value-added products such as oligosaccharides [23]. At present, the accumulation of research results on the structure and mechanism of chitin degradation enzymes has significantly increased the possibility of controlling the depolymerization process and the polymerization degree of products in the process of oligosaccharide production by enzymatic hydrolysis [24]. Theoretically, various forms of oligosaccharides can be prepared by selecting different substrates and hydrolases (Figure 3) [25]. Although chitin enzymes can effectively degrade amorphous regions of chitin, their degradation efficiency of crystalline chitin is very low. There are still some problems, such as poor degradation activity and low production efficiency of a single enzyme, which make it difficult to realize industrialisation. Therefore, it is urgent to study chitin-degrading enzymes. The raw materials used in enzymatic hydrolysis are mostly amorphous chitin, such as colloidal chitin and swelling chitin after processing in chitin. Therefore, no matter what production method is used, the extraction and post-treatment of chitin are necessary links, and there are many problems, such as high pollution and low yield. Although biosynthesis can alleviate the above problems, it still needs to break through the production cost [22,26]. How to realize the direct enzymatic utilization of crystal chitin and even natural chitin materials, such as shrimp and crab shells, has become a hot issue in the green and efficient utilization of resources in chitin.
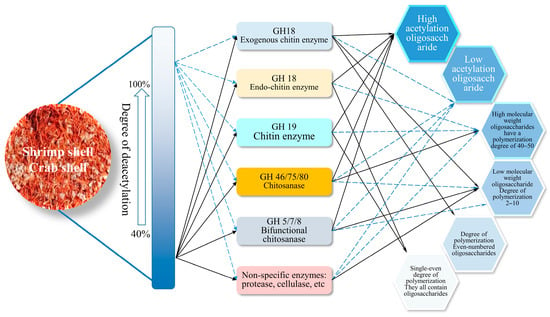
Figure 3.
Theoretical prediction diagram of oligosaccharide preparation with different characteristics by enzymatic method [25].
3. Research History and Classification of LPMO
Lytic polysaccharide monooxygenase (LPMOs) is a kind of copper ion-dependent oxidase, which can cut glycosidic bonds in carbohydrates (cellulose or chitin) by oxidation so that refractory biomass can be depolymerized, and more binding sites of glycoside hydrolases can be exposed, thus synergistically enhancing the hydrolysis of polysaccharides by glycoside hydrolases [2,27,28,29]. The pre-LPMO times can be traced back to 1954 at the earliest [29]. Reese and Gilligan put forward the “C1 factor”, which can target and destroy cellulose crystals when studying the “swelling factor in cellulose hydrolysis” [30]. In 1974, Eriksson and others found that Sporotrichum pulverulentum has unknown oxidative activity, which can lead to the swelling of cotton crystalline cellulose [31]. In the late 1990s, chitin-binding protein (CBP) with a high affinity for chitin was discovered and classified as a carbohydrate-binding module family (CBM33) [32]. Another protein with weak endoglucanase activity (EGIV) is classified into the glycoside hydrolases family (GH61) [33]. Further structural studies show that CBM33 and GH61 are far from other sibling families, but both have conservative metal binding sites [34].
In 2010, Vaaje-Kolstad et al. [35] confirmed that CBP21 can crack glycosidic bonds in β-chitin through redox reaction. Vaaje-Kolstad believed that the discovery of LPMOs not only proved the existence of a new enzyme action mode but also provided a brand-new way for chitin and efficient enzyme transformation of cellulose resources. It marked a turning point in the field of glycan processing enzymes and stimulated researchers’ enthusiasm in this direction. In 2012, GH61 and CBM33 enzymes were uniformly classified as “LPMO” [36]. In 2013, Antyony et al. [37] decided to merge the lignin-degrading enzyme family into the LPMOs family and introduced a new CAZy category, Auxiliary Activity (AA) family. CBM33 was reclassified as an AA10 family, and GH61 was reclassified as an AA9 family. Subsequently, more and more new AA families were discovered, and the substrate spectrum of LPMOs expanded from chitin and cellulose to biological polysaccharides such as xylan, starch and pectin.
LPMOs exist widely in nature and are found in viruses, microorganisms, animals and plants. At present, according to the information on LPMOs in the CAZy database (), LPMOs are divided into eight AA families, which are AA9, AA10, AA11, AA13, AA14, AA15, AA16 and AA17 (Figure 4) [2]. LPMOs of the AA9 family are mainly derived from fungi, and LPMOs of the AA9 family contain several substrate specificities, mainly targeting cellulose, cellulooligosaccharides and hemicellulose [38]. LPMOs of the AA10 family come from a wide range of sources, mainly bacteria, but there are also LPMOs of the AA10 family in a few eukaryotes and viruses [39]. LPMOs of this family can act on chitin or cellulose, or both [27]. LPMOs of the AA11 family are a relatively small family, which is only found in the fungal family (especially ascomycete fungi) at present, and a few LPMOs of the AA11 family have effects on chitin [40,41]. LPMOs of the AA13 family were produced in LPMOs containing starch-acting substrate [42,43]. LPMOs of the AA14 family mainly come from white rot fungi and have oxidative activity on wood fibre [44]. LPMOs of the AA15 family are the first family found in insects, which can act on chitin or cellulose or both [45]. LPMOs of the AA16 family mainly exist in fungi and some oomycetes and have activity on cellulose and a synergistic effect on cellulase to promote cellulose degradation [46]. LPMOs of the AA17 family are the newest LPMOs family, which is only found in oomycetes and can oxidize pectin [47]. LPMOs with chitin activity have been found in bacteria, fungi and insects. Although LCMs have the same Enzyme Commission Number (EC 1.14. 99.53), they are classified into AA10, AA11 and AA15 families in the CAZy database. A large number of studies have shown that the multi-enzyme synergy between LCMs and glycoside hydrolase has important application potential in biorefining, and the research on multi-enzyme degradation systems under the cooperation of LCMs has become a new direction of resource development in chitin.
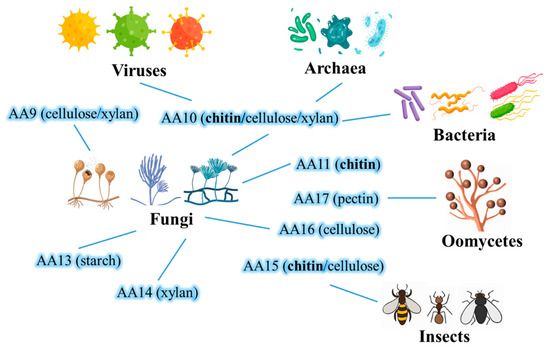
Figure 4.
Distribution and classification of LPMOs.
4. LCMs with Chitin Activity
4.1. Phylogenetic Analysis of LCMs
In order to study the phylogenetic relationship of LCMs, 24 amino acid sequences were selected from AA10, AA11 and AA15 families in the CAZy database for analysis (Figure 5) [8,48]. The minimum evolutionary algorithm is used to analyze the phylogeny of LCMs sequences, and the analysis results are shown in Figure 3. The three families formed distinct clustering branches, and AA10, which mainly originated from bacteria, showed complete differences from AA11 and AA15. AA11 from fungi and AA15 from insects are clustered together, which indicates that the two AA families have sequence similarity. As the largest AA family of LPMOs, AA10 is mainly divided into two branches, clade I and clade II, and each branch has two subclades [49]. Subclades A and subclades B with cellulose-bound CBM (CBM 2 and CBM 3) and cellulose-based activity belong to clade II. The subclades C and subclades D with chitin-binding CBM (CBM 5 and CBM 12) and mainly chitin activity belong to clade II. In terms of species distribution, subclade D mainly contains Firmicutes and Proteobacteria, while the other three subclades mainly come from actinomycetes. LCMs perform similar chitin oxidation processes across a wide range of species and have potential as molecular markers for biological evolution research [8].
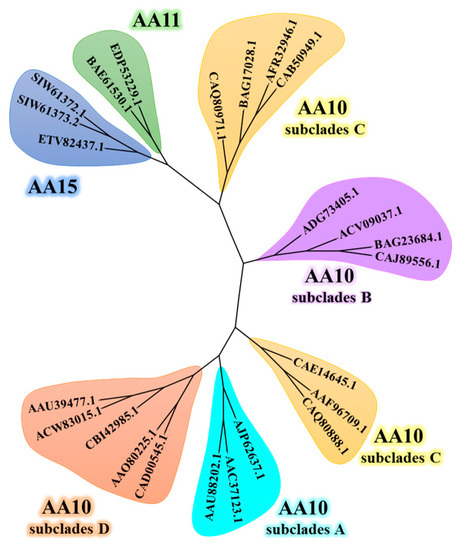
Figure 5.
Phylogenetic tree construction of LCMs.
4.2. Structural Characteristics of LCMs
In total, 27 LCMs from different species and AA families were analyzed in the CAZy database. The modular characteristics of representative LCMs of each family are shown in Figure 6. Generally speaking, LCMs are 90–200 residues in length and have 17–42 residues of signal peptides to facilitate the transport and secretion of LCMs. When LCMs are transported to periplasmic space, the signal peptides will be hydrolyzed and removed. More than 30% of LCMs are composed of multiple catalytic modules, including one or more CBM modules besides catalytic module, and a few LCMs also have GH modules, which play an important role in the activity, stability, substrate affinity and specificity of LCMs [9,50].
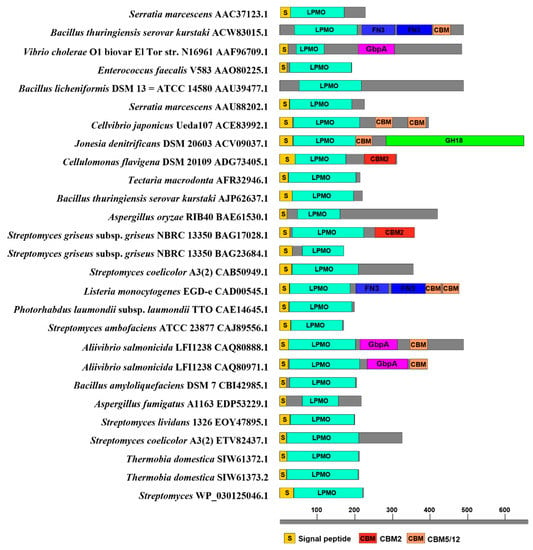
Figure 6.
Signal peptide, catalytic domain and CBM of LCMs.
4.3. LCMs Active Sites and Their Catalytic Mechanism for Chitin
Up to now, all LPMOs have a highly conserved planar structure similar to the β-sandwich folding of immunoglobulin G (IgG) and histidine scaffold [51]. Usually, β-sandwich folding consists of 8–10 reverse parallel β-folds interconnected by α-helices. The histidine scaffold consists of an N-terminal histidine and an internal histidine, and the two histidines coordinate the Cu (II) ion through three nitrogen atoms, forming the copper ion active centre [52]. The main difference between AA9 and AA10 active sites lies in the third non-coordination active site residue, where AA9 is mainly tyrosine, while AA10 family is mainly phenylalanine (Phe) [49]. Multiple extended loops (L2, L3, LS and LC) connecting beta chains have various sequences and topological structures. The changes in these loops, especially the high variability of the L2 region, may have an important relationship with substrate specificity and regioselectivity [53]. Zhou et al. [50] extended loops study on LPMOs with cellulose and chitin activity in the AA10 family shows that more than 70% residues of the motif (Y (W) EPQSVE) are polar amino acids in LCMs, which makes the substrate-binding surface negatively charged. In cellulose-active LPMOs, the proportion of hydrophobic amino acids in the L2 core motif is more than 70%, and its substrate-binding surface is uncharged or positively charged. Therefore, the amino acid residue composition of the L2 core motif determines whether LPMOs are suitable for binding to positively charged chitin substrate or uncharged cellulose substrate.
Since it was found that LPMOs can break the glycosidic bond of chitin by oxidation, its catalytic mechanism has been widely studied. Initially, Vaaje-Kolstad et al. [35] used mass spectrometry and chromatographic separation methods to confirm the activity mechanism of LPMOs in chitin in the absence or existence of H218O or 18O2 and found that one oxygen atom introduced at the end of oxidation chain came from water and the second oxygen atom came from molecular oxygen. They also found that in the presence of reducing agents such as ascorbic acid, the C-H bond of C1 carbon in chitin can be specifically oxidized to C-OH, and then its glycosidic bond can be broken. At the same time, it is revealed that the metal binding sites of LPMOs have considerable structural plasticity. Soon, Phillips et al. [54] concluded that copper is a natural metal of LPMOs with cellulose as a substrate. In the same year, this conclusion was confirmed by Vaaje-Kolstad et al. [55] in their research with chitin as a substrate. Mikael Gudmundsson et al. [56] used the diffraction data collection strategy of minimum deposition X-ray dose to analyze the oxidation mode and crystal structure of chitin-specific LPMO (EfaCBM33A) from Enterococcus faecalis and the structural change reflected the photoreduction of Cu (II) to Cu (I). Based on the reported catalytic characteristics of LPMOs and other catalytic mechanisms of copper monooxygenase, Walton and Davies [57] reviewed the catalytic mechanisms of LPMOs, and proposed two possible catalytic mechanisms of LPMOs: one is that LPMOs bind to O2 before binding to substrate, and dissociate from the substrate at the beginning of catalysis, which may be short-lived, but the premise of this scheme is that the active site must be exposed to the solution, otherwise O2 will not bind to enzyme; another mechanism is that LPMOs first bind to the substrate, but there are active sites between the binding surface of LPMOs and the substrate that can contact O2. Most of the reported studies on the catalytic mechanism of LPOMs are AA9 family or cellulose activity, but their catalytic mechanisms are still controversial. Because the active centre configuration of AA10 family chitin LPMOs is different from AA9 LPMOs (and some cellulose-active AA10 LPMOs), the detailed mechanism of AA10 family chitin LPMOs needs further study [58].
4.4. Types and Functions of CBMs in LCMs
During the enzymatic hydrolysis of biomass, CBMs of degrading enzymes play a role in binding enzymes to substrate [59]. CBMs are helpful in improving the affinity and stability of substrates, and the truncation of these domains will lead to the degradation of enzyme performance [53]. The correct coordination of His and Cu2+ at the N terminal of LCMs determines the activity of LCMs. Therefore, CBM modules are mostly located on the C side of LCMs [37]. CBM has rich structural diversity, which can be divided into three types: A, B and C, according to substrate binding characteristics [60]. The CBM of LCMs is mostly type A CBM, including CBM1, CBM2, CBM5/12, CBM14 and CBM73. The substrate-binding surfaces of CBM2, CBM5, CBM14 and CBM73 are flat and rich in aromatic residues and have a good affinity for insoluble and highly crystalline chitin [61,62,63]. Point mutation, deletion or addition of CBMs will affect the binding of LCMs to substrate and the yield of oxidation products [58,59]. CjLPMO10A (from Cellvibrio japonicus) containing CBM5 and CBM73 decreased its activity to α-chitin after removing CBM module [63].
Madland et al. [64] showed that the affinity of CBM73 to crystalline chitin was higher than that of CBM5. The activity of four-module LCMs (named BCLPMO10A) derived from Bacillus cereus against chitin depends on CBM5, which can promote substrate binding and protect enzymes from inactivation [65]. The results of the research on the multi-modular LPMO from Bacillus thuringiensis serovar kurstaki (BtLPMO10A) show that BtAA10 alone can not bind α-chitin, but only has weak binding to β-chitin and CBM5 can improve the chitin-binding efficiency of BtLPMO10A [66]. The comparison of structure and biochemical characteristics between BtLPMO10A and BtLPMO10B shows that CBM5 at the C terminal of LCMs is the main reason why two enzymes with highly conserved catalytic domains show different substrate preferences [67]. SgLPMO10A containing CBM2 from Streptomyces griseus HUT6037 has activity on cellulose and chitin. CBM2 module deletion studies show that CBM2 plays a role in substrate binding, but excessive CBM2 will inhibit the entry of enzymes and lead to the decrease of products [68]. In addition, many studies have shown that CBM prevents nonproductive reactions between a reduced LPMO and its co-substrate, which could lead to enzyme inactivation by tightly integrating LPMOs with substrates [65,69,70]. At present, the mechanism of CBM enhancing activity in LCMs and the interaction among CBM, linking region, and catalytic domain still need further study.
4.5. Roles of FnIII and Linker in LPMOs
Different modules/domains in multi-module LPMOs are usually connected by flexible linkers, and fibronectin-type III (FnIII)-like domains in some LPMOs also play a role in connection [8]. Different from the core catalytic region with abundant hydrophobic residues, the low-complexity regions (LCRs) are the lower hydrophobic amino acids in the linker. The linker is ubiquitous in cellulase and chitin enzymes. In these carbohydrate-active enzymes, the linker produces a longer carbohydrate binding surface by allowing close contact between the catalytic domain and CBM [71]. By characterizing ScLPMO10C of Streptomyces coelicolor, it is found that the linker is disordered and extended, and the distance between the catalytic domain and CBM allows them to move independently of each other [72]. BcLPMO10A, derived from Bacillus cereus, has an AA10 and a CBM5 connected by two FnIII domains. Through domain deletion and characterization, it was found that the FnIII domain acted as a linker in BtLPMO10A, which neither bound chitin nor assisted the binding of BtLPMO10A to chitin, and the chitin activity of BcLPMO10A strongly depended on CBM5 [66]. To further study the role of the linker in LPMOs, Aishwarya et al. [73] selected BcAA9C, which can be induced by cellulose, among the nine LPMOs contained in Botrytis cinerea genome for further study. The results showed that linker deletion did not affect the catalyzing the oxidation of Amplex Red of BcAA9C but affected the degradation ability of BcAA9C to refractory polysaccharide substrates. In addition, the linker also contributes to the combination of BcAA9C with polysaccharide and its thermal stability. Therefore, the linker is not only a spacer in LPMOs but also has many functions. At present, there are few studies on linker in LCMs, and we speculate that linker plays a similar role in LCMs as in cellulose-active LPMOs. Its possible effects on promoting enzyme activity, substrate binding and thermal stability need further study.
4.6. Multifunctional LCMs
With the further study of LCMs, LCMs with various active functions have been excavated. The multifunction of LCMs is mainly manifested in the degradation ability of various substrates and the superimposed glycoside hydrolase activity. At present, there are three kinds of LCMs with cellulose and chitin activity in the CAZy database. They are KpLPMO10A (WP_030125046.1) from Kitasatospora papulosa DSM41643 and SgLPMO10A (BAG17028.1) from Streptomyces griseus subsp. griseus NBRC 13350 belonging to AA10 family, and TDAA15A from Thermobia domestica (SIW61372.1) belonging to AA15 family. KpLPMO10A has a high similarity with members of AA clade II subclade A, which can release C1 and C4 oxidation products with chitin and cellulose as substrates. In addition, KpLPMO10A also has rare xylan activity (C4 oxidation) [39]. SgLPMO10A is an LPMO with a CBM2 domain have activities on both chitin and cellulose. Quantitative analysis of transcripts showed that transcription was found on glucose, chitin, cellulose and filter paper, and the highest transcription level was found on filter paper except for glucose. Substrate binding and degradation studies show that the weak chitin activity of SgLPMO10A is difficult to boost chitinolytic activity. CBM2 provides a substrate-binding function for SgLPMO10A, but excessive CBM2 will inhibit and slow down the enzyme reaction [68].
Thermobia domestica can decompose and digest crystalline cellulose without the help of microorganisms, and its digestive proteome contains more than 20 LPMOs. Among them, TDAA15A, which has the most abundant content, has been fully characterized. TDAA15A has produced C1 oxidation and C1 aldose acid mode products with microcrystalline cellulose and β-chitin as substrates. TDAA15A has a synergistic effect with cellobiohydrolase (GH6), endoglucanase (GH7), endoglucanase (GH9), β-glucosidase (GH1) and endochitinase (GH18), which indicates that TDAA15A plays an important role in the development and metamorphosis of T. domestica and its digestion of non-symbiotic cellulose. In addition, when a novel AA10 LPMO (BaLPMO10) was found in B. amyloliquefaciens and glycoside hydrolase synergistically degraded microcrystalline cellulose and colloidal chitin, the sugar-reducing content increased by 7% and 23%, respectively. The results of molecular docking and molecular dynamics simulation showed that the distance between BaLPMO10 and cellulose is much longer than that between chitosan and cellulose, and the number of hydrogen bonds between BaLPMO10 and cellulose is also low [74]. It is worth noting that only SgLPMO10A has the CBM module in multi-substrate active LCMs, which further supports the hypothesis that the substrate specificity of LPMOs is mainly determined by the catalytic domain.
The only chitin enzyme (Jd1381) from Jonesia denitrificans contains three domains of AA10, CBM5 and chitin enzyme of GH18 family (Chi18). The enzymatic characterization and crystalline structure of a chitin-active LPMO domain of Jd1381 (JdLPMO10A) showed that JdLPMO10A contains 142 residues, which is the smallest LPMO characterized so far, and it has C1 oxidation activity for α-and β-chitin [75,76]. Sophanit et al. [77] used bioinformatics and biochemical analysis to identify the enzymatic hydrolysis mechanism of Jd1381. Activity analysis of the full-length and truncated versions of JD1381 showed that LPMO and GH18 domains alone showed corresponding chitin activity and had a synergistic effect, which played a decisive role in the degradation efficiency of chitin by JD1381. The enzymatic hydrolysis of α-chitin was not as good as that of Jd1381 alone. The optimum activity of Jd1381 requires that the AA10 and Chi18 domains are close to each other in space. The study of LCMs with multiple active functions provides a theoretical basis for the study of substrate specificity and the synergistic effect between LPMOs and hydrolases and opens up a new way for the development of effective catalysts for the transformation of chitin resources.
5. Activity Determination and Screening of LCMs
5.1. Detection Methods for LCMs
Enzyme activity analysis and determination methods play a decisive role in enzyme screening, enzyme properties characterization and enzyme application. As a monooxygenase targeting insoluble polysaccharides, only a small amount of soluble products (aldose/ketoaldose oligomer) were released after LPMOs were catalyzed, and most of the insoluble oxidation products and substrates remained intact. The low amount of soluble products and insoluble products brought difficulties to qualitative and quantitative analysis. Wang et al. [78] reviewed the screening methods in LPMO function research and summarized the advantages, disadvantages and applicability of different methods. At present, there are many methods to detect LPMO activity, including mass spectrometry, chromatography, labelling and indirect measurement. These methods have different advantages, disadvantages and applicability. Therefore, for the research and application of LPMO, it is very important to choose and use reasonable detection methods [79].
Matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF-MS) is the most widely used method for detecting LPMO activity. The high sensitivity of this method makes it a gold standard for detecting LPMO activity, but it can only be qualitatively analyzed but not quantitatively [79]. In addition, the complex sample preparation process and expensive inspection equipment also limit the application of this method in large-scale enzyme screening and characterization. X-ray photoelectron spectroscopy (XPS), fluorescence labelling and isotope labelling can all detect insoluble substrates, but these methods are cumbersome to operate, need special instruments and require high laboratory conditions, so they are not universal. Both HPAEC-PAD and HPLC methods can be used for qualitative analysis and quantitative analysis with high detection accuracy, but they can only detect soluble products. The nickel ion adsorption method can detect insoluble substrates effectively and can be used for quantitative analysis. It is easy to operate and suitable for various experimental conditions. However, this method requires high purity of samples and relatively low sensitivity. The FRET quenching method is suitable for the detection of LPMO activity in the solution environment. It has a good effect in detecting the interaction between LPMO and other proteins or molecules and the inactivation mechanism of LPMO. However, this method needs to be equipped with corresponding fluorescence lenses, lasers and other equipment, and the setting of experimental conditions is also complicated. Electron paramagnetic response (EPR), viscosity detection and 2,6-dimethoxyphenol (2,6-DMP) are the main indirect methods to measure the changes of other parameters caused by LPMO reaction. Among them, the 2,6-DMP method has been widely used because of its high sensitivity, simple operation, easy reading and no need for complicated instruments and equipment. But it also has some disadvantages, such as limited specificity, being affected by reaction conditions and not being suitable for all LPMO enzymes. When choosing the enzyme activity detection method, it needs to be comprehensively considered according to the specific experimental needs and the characteristics of the research objects.
5.2. LCMs Screening Obtained
Oxidative cracking of chitin by LCMs can directly destroy the crystal structure of chitin and increase the substrate concentration of chitin enzyme, thus reducing the enzymatic hydrolysis resistance of the matrix and promoting the enzymatic hydrolysis of crystal chitin. The enzymatic hydrolysis characteristics of LCMs make them play an irreplaceable role in promoting the transformation of chitin resources, and it has become a new challenge for LCMs research to maximize their commercial application potential. Screening for stable and efficient LCMs is one of the research hotspots. LCMs, as a coenzyme of chitin degradation enzyme, are mostly obtained from the genome of strains with chitin degradation ability through bioinformatics mining (Table 1). CBP21 was found in the Gram-negative soil bacterium Serratia marcescens [80]. CjLPMO10A is derived from the Gram-negative soil bacterium Cellvibrio japonicus [63]. The versatile JdLPMO10A exists in the Gram-positive bacterium Jonesia denitrificans [75]. Li et al. [81] mined NaLPMO10A in Natrialbaceae archaeon by sequence alignment. NaLPMO10A not only shows high thermal stability and pH stability under alkaline conditions but also has high tolerance and stability to high concentrations of metal ions. Identified LPMO from Photorhabdus luminescen called PlAA10 can act on both cellulose and chitin [82]. AoLpmo11 with chitin activity excavated from the Aspergillus oryzae genome is the key LCMs for the establishment of the AA11 family [40]. Federico et al. [45] discovered TdAA15A with cellulose and chitin activity and TdAA15B with chitin activity by studying the secretory proteome binding genome of Thermobia domestica, which are the initial members of the AA15 family. Subsequently, LCMs in insect genomes were excavated one after another [83].

Table 1.
Overview of characterized LCMs.
With the rapid development of omics technology, there is a wider choice for obtaining LCMs. Sivasamy et al. [84] summarize the role of secretomics in identifying enzymes involved in lignocellulose deconstruction, the development of enzyme cocktails and the construction of synthetic microbial consortia for biomass valorization. Several examples of identifying LPMO from secretomics are given. Jiang et al. [85] used metagenomic and genomic analyses to locate the contain at last one AA10 LPMO-encoding gene localized in the chitin degradation cluster in Pseudoalteromonas and discovered a new metabolic model of microbial degradation and utilization of chitin. Genomic and metagenomic analysis shows that this LCMs-dependent chitin oxidation utilization pathway may be ubiquitous in marine Gammaproteobacteria. Recently, Zhang et al. used selective gradient domestication technology and high-throughput sequencing technology to identify a new LPMO (M2822) from the metagenome of domesticated flora [43]. Firstly, soil samples were screened according to soil bacterial species and chitin enzyme biodiversity. Then, chitin powder was used as the substrate for the gradient culture of soil samples to enrich chitin-degrading bacteria. M2822 was excavated by sequencing and analyzing the metagenome of the enriched microbial community. This work is the first time to mine LCM in metagenome enriched by microbial community, and the obtained M2822 shows a good application prospect in efficient COS production.
6. Preparation of LPMOs
At present, a large number of LCMs have been characterized and showed great potential for application, and a large number of enzymes need to be obtained in both enzymatic properties and applied research. Most of the LCMs come from bacteria, and their main heterologous expression hosts are Escherichia coli and Bacillus [86]. The most commonly used E. coli hosts are E. coli BL21 (DE3) and E. coli BL21 Star (DE3). The plasmids used are mainly pET-11a, pET-22b, pET-26b(+), pET-28a(+) and pRSETB. Most LCMs have signal peptides, and LCMs can still be successfully expressed in Escherichia coli after removal of the signal peptide. Although the loss of the signal peptide renders LCMs unable to be secreted into the periplasmic space, it is beneficial to increase the yield, although inclusion bodies are generated at the same time that they impede the bulk acquisition of active protein [87,88]. Therefore, the selection of appropriate signal peptides to improve the efficiency of recombinant LCMs to reach periplasmic space is an important factor to be considered in the heterologous expression of LCMs. Some LCMs can be expressed by using their native signal peptides. However, the use of exogenous signal peptides sometimes improves the expression efficiency of LCMs. Courtade et al. [89] compared five signal peptides and found that the signal peptide of SmAA10A (CBP21) was the best. It can also achieve high expression of various proteins [58]. By comparing 12 different signal peptides with the original signal peptide of SmAA10A, Yang Y et al. found that pelB is the best signal peptide to increase the yield of SmAA10A [90]. Subsequently, AA10 LPMOs from various sources were successfully expressed using pelB [91,92]. The T7 expression system of E. coli has the advantages of mature technology, simple operation and high expression level. However, its disadvantages include protein misfolding and aggregation to form inclusion bodies when expressing specific proteins, and molecular chaperones are needed to help proteins fold correctly [92,93]. In addition, due to E. coli has almost no secretion ability, and the expressed protein usually needs to be purified after cell fragmentation, while other proteins and metabolites in cells further increase the difficulty of purification [93].
Bacillus subtilis has the ability to protein secretion, and LCMs expressed by Bacillus subtilis as a host can be secreted extracellularly, which reduces the steps and costs of isolation and purification [94,95]. The BatLPMO10 with the signal peptide allows stable overexpression without induction in the protease-deficient strain B. subtilis LKS87 [95]. The use of non-native signal peptides will make the expression of LPMO more effective in E. coli or Bacillus [89,90,96]. TfAA10A from Thermobifida fusca was successfully expressed in Synechococcus elongatus [97]. As an expression host, Elongatus has the advantages of rapid growth and simple gene manipulation, and it can use carbon dioxide to grow on cheap inorganic nutrients, so it has great application potential in the heterologous expression of LCMs. The S. coelicolor A3(2) has a powerful LPMOs system, which has four groups with typical chitin-oxidizing LPMOs, two with typical cellulose-active LPMOs, and one with chitin active non-chartered enzymes [98]. Li et al. [99] overexpressed Sclpmo10G in E. coli Rosetta (DE3) and S. coelicolor A3(2), respectively, after confirming that the Sclpmo10G gene was strongly induced by chitin. ScLPMO10G showed chitin oxidation activity in vitro, and synergistic action with chitin enzyme could significantly increase reducing sugar levels. Overexpression of ScLPMO10G in vivo made the strain have stronger chitin degradation ability, and the reducing sugar level produced increased by 2.97 times. The first AA11 family LCMs (AoLpmo11) from Aspergillus oryzae RIB40 were successfully expressed in E. coli BL21 Star (DE3) by pET-26a after codon optimization and adding pelB signal peptide [40]. The single-domain AA11 LPMO from A. fumigatus achieved high expression using Pichia pastoris yeast expression system [100]. TdAA15A and TdAA15B (Thermobia domestica) [45], CgAA15a and CgAA15b (Coptotermes gestroi) [83], AaAA15A (Aphanomyces astaci) [101], which belong to AA15 family, were successfully expressed in E. coli Rosetta 2 (DE3) pLysS. The expression and catalytic activity of OfLPMO15-1 from Ostrinia furnacalis were studied by using pPIC9 plasmid in Pichia pastoris strain GS115, which provided new insight into the enzymatic mechanism of epidermal chitin transformation during insect moulting [102]. Recently, Hernández-Rollán et al. [103] built an open-source lytic polysaccharide monooxygenase golden gate cloning (LyGo) platform. The open-source platform has an easy-to-follow online protocol for cloning and expression, which allows users to combine gene fragments with 14 different expression vectors in a simple 15 min response, thus realizing parallel and rapid exploration of multiple gene backgrounds, hosts and expression strategies.
7. Engineering of LCMs
Different from the degradation of biomass under natural conditions, the conditions used in the industrial biocatalysis process are relatively harsh. If LCMs can play a great role in industrial biocatalysis, a large number of cheap enzymes and excellent enzymatic characteristics are indispensable. To make LCMs become powerful industrial biocatalysts, improving the catalytic activity, stability, temperature and pH tolerance of enzymes and reducing the sensitivity of LCMs to reactive oxygen species have become the main goals of rational engineering transformation of LCMs. Forsberg et al. [70] mutated the active centre residues in the C1/C4-oxidizing LPMO10 from Micromonospora aurantiaca (MaLPMO10B) into corresponding residues in strictly C1 oxidizing LPMO10s, in which W82Y/N85F/Q141W and W82Y/N85F/Y116F/Q141W mutations made MaLPMO10B lose chitin activity, which confirmed the importance of residues on substrate-binding surface in substrate specificity. In addition, W82Y/N85F mutant has better cellulose activity than the wild type, and its specificity has changed from C1/C4 to almost completely C1. Loose et al. [104] used point mutation to study the action of 13 conserved amino acids on the binding surface of SMLPMO10A flat substrate. Point mutations of residues that do not interact with catalytic copper sites but participate in substrate binding have a significant impact on the overall enzymatic properties, especially the stability. The main reason is that point mutation leads to the decrease of chitin-binding level, which increases the oxidation of essential residues in active sites and accelerates the loss of enzyme activity.
Jensen et al. [105] used ScLPMO10C (LPMO of S. coelicolor A3 (2)) to construct a mutation library to transform the chitin activity of LPMOs into cellulose activity. Four mutants (D80/X82/F111/Q141) were screened from the library using a newly developed MS-based high-throughput screening method. These mutants showed a significant decrease in activity and thermal stability and were deactivated faster during the catalytic process, which may be related to autocatalytic oxidative damage caused by weakened substrate binding. Recently, Zhou et al. [106] Mutated CjLPMO10A with chitin activity from Cellvibrio japonicus through a reasonable design of disulfide bond. The thermal stability and resistance to chemical denaturation of the variant M1 (N78C/H116C) were improved. The half-life of M1 at 60 °C increased 3 times, the apparent Tm increased by 7 °C, and the specific activity of M1 was 1.5 times that of the wild type. Therefore, through rational engineering transformation, the stability and activity of LPMO can be improved at the same time, thus improving the industrial adaptability of enzymes, which is of great significance to application. Forsberg et al. [107] summarized the engineering transformation work of LPMO in a summary entitled Engineering lytic polysaccharide monooxygenases (LPMOs).
LPMO reaction requires binding copper ion, reducing power and oxygen co-substrate at the active site of the enzyme, which will produce many possible side reactions, thus increasing the unproductive consumption of reaction components. The engineering modification of LPMO is still in its infancy, and the challenges of engineering LPMO are the complexity of evaluating and quantifying the activities of wild-type enzymes and their engineering variants due to the occurrence of side reactions. The sensitivity of LPMO to substrate concentration and transition metal ions in the reaction mixture makes the engineering of LPMO more difficult. However, the lack of in-depth understanding of the structure-function relationship in LPMOs, especially the characteristic information except for thermal stability, makes rational transformation difficult [108,109]. If the complexity of enzymes can be solved, the construction of an easy-to-use and high-throughput LPMO activity detection method will greatly promote the engineering transformation of LPMOs.
8. Study on the Synergistic Effect of LCMs on Chitin Enzyme and Its Application
Although the chitin enzyme can degrade chitin, it is impossible to degrade crystalline chitin efficiently by using a single pure enzyme. LCMs provide a new way to solve this problem. The multi-enzyme degradation system composed of LCMs, chitin enzyme and chitosanase has high degradation efficiency for crystal chitin. The efficient degradation mode of crystal chitin under the cooperation of multiple enzymes is that LCMs bind to the surface of crystal chitin (and peel them off from the crystal structure by oxidizing chitin chain glycosidic bonds) to reduce the crystallinity near the oxidation site and providing sufficient substrates for other chitin degrading enzymes (ChiA and ChiB). ChiA and ChiB hydrolyze chitin at the non-reducing end and the reducing end, respectively, to produce oligosaccharides, and chitosanase converts oligosaccharides into monosaccharides [28]. So far, no impediment of GHs with LPMO action on chitin has been observed [110].
Although LCMs have not been commercialized at present, LCMs have great potential for development and application because they can promote the degradation of chitin, especially crystal chitin, by chitin enzyme (Table 2). LPMOs with chitin activity (CBP21) are the first LPMOs to be studied [80]. Under the synergistic effect of CBP21, the time required for ChiA and ChiC to completely degrade chitin is reduced by about 7 times, and the yield of chitobiose increased from 127 to 162 μM by the synergistic action of chiABC and CBP21 [111]. The BCP21 can increase the initial reaction rate of Serratia marcescens chiA and chiB by 6 and 9 times [112]. Four different LPMOs (SgLPMO10B, -C, -D, and -F) from S. griseus all showed promotive effects on chitin degradation by three chitin enzymes (smChiA, -B, -C) of S. marcescens [113,114]. The chitin degradation system of Aeromonas salmonicida showed that it contained two chitin enzymes of GH18 and GH19, respectively, and LCM of the AA10 family (AsLPMO10A). When AsLPMO10A cooperated with all chitin enzymes to treat powdered chitin, the degradation efficiency increased by more than two times [114]. A novel LCM (NaLPMO10A) from Natrialbaceae archaeon can not only significantly improve the degradation efficiency of chitin but also show good stability under alkaline and high concentration of metal ions. Therefore, archaeal NaLPMO10A could be a promising enzyme for improving saccharification under extreme conditions [81]. A novel AA10 LPMO derived from B. subtilis (BsLPMO10A) has a wide substrate spectrum, and its synergistic effect with corresponding GHs can significantly increase the reducing sugar yield of dextran, xylan, cellulose and chitin as substrates [115]. Ma et al. [116] using TgAA11 of Trichoderma guizhouense NJAU 4742, the degradation efficiency of chitinase (Sg-chi from Streptomyces griseus) for α-and β-chitin was improved by 39.9% and 288.2%, respectively. M2822 and chitin-degrading enzymes secreted by Chitiniphilus sp. were found in the degrading and enriching flora in chitin. LZ32 could efficiently hydrolyze shrimp shell powder. After 12 h of enzymatic hydrolysis, COS yield reached 4724 μg/mL [117].

Table 2.
Synergistic effects of LCMs and chitinase on the degradation of chitin.
9. Current Challenges and Future Perspective
The discovery and research of LPMOs have opened up a new world for the large-scale development and utilization of poorly degradable polysaccharides biomass. Although LPMOs have made exciting progress in the utilization of cellulose resources, the development and utilization of LCMs resources in chitin is still in its infancy, and its industrial application still faces many challenges. For example, the catalytic efficiency and stability of enzymes are low under severe conditions such as high temperature and extreme pH, the output of LCMs is not suitable for actual biorefining, and the synergistic ratio of LCMs and other chitin enzymes. However, the unclear catalytic mechanism of LCMs makes it difficult to solve these problems. Fortunately, with the efforts of many scientists, LPMOs with cellulose activity have made gratifying progress in three-dimensional structure analysis, catalytic mechanism and multi-enzyme synergy, as well as improving enzyme activity and reducing application cost [106,107,118]. This provides a good reference for the research and application of LCMs. Therefore, future research on LCMs should focus on solving the key problems and challenges such as catalytic mechanism analysis, high activity and strong stability transformation, high expression and low-cost preparation. For example, more LCMs with excellent performance can be obtained by mining metagenomes of specific ecological environment samples. The interaction and electron transfer between the residues and substrates near the active sites of LCMs were further analyzed by quantitative calculation and molecular dynamics simulation. The catalytic efficiency and thermal stability of LCMs were improved by amino acid mutation. Construct a suitable expression system and regulate transcription, translation and secretion systematically, to realize efficient heterologous expression of LCMs. In-depth study on the synergistic mechanism of LPMO and chitin-degrading enzymes, and construction of complex enzyme mixture for chitin degradation, so that it is possible to achieve efficient and low-cost production of chitin derivatives.
Recently, Vandhana et al. [29] summarized the role of LPMO and LPMO-like proteins in fungal and oomycete plant pathogenesis (e.g., potato late blight) and mutualism/commensal symbiosis (e.g., ectomycorrhizae) and further reviewed the potential importance of LPMO in diseases caused by bacteria (e.g., pneumonia), fungi (e.g., human meningitis), oomycetes and viruses (e.g., entomopox). Jagadeeswaran et al. [119] summarized the role of LPMOs in the occurrence of plant diseases and insect pests from the perspective of plant defence. Loose et al. [120] proved that Melolontha melolontha entomopoxvirus fusolin protein released from spindles is chitin-degrading LPMOs. The fusolin is still active after being stored at 4 °C for 10 years, and it can withstand high temperatures and oxidative stress in the crystalline state. This characteristic of being able to tolerate extreme environments makes fusolin have great application potential in insect disease control. The above research has opened up a new research idea for LCMs in the control of plant diseases and insect pests. Therefore, LCMS have great potential in chitin resources and plant pest control. Novel LCMS still need to be explored, and the catalytic mechanism needs to be deeply investigated. Through rational modification of LCMS to solve the problems such as poor stability and cost of use of LCMS in practical applications. By solving these problems and challenges, we can promote the high-value and resource application of chitin resources in the development of the bioenergy and bio-refining industry.
Author Contributions
Conceptualization, Y.Z. and X.Z.; methodology, Y.Z., D.P. and J.L.; software, Y.Z., D.P., Y.X. and J.L.; formal analysis, Y.Z., D.P., Y.X. and J.L.; investigation, J.L., Y.X., D.P. and P.X.; resources, X.Z.; data curation, Y.Z.; writing—original draft preparation, D.P. and Y.Z.; writing—review and editing, D.P. and X.Z.; visualization, Y.Z. and X.Z.; supervision, X.Z.; project administration, Y.Z.; funding acquisition, X.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Grant No. 32100065), the Natural Science Foundation of Shandong Province of China (No. ZR2023MB095) and the Doctoral Research Startup Foundation of Liaocheng University (No. 318052041).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cho, E.J.; Trinh, L.T.P.; Song, Y.; Lee, Y.G.; Bae, H.-J. Bioconversion of Biomass Waste into High Value Chemicals. Bioresour. Technol. 2020, 298, 122386. [Google Scholar] [CrossRef]
- Guo, X.; An, Y.; Liu, F.; Lu, F.; Wang, B. Lytic Polysaccharide Monooxygenase—A New Driving Force for Lignocellulosic Biomass Degradation. Bioresour. Technol. 2022, 362, 127803. [Google Scholar] [CrossRef]
- Mohan, K.; Ganesan, A.R.; Muralisankar, T.; Jayakumar, R.; Sathishkumar, P.; Uthayakumar, V.; Chandirasekar, R.; Revathi, N. Recent Insights into the Extraction, Characterization, and Bioactivities of Chitin and Chitosan from Insects. Trends Food Sci. Technol. 2020, 105, 17–42. [Google Scholar] [CrossRef]
- Shi, X.; Ye, X.; Zhong, H.; Wang, T.; Jin, F. Sustainable Nitrogen-Containing Chemicals and Materials from Natural Marine Resources Chitin and Microalgae. Mol. Catal. 2021, 505, 111517. [Google Scholar] [CrossRef]
- Zhou, D.; Shen, D.; Lu, W.; Song, T.; Wang, M.; Feng, H.; Shentu, J.; Long, Y. Production of 5-Hydroxymethylfurfural from Chitin Biomass: A Review. Molecules 2020, 25, 541. [Google Scholar] [CrossRef]
- Kamal, M.; Adly, E.; Alharbi, S.A.; Khaled, A.S.; Rady, M.H.; Ibrahim, N.A. Exploring Simplified Methods for Insect Chitin Extraction and Application as a Potential Alternative Bioethanol Resource. Insects 2020, 11, 788. [Google Scholar] [CrossRef]
- Liaqat, F.; Eltem, R. Chitooligosaccharides and Their Biological Activities: A Comprehensive Review. Carbohydr. Polym. 2018, 184, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Jhariya, U.; Purohit, H.J.; Dafale, N.A. Synergistic Action of Lytic Polysaccharide Monooxygenase with Gly-coside Hydrolase for Lignocellulosic Waste Valorization: A Review. Biomass Conv. Bioref. 2021, 13, 8272–8745. [Google Scholar] [CrossRef]
- Yu, X.; Zhao, Y.; Yu, J.; Wang, L. Recent Advances in the Efficient Degradation of Lignocellulosic Metabolic Networks by Lytic Polysaccharide Monooxygenase. Acta Biochim. Biophys. Sin. 2023, 55, 529. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Chen, X. Don’t Waste Seafood Waste. Nature 2015, 524, 155–157. [Google Scholar] [CrossRef] [PubMed]
- Rovaletti, A.; De Gioia, L.; Fantucci, P.; Greco, C.; Vertemara, J.; Zampella, G.; Arrigoni, F.; Bertini, L. Recent Theoretical Insights into the Oxidative Degradation of Biopolymers and Plastics by Metalloenzymes. Int. J. Mol. Sci. 2023, 24, 6368. [Google Scholar] [CrossRef] [PubMed]
- Merzendorfer, H. Insect Chitin Synthases: A Review. J. Comp. Physiol. B 2005, 176, 1–15. [Google Scholar] [CrossRef]
- Santos, V.P.; Marques, N.S.S.; Maia, P.C.S.V.; de Lima, M.A.B.; de Oliveira Franco, L.; de Campos-Takaki, G.M. Seafood Waste as Attractive Source of Chitin and Chitosan Production and Their Applications. Int. J. Mol. Sci. 2020, 21, 4290. [Google Scholar] [CrossRef] [PubMed]
- Arnold, N.D.; Brück, W.M.; Garbe, D.; Brück, T.B. Enzymatic Modification of Native Chitin and Conversion to Specialty Chemical Products. Mar. Drug 2020, 18, 93. [Google Scholar] [CrossRef]
- Liu, Y.; Qin, Z.; Wang, C.; Jiang, Z. N-Acetyl-d-Glucosamine-Based Oligosaccharides from Chitin: Enzymatic Production, Characterization and Biological Activities. Carbohydr. Polym. 2023, 315, 121019. [Google Scholar] [CrossRef]
- Wani, A.K.; Akhtar, N.; Mir, T.u.G.; Rahayu, F.; Suhara, C.; Anjli, A.; Chopra, C.; Singh, R.; Prakash, A.; El Messaoudi, N.; et al. Eco-Friendly and Safe Alternatives for the Valorization of Shrimp Farming Waste. Environ. Sci. Pollut. Res. 2023, 30, 1–30. [Google Scholar] [CrossRef]
- Cao, S.; Liu, Y.; Shi, L.; Zhu, W.; Wang, H. N-Acetylglucosamine as a Platform Chemical Produced from Renewable Resources: Opportunity, Challenge, and Future Prospects. Green Chem. 2022, 24, 493–509. [Google Scholar] [CrossRef]
- Anil, S. Potential Medical Applications of Chitooligosaccharides. Polymers 2022, 14, 3558. [Google Scholar] [CrossRef]
- Kumar, M.; Brar, A.; Vivekanand, V.; Pareek, N. Bioconversion of Chitin to Bioactive Chitooligosaccharides: Amelioration and Coastal Pollution Reduction by Microbial Resources. Mar. Biotechnol. 2018, 20, 269–281. [Google Scholar] [CrossRef]
- Gonçalves, C.; Ferreira, N.; Lourenço, L. Production of Low Molecular Weight Chitosan and Chitooligosaccharides (COS): A Review. Polymers 2021, 13, 2466. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Mo, X.; Zhou, N.; Wang, Y.; Wei, G.; Hao, Z.; Chen, K. Identification of Chitinolytic Enzymes in Chitinolyticbacter Meiyuanensis and Mechanism of Efficiently Hydrolyzing Chitin to N-Acetyl Glucosamine. Front. Microbiol. 2020, 11, 572053. [Google Scholar] [CrossRef]
- Ling, M.; Li, J.; Du, G.; Liu, L. Metabolic Engineering for the Production of Chitooligosaccharides: Advances and Perspectives. Emerg. Top. Life Sci. 2018, 2, 377–388. [Google Scholar] [CrossRef]
- Sharma, A.; Arya, S.K.; Singh, J.; Kapoor, B.; Bhatti, J.S.; Suttee, A.; Singh, G. Prospects of Chitinase in Sustainable Farming and Modern Biotechnology: An Update on Recent Progress and Challenges. Biotechnol. Genet. Eng. Rev. 2023, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Chakdar, H.; Pandiyan, K.; Thapa, S.; Shahid, M.; Singh, A.; Srivastava, A.K.; Saxena, A.K. Bacterial Chitinases: Genetics, Engineering and Applications. World J. Microbiol. Biotechnol. 2022, 38, 252. [Google Scholar] [CrossRef]
- Aktuganov, G.E.; Melent’ev, A.I. Specific Features of Chitosan Depolymerization by Chitinases, Chitosanases, and Nonspe-cific Enzymes in the Production of Bioactive Chitooligosaccharides (Review). Appl. Biochem. Microbiol. 2017, 53, 611–627. [Google Scholar] [CrossRef]
- Ahuja, V.; Bhatt, A.; Sharma, V.; Rathour, R.; Rana, N.; Bhatia, R.; Varjani, S.; Kumar, M.; Magdouli, S.; Yang, Y.-H.; et al. Advances in Glucosamine Production from Waste Biomass and Microbial Fermentation Technology and Its Applications. Biomass Convers. Biorefinery 2021, 1–23. [Google Scholar] [CrossRef]
- Hemsworth, G.R.; Johnston, E.M.; Davies, G.J.; Walton, P.H. Lytic Polysaccharide Monooxygenases in Biomass Conversion. Trends Biotechnol. 2015, 33, 747–761. [Google Scholar] [CrossRef] [PubMed]
- Courtade, G.; Aachmann, F.L. Chitin-active lytic polysaccharide monooxygenases. In Targeting Chitin-Containing Organisms; Springer: Singapore, 2019; pp. 115–129. [Google Scholar]
- Vandhana, T.M.; Reyre, J.; Sushmaa, D.; Berrin, J.; Bissaro, B.; Madhuprakash, J. On the Expansion of Biological Functions of Lytic Polysaccharide Monooxygenases. New Phytol. 2022, 233, 2380–2396. [Google Scholar] [CrossRef]
- Reese, E.T.; Gilligan, W. The Swelling Factor in Cellulose Hydrolysis. Text. Res. J. 1954, 24, 663–669. [Google Scholar] [CrossRef]
- Eriksson, K.-E.; Pettersson, B.; Westermark, U. Oxidation: An Important Enzyme Reaction in Fungal Degradation of Cellulose. FEBS Lett. 1974, 49, 282–285. [Google Scholar] [CrossRef]
- Sunna, A.; Gibbs, M.D.; Bergquist, P.L. A Novel Thermostable Multidomain 1,4-β-Xylanase from ‘Caldibacillus Cellulovorans’ and Effect of Its Xylan-Binding Domain on Enzyme Activity. Microbiology 2000, 146, 2947–2955. [Google Scholar] [CrossRef] [PubMed]
- Saloheimo, M.; Nakari-SetaLa, T.; Tenkanen, M.; Penttila, M. CDNA Cloning of a Trichoderma Reesei Cellulase and Demonstration of Endoglucanase Activity by Expression in Yeast. Eur. J. Biochem. 1997, 249, 584–591. [Google Scholar] [CrossRef]
- Harris, P.V.; Welner, D.; McFarland, K.C.; Re, E.; Navarro Poulsen, J.-C.; Brown, K.; Salbo, R.; Ding, H.; Vlasenko, E.; Merino, S.; et al. Stimulation of Lignocellulosic Biomass Hydrolysis by Proteins of Glycoside Hydrolase Family 61: Structure and Function of a Large, Enigmatic Family. Biochemistry 2010, 49, 3305–3316. [Google Scholar] [CrossRef]
- Vaaje-Kolstad, G.; Westereng, B.; Horn, S.J.; Liu, Z.; Zhai, H.; Sørlie, M.; Eijsink, V.G.H. An Oxidative Enzyme Boosting the Enzymatic Conversion of Recalcitrant Polysaccharides. Science 2010, 330, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Horn, S.; Vaaje-Kolstad, G.; Westereng, B.; Eijsink, V.G. Novel Enzymes for the Degradation of Cellulose. Biotechnol. Biofuels 2012, 5, 45. [Google Scholar] [CrossRef]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the Enzymatic Repertoire of the CAZy Database to Integrate Auxiliary Redox Enzymes. Biotechnol. Biofuels 2013, 6, 41. [Google Scholar] [CrossRef]
- Long, L.; Hu, Y.; Sun, F.; Gao, W.; Hao, Z.; Yin, H. Advances in Lytic Polysaccharide Monooxygenases with the Cellu-lose-Degrading Auxiliary Activity Family 9 to Facilitate Cellulose Degradation for Biorefinery. Int. J. Biol. Macromol. 2022, 219, 68–83. [Google Scholar] [CrossRef] [PubMed]
- Corrêa, T.L.R.; Júnior, A.T.; Wolf, L.D.; Buckeridge, M.S.; dos Santos, L.V.; Murakami, M.T. An Actinobacteria Lytic Poly-saccharide Monooxygenase Acts on Both Cellulose and Xylan to Boost Biomass Saccharification. Biotechnol. Biofuels 2019, 12, 117. [Google Scholar] [CrossRef]
- Hemsworth, G.R.; Henrissat, B.; Davies, G.J.; Walton, P.H. Discovery and Characterization of a New Family of Lytic Poly-saccharide Monooxygenases. Nat. Chem. Biol. 2013, 10, 122–126. [Google Scholar] [CrossRef]
- Wang, D.; Li, J.; Wong, A.C.Y.; Aachmann, F.L.; Hsieh, Y.S.Y. A Colorimetric Assay to Rapidly Determine the Activities of Lytic Polysaccharide Monooxygenases. Biotechnol. Biofuels 2018, 11, 215. [Google Scholar] [CrossRef] [PubMed]
- Vu, V.V.; Beeson, W.T.; Span, E.A.; Farquhar, E.R.; Marletta, M.A. A Family of Starch-Active Polysaccharide Monooxygen-ases. Proc. Natl. Acad. Sci. USA 2014, 111, 13822–13827. [Google Scholar] [CrossRef] [PubMed]
- Haddad Momeni, M.; Leth, M.L.; Sternberg, C.; Schoof, E.; Nielsen, M.W.; Holck, J.; Workman, C.T.; Hoof, J.B.; Abou Hachem, M. Loss of AA13 LPMOs Impairs Degradation of Resistant Starch and Reduces the Growth of Aspergillus Nidulans. Biotechnol. Biofuels 2020, 13, 135. [Google Scholar] [CrossRef] [PubMed]
- Couturier, M.; Ladevèze, S.; Sulzenbacher, G.; Ciano, L.; Fanuel, M.; Moreau, C.; Villares, A.; Cathala, B.; Chaspoul, F.; Frandsen, K.E.; et al. Lytic Xylan Oxidases from Wood-Decay Fungi Unlock Biomass Degradation. Nat. Chem. Biol. 2018, 14, 306–310. [Google Scholar] [CrossRef]
- Sabbadin, F.; Hemsworth, G.R.; Ciano, L.; Henrissat, B.; Dupree, P.; Tryfona, T.; Marques, R.D.S.; Sweeney, S.T.; Besser, K.; Elias, L.; et al. An Ancient Family of Lytic Polysaccharide Monooxygenases with Roles in Arthropod Development and Bio-mass Digestion. Nat. Commun. 2018, 9, 756. [Google Scholar] [CrossRef]
- Filiatrault-Chastel, C.; Navarro, D.; Haon, M.; Grisel, S.; Herpoël-Gimbert, I.; Chevret, D.; Fanuel, M.; Henrissat, B.; Heiss-Blanquet, S.; Margeot, A.; et al. AA16, a New Lytic Polysaccharide Monooxygenase Family Identified in Fungal Se-cretomes. Biotechnol. Biofuels 2019, 12, 55. [Google Scholar] [CrossRef]
- Sabbadin, F.; Urresti, S.; Henrissat, B.; Avrova, A.O.; Welsh, L.R.J.; Lindley, P.J.; Csukai, M.; Squires, J.N.; Walton, P.H.; Davies, G.J.; et al. Secreted Pectin Monooxygenases Drive Plant Infection by Pathogenic Oomycetes. Science 2021, 373, 774–779. [Google Scholar] [CrossRef]
- Nakagawa, Y.S.; Kudo, M.; Loose, J.S.M.; Ishikawa, T.; Totani, K.; Eijsink, V.G.H.; Vaaje-Kolstad, G. A Small Lytic Polysac-charide Monooxygenase FromStreptomyces Griseustargeting α- and β-Chitin. FEBS J. 2015, 282, 1065–1079. [Google Scholar] [CrossRef]
- Book, A.J.; Yennamalli, R.M.; Takasuka, T.E.; Currie, C.R.; Phillips, G.N.; Fox, B.G. Evolution of Substrate Specificity in Bac-terial AA10 Lytic Polysaccharide Monooxygenases. Biotechnol. Biofuels 2014, 7, 109. [Google Scholar] [CrossRef]
- Zhou, X.; Qi, X.; Huang, H.; Zhu, H. Sequence and Structural Analysis of AA9 and AA10 LPMOs: An Insight into the Basis of Substrate Specificity and Regioselectivity. Int. J. Mol. Sci. 2019, 20, 4594. [Google Scholar] [CrossRef]
- Frandsen, K.E.H.; Lo Leggio, L. Lytic Polysaccharide Monooxygenases: A Crystallographer’s View on a New Class of Bio-mass-Degrading Enzymes. Int. Union Crystallogr. J. 2016, 3, 448–467. [Google Scholar] [CrossRef] [PubMed]
- Tandrup, T.; Frandsen, K.E.H.; Johansen, K.S.; Berrin, J.-G.; Lo Leggio, L. Recent Insights into Lytic Polysaccharide Monooxygenases (LPMOs). Biochem. Soc. Trans. 2018, 46, 1431–1447. [Google Scholar] [CrossRef] [PubMed]
- Vaaje-Kolstad, G.; Forsberg, Z.; Loose, J.S.; Bissaro, B.; Eijsink, V.G. Structural Diversity of Lytic Polysaccharide Monooxy-genases. Curr. Opin. Struct. Biol. 2017, 44, 67–76. [Google Scholar] [CrossRef]
- Phillips, C.M.; Beeson, W.T.; Cate, J.H.; Marletta, M.A. Cellobiose Dehydrogenase and a Copper-Dependent Polysaccharide Monooxygenase Potentiate Cellulose Degradation by Neurospora Crassa. ACS Chem. Biol. 2011, 6, 1399–1406. [Google Scholar] [CrossRef]
- Vaaje-Kolstad, G.; Bøhle, L.A.; Gåseidnes, S.; Dalhus, B.; Bjørås, M.; Mathiesen, G.; Eijsink, V.G.H. Characterization of the Chitinolytic Machinery of Enterococcus Faecalis V583 and High-Resolution Structure of Its Oxidative CBM33 Enzyme. J. Mol. Biol. 2012, 416, 239–254. [Google Scholar] [CrossRef] [PubMed]
- Gudmundsson, M.; Kim, S.; Wu, M.; Ishida, T.; Momeni, M.H.; Vaaje-Kolstad, G.; Lundberg, D.; Royant, A.; Ståhlberg, J.; Eijsink, V.G.H.; et al. Structural and Electronic Snapshots during the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photoreduction. J. Biol. Chem. 2014, 289, 18782–18792. [Google Scholar] [CrossRef] [PubMed]
- Walton, P.H.; Davies, G.J. On the Catalytic Mechanisms of Lytic Polysaccharide Monooxygenases. Curr. Opin. Chem. Biol. 2016, 31, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, Z.; Røhr, Å.K.; Mekasha, S.; Andersson, K.K.; Eijsink, V.G.H.; Vaaje-Kolstad, G.; Sørlie, M. Comparative Study of Two Chitin-Active and Two Cellulose-Active AA10-Type Lytic Polysaccharide Monooxygenases. Biochemistry 2014, 53, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, Z.; Courtade, G. On the Impact of Carbohydrate-Binding Modules (CBMs) in Lytic Polysaccharide Monooxygenases (LPMOs). Essays Biochem. 2023, 67, 561–574. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, H.J.; Knox, J.P.; Boraston, A.B. Advances in Understanding the Molecular Basis of Plant Cell Wall Polysaccharide Recognition by Carbohydrate-Binding Modules. Curr. Opin. Struct. Biol. 2013, 23, 669–677. [Google Scholar] [CrossRef]
- Crasson, O.; Courtade, G.; Léonard, R.R.; Aachmann, F.L.; Legrand, F.; Parente, R.; Baurain, D.; Galleni, M.; Sørlie, M.; Vandevenne, M. Human Chitotriosidase: Catalytic Domain or Carbohydrate Binding Module, Who’s Leading HCHT’s Bio-logical Function. Sci. Rep. 2017, 7, 2768. [Google Scholar] [CrossRef]
- Boraston, A.B.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Carbohydrate-Binding Modules: Fine-Tuning Polysaccharide Recognition. Biochem. J. 2004, 382, 769–781. [Google Scholar] [CrossRef]
- Forsberg, Z.; Nelson, C.E.; Dalhus, B.; Mekasha, S.; Loose, J.S.M.; Crouch, L.I.; Røhr, Å.K.; Gardner, J.G.; Eijsink, V.G.H.; Vaaje-Kolstad, G. Structural and Functional Analysis of a Lytic Polysaccharide Monooxygenase Important for Efficient Uti-lization of Chitin in Cellvibrio japonicus. J. Biol. Chem. 2016, 291, 7300–7312. [Google Scholar] [CrossRef]
- Madland, E.; Forsberg, Z.; Wang, Y.; Lindorff-Larsen, K.; Niebisch, A.; Modregger, J.; Eijsink, V.G.H.; Aachmann, F.L.; Courtade, G. Structural and Functional Variation of Chitin-Binding Domains of a Lytic Polysaccharide Monooxygenase from Cellvibrio japonicus. J. Biol. Chem. 2021, 297, 101084. [Google Scholar] [CrossRef]
- Mutahir, Z.; Mekasha, S.; Loose, J.S.M.; Abbas, F.; Vaaje-Kolstad, G.; Eijsink, V.G.H.; Forsberg, Z. Characterization and Synergistic Action of a Tetra-modular Lytic Polysaccharide Monooxygenase from Bacillus cereus. FEBS Lett. 2018, 592, 2562–2571. [Google Scholar] [CrossRef] [PubMed]
- Manjeet, K.; Madhuprakash, J.; Mormann, M.; Moerschbacher, B.M.; Podile, A.R. A Carbohydrate Binding Module-5 Is Es-sential for Oxidative Cleavage of Chitin by a Multi-Modular Lytic Polysaccharide Monooxygenase from Bacillus thuringiensis Serovar Kurstaki. Int. J. Biol. Macromol. 2019, 127, 649–656. [Google Scholar] [CrossRef]
- Zhang, H.; Zhou, H.; Zhao, Y.; Li, T.; Yin, H. Comparative Studies of Two AA10 Family Lytic Polysaccharide Monooxygenases from Bacillus thuringiensis. PeerJ 2023, 11, e14670. [Google Scholar] [CrossRef]
- Sato, K.; Chiba, D.; Yoshida, S.; Takahashi, M.; Totani, K.; Shida, Y.; Ogasawara, W.; Nakagawa, Y.S. Functional Analysis of a Novel Lytic Polysaccharide Monooxygenase from Streptomyces Griseus on Cellulose and Chitin. Int. J. Biol. Macromol. 2020, 164, 2085–2091. [Google Scholar] [CrossRef] [PubMed]
- Bissaro, B.; Røhr, Å.K.; Müller, G.; Chylenski, P.; Skaugen, M.; Forsberg, Z.; Horn, S.J.; Vaaje-Kolstad, G.; Eijsink, V.G.H. Oxidative Cleavage of Polysaccharides by Monocopper Enzymes Depends on H2O2. Nat. Chem. Biol. 2017, 13, 1123–1128. [Google Scholar] [CrossRef]
- Forsberg, Z.; Bissaro, B.; Gullesen, J.; Dalhus, B.; Vaaje-Kolstad, G.; Eijsink, V.G.H. Structural Determinants of Bacterial Lytic Polysaccharide Monooxygenase Functionality. J. Biol. Chem. 2018, 293, 1397–1412. [Google Scholar] [CrossRef] [PubMed]
- Meier, K.K.; Jones, S.M.; Kaper, T.; Hansson, H.; Koetsier, M.J.; Karkehabadi, S.; Solomon, E.I.; Sandgren, M.; Kelemen, B. Oxygen Activation by Cu LPMOs in Recalcitrant Carbohydrate Polysaccharide Conversion to Monomer Sugars. Chem. Rev. 2017, 118, 2593–2635. [Google Scholar] [CrossRef]
- Courtade, G.; Forsberg, Z.; Heggset, E.B.; Eijsink, V.G.H.; Aachmann, F.L. The Carbohydrate-Binding Module and Linker of a Modular Lytic Polysaccharide Monooxygenase Promote Localized Cellulose Oxidation. J. Biol. Chem. 2018, 293, 13006–13015. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Nagar, P.; Rathore, S.; Adlakha, N. The Linker Region Promotes Activity and Binding Efficiency of Modular LPMO towards Polymeric Substrate. Microbiol. Spectr. 2022, 10, 1. [Google Scholar] [CrossRef]
- Guo, X.; An, Y.; Jiang, L.; Zhang, J.; Lu, F.; Liu, F. The Discovery and Enzymatic Characterization of a Novel AA10 LPMO from Bacillus amyloliquefaciens with Dual Substrate Specificity. Int. J. Biol. Macromol. 2022, 203, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Mekasha, S.; Forsberg, Z.; Dalhus, B.; Bacik, J.-P.; Choudhary, S.; Schmidt-Dannert, C.; Vaaje-Kolstad, G.; Eijsink, V.G.H. Structural and Functional Characterization of a Small Chitin-Active Lytic Polysaccharide Monooxygenase Domain of a Multi-Modular Chitinase FromJonesia Denitrificans. FEBS Lett. 2015, 590, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Bacik, J.-P.; Mekasha, S.; Forsberg, Z.; Kovalevsky, A.Y.; Vaaje-Kolstad, G.; Eijsink, V.G.H.; Nix, J.C.; Coates, L.; Cuneo, M.J.; Unkefer, C.J.; et al. Neutron and Atomic Resolution X-Ray Structures of a Lytic Polysaccharide Monooxygenase Reveal Copper-Mediated Dioxygen Binding and Evidence for N-Terminal Deprotonation. Biochemistry 2017, 56, 2529–2532. [Google Scholar] [CrossRef]
- Mekasha, S.; Tuveng, T.R.; Askarian, F.; Choudhary, S.; Schmidt-Dannert, C.; Niebisch, A.; Modregger, J.; Vaaje-Kolstad, G.; Eijsink, V.G.H. A Trimodular Bacterial Enzyme Combining Hydrolytic Activity with Oxidative Glycosidic Bond Cleavage Efficiently Degrades Chitin. J. Biol. Chem. 2020, 295, 9134–9146. [Google Scholar] [CrossRef]
- Wang, D.; Li, Y.; Zheng, Y.; Hsieh, Y.S.Y. Recent Advances in Screening Methods for the Functional Investigation of Lytic Polysaccharide Monooxygenases. Front. Chem. 2021, 9, 653754. [Google Scholar] [CrossRef]
- Westereng, B.; Arntzen, M.Ø.; Østby, H.; Agger, J.W.; Vaaje-Kolstad, G.; Eijsink, V.G.H. Analyzing activities of lytic poly-saccharide monooxygenases by liquid chromatography and mass spectrometry. In Protein-Carbohydrate Interactions: Methods and Protocols; Springer: New York, NY, USA, 2023; pp. 27–51. [Google Scholar]
- Vaaje-Kolstad, G.; Horn, S.J.; van Aalten, D.M.F.; Synstad, B.; Eijsink, V.G.H. The Non-Catalytic Chitin-Binding Protein CBP21 from Serratia marcescens Is Essential for Chitin Degradation. J. Biol. Chem. 2005, 280, 28492–28497. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Liu, Y.; Liu, Y.; Li, Y.; Yu, H. Heterologous Expression and Characterization of a Novel Lytic Polysaccharide Monooxygenase from Natrialbaceae Archaeon and Its Application for Chitin Biodegradation. Bioresour. Technol. 2022, 354, 127174. [Google Scholar] [CrossRef]
- Munzone, A.; El Kerdi, B.; Fanuel, M.; Rogniaux, H.; Ropartz, D.; Réglier, M.; Royant, A.; Simaan, A.J.; Decroos, C. Char-acterization of a Bacterial Copper-dependent Lytic Polysaccharide Monooxygenase with an Unusual Second Coordination Sphere. FEBS J. 2020, 287, 3298–3314. [Google Scholar] [CrossRef] [PubMed]
- Franco Cairo, J.P.L.; Cannella, D.; Oliveira, L.C.; Gonçalves, T.A.; Rubio, M.V.; Terrasan, C.R.F.; Tramontina, R.; Mofatto, L.S.; Carazzolle, M.F.; Garcia, W.; et al. On the Roles of AA15 Lytic Polysaccharide Monooxygenases Derived from the Termite Coptotermes gestroi. J. Inorg. Biochem. 2021, 216, 111316. [Google Scholar] [CrossRef]
- Sethupathy, S.; Morales, G.M.; Li, Y.; Wang, Y.; Jiang, J.; Sun, J.; Zhu, D. Harnessing Microbial Wealth for Lignocellulose Biomass Valorization through Secretomics: A Review. Biotechnol. Biofuels 2021, 14, 154. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.-X.; Li, P.-Y.; Chen, X.-L.; Zhang, Y.-S.; Wang, J.-P.; Wang, Y.-J.; Sheng, Q.; Sun, Z.-Z.; Qin, Q.-L.; Ren, X.-B.; et al. A Pathway for Chitin Oxidation in Marine Bacteria. Nat. Commun. 2022, 13, 5889. [Google Scholar] [CrossRef] [PubMed]
- Song, X.-F.; Feng, C. Lytic polysaccharide monooxygenase and its application. Acta Microbiol. Sin. 2023, 63, 2534–2551. [Google Scholar] [CrossRef]
- Gregory, R.C.; Hemsworth, G.R.; Turkenburg, J.P.; Hart, S.J.; Walton, P.H.; Davies, G.J. Activity, Stability and 3-D Structure of the Cu(<scp>ii</Scp>) Form of a Chitin-Active Lytic Polysaccharide Monooxygenase from Bacillus amyloliquefaciens. Dalton Trans. 2016, 45, 16904–16912. [Google Scholar] [CrossRef]
- Ghatge, S.S.; Telke, A.A.; Waghmode, T.R.; Lee, Y.; Lee, K.-W.; Oh, D.-B.; Shin, H.-D.; Kim, S.-W. Multifunctional Cellulolytic Auxiliary Activity Protein HcAA10-2 from Hahella Chejuensis Enhances Enzymatic Hydrolysis of Crystalline Cellulose. Appl. Microbiol. Biotechnol. 2014, 99, 3041–3055. [Google Scholar] [CrossRef]
- Courtade, G.; Le, S.B.; Sætrom, G.I.; Brautaset, T.; Aachmann, F.L. A Novel Expression System for Lytic Polysaccharide Monooxygenases. Carbohydr. Res. 2017, 448, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, J.; Liu, X.; Pan, X.; Hou, J.; Ran, C.; Zhou, Z. Improving Extracellular Production of Serratia marcescens Lytic Polysaccharide Monooxygenase CBP21 and Aeromonas Veronii B565 Chitinase Chi92 in Escherichia coli and Their Syner-gism. AMB Expr. 2017, 7, 170. [Google Scholar] [CrossRef] [PubMed]
- Chaplin, A.K.; Wilson, M.T.; Hough, M.A.; Svistunenko, D.A.; Hemsworth, G.R.; Walton, P.H.; Vijgenboom, E.; Worrall, J.A.R. Heterogeneity in the Histidine-Brace Copper Coordination Sphere in Auxiliary Activity Family 10 (AA10) Lytic Poly-saccharide Monooxygenases. J. Biol. Chem. 2016, 291, 12838–12850. [Google Scholar] [CrossRef]
- Fowler, C.A.; Sabbadin, F.; Ciano, L.; Hemsworth, G.R.; Elias, L.; Bruce, N.; McQueen-Mason, S.; Davies, G.J.; Walton, P.H. Discovery, Activity and Characterisation of an AA10 Lytic Polysaccharide Oxygenase from the Shipworm Symbiont Teredinibacter Turnerae. Biotechnol. Biofuels 2019, 12, 232. [Google Scholar] [CrossRef] [PubMed]
- Gaber, Y.; Rashad, B.; Hussein, R.; Abdelgawad, M.; Ali, N.S.; Dishisha, T.; Várnai, A. Heterologous Expression of Lytic Polysaccharide Monooxygenases (LPMOs). Biotechnol. Adv. 2020, 43, 107583. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.-J.; Yoon, S.-H.; Kim, Y.-W. Overproduction and Characterization of a Lytic Polysaccharide Monooxygenase in Bacillus Subtilis Using an Assay Based on Ascorbate Consumption. Enzym. Microb. Technol. 2016, 93–94, 150–156. [Google Scholar] [CrossRef]
- Schallmey, M.; Singh, A.; Ward, O.P. Developments in the Use of Bacillus Species for Industrial Production. Can. J. Microbiol. 2004, 50, 1–17. [Google Scholar] [CrossRef]
- Guo, X.; Chai, C.; An, Y.; Peng, C.; Shi, N.; Wang, W.; Lu, F.; Dai, Y.; Liu, F. Rational Design of Signal Peptides for Improved MtC1LPMO Production in Bacillus amyloliquefaciens. Int. J. Biol. Macromol. 2021, 175, 262–269. [Google Scholar] [CrossRef]
- Russo, D.A.; Zedler, J.A.Z.; Wittmann, D.N.; Möllers, B.; Singh, R.K.; Batth, T.S.; van Oort, B.; Olsen, J.V.; Bjerrum, M.J.; Jensen, P.E. Expression and Secretion of a Lytic Polysaccharide Monooxygenase by a Fast-Growing Cyanobacterium. Biotechnol. Biofuels 2019, 12, 74. [Google Scholar] [CrossRef]
- Votvik, A.K.; Røhr, Å.K.; Bissaro, B.; Stepnov, A.A.; Sørlie, M.; Eijsink, V.G.H.; Forsberg, Z. Structural and Functional Characterization of the Catalytic Domain of a Cell-Wall Anchored Bacterial Lytic Polysaccharide Monooxygenase from Streptomyces coelicolor. Sci. Rep. 2023, 13, 5145. [Google Scholar] [CrossRef]
- Li, F.; Zhao, H.; Liu, Y.; Zhang, J.; Yu, H. Chitin Biodegradation by Lytic Polysaccharide Monooxygenases from Streptomyces coelicolor In Vitro and In Vivo. Int. J. Mol. Sci. 2022, 24, 275. [Google Scholar] [CrossRef] [PubMed]
- Støpamo, F.G.; Røhr, Å.K.; Mekasha, S.; Petrović, D.M.; Várnai, A.; Eijsink, V.G.H. Characterization of a Lytic Polysaccharide Monooxygenase from Aspergillus fumigatus Shows Functional Variation among Family AA11 Fungal LPMOs. J. Biol. Chem. 2021, 297, 101421. [Google Scholar] [CrossRef]
- Sabbadin, F.; Henrissat, B.; Bruce, N.C.; McQueen-Mason, S.J. Lytic Polysaccharide Monooxygenases as Chitin-Specific Vir-ulence Factors in Crayfish Plague. Biomolecules 2021, 11, 1180. [Google Scholar] [CrossRef]
- Qu, M.; Guo, X.; Tian, S.; Yang, Q.; Kim, M.; Mun, S.; Noh, M.Y.; Kramer, K.J.; Muthukrishnan, S.; Arakane, Y. AA15 Lytic Polysaccharide Monooxygenase Is Required for Efficient Chitinous Cuticle Turnover during Insect Molting. Commun. Biol. 2022, 5, 518. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Rollán, C.; Falkenberg, K.B.; Rennig, M.; Bertelsen, A.B.; Ipsen, J.Ø.; Brander, S.; Daley, D.O.; Johansen, K.S.; Nørholm, M.H.H. LyGo: A Platform for Rapid Screening of Lytic Polysaccharide Monooxygenase Production. ACS Synth. Biol. 2021, 10, 897–906. [Google Scholar] [CrossRef] [PubMed]
- Loose, J.S.M.; Arntzen, M.Ø.; Bissaro, B.; Ludwig, R.; Eijsink, V.G.H.; Vaaje-Kolstad, G. Multipoint Precision Binding of Substrate Protects Lytic Polysaccharide Monooxygenases from Self-Destructive Off-Pathway Processes. Biochemistry 2018, 57, 4114–4124. [Google Scholar] [CrossRef]
- Jensen, M.S.; Klinkenberg, G.; Bissaro, B.; Chylenski, P.; Vaaje-Kolstad, G.; Kvitvang, H.F.; Nærdal, G.K.; Sletta, H.; Forsberg, Z.; Eijsink, V.G.H. Engineering Chitinolytic Activity into a Cellulose-Active Lytic Polysaccharide Monooxygenase Provides Insights into Substrate Specificity. J. Biol. Chem. 2019, 294, 19349–19364. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Xu, Z.; Li, Y.; He, J.; Zhu, H. Improvement of the Stability and Activity of an LPMO Through Rational Disulfide Bonds Design. Front. Bioeng. Biotechnol. 2022, 9, 815990. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, Z.; Stepnov, A.A.; Nærdal, G.K.; Klinkenberg, G.; Eijsink, V.G.H. Engineering Lytic Polysaccharide Monooxygenases (LPMOs). In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2020; pp. 1–34. [Google Scholar]
- Lo Leggio, L.; Weihe, C.D.; Poulsen, J.-C.N.; Sweeney, M.; Rasmussen, F.; Lin, J.; De Maria, L.; Wogulis, M. Structure of a Lytic Polysaccharide Monooxygenase from Aspergillus fumigatus and an Engineered Thermostable Variant. Carbohydr. Res. 2018, 469, 55–59. [Google Scholar] [CrossRef]
- Tanghe, M.; Danneels, B.; Last, M.; Beerens, K.; Stals, I.; Desmet, T. Disulfide Bridges as Essential Elements for the Thermo-stability of Lytic Polysaccharide Monooxygenase LPMO10C from Streptomyces coelicolor. Protein Eng. Des. Sel. 2017, 30, 401–408. [Google Scholar] [CrossRef]
- Sørlie, M.; Keller, M.B.; Westh, P. The Interplay between Lytic Polysaccharide Monooxygenases and Glycoside Hydrolases. Essays Biochem. 2023, 67, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Suzuki, M.; Taiyoji, M.; Nikaidou, N.; Watanabe, T. Chitin Binding Protein (CBP21) in the Culture Supernatant of Serratia marcescens 2170. Biosci. Biotechnol. Biochem. 1998, 62, 128–135. [Google Scholar] [CrossRef]
- Hamre, A.G.; Eide, K.B.; Wold, H.H.; Sørlie, M. Activation of Enzymatic Chitin Degradation by a Lytic Polysaccharide Monooxygenase. Carbohydr. Res. 2015, 407, 166–169. [Google Scholar] [CrossRef]
- Nakagawa, Y.S.; Kudo, M.; Onodera, R.; Ang, L.Z.P.; Watanabe, T.; Totani, K.; Eijsink, V.G.; Vaaje-Kolstad, G. Analysis of Four Chitin-Active Lytic Polysaccharide Monooxygenases from Streptomyces Griseus Reveals Functional Varia-tion. J. Agric. Food Chem. 2020, 68, 13641–13650. [Google Scholar] [CrossRef]
- Pentekhina, I.; Hattori, T.; Tran, D.M.; Shima, M.; Watanabe, T.; Sugimoto, H.; Suzuki, K. Chitinase System of Aeromonas Salmonicida, and Characterization of Enzymes Involved in Chitin Degradation. Biosci. Biotechnol. Biochem. 2020, 84, 1936–1947. [Google Scholar] [CrossRef]
- Sun, X.-B.; Gao, D.-Y.; Cao, J.-W.; Liu, Y.; Rong, Z.-T.; Wang, J.-K.; Wang, Q. BsLPMO10A from Bacillus Subtilis Boosts the Depolymerization of Diverse Polysaccharides Linked via β-1,4-Glycosidic Bonds. Int. J. Biological Macromol. 2023, 230, 123133. [Google Scholar] [CrossRef]
- Ma, L.; Liu, Z.; Kong, Z.; Wang, M.; Li, T.; Zhu, H.; Wan, Q.; Liu, D.; Shen, Q. Functional Characterization of a Novel Copper-Dependent Lytic Polysaccharide Monooxygenase TgAA11 from Trichoderma Guizhouense NJAU 4742 in the Oxi-dative Degradation of Chitin. Carbohydr. Polym. 2021, 258, 117708. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Pan, D.; Xiao, P.; Xu, Q.; Geng, F.; Zhang, X.; Zhou, X.; Xu, H. A Novel Lytic Polysaccharide Monooxygenase from Enrichment Microbiota and Its Application for Shrimp Shell Powder Biodegradation. Front. Microbiol. 2023, 14, 1097492. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Haider, J.; Liu, P.; Yang, J.; Tan, Z.; Huang, T.; Lin, J.; Jiang, M.; Liu, H.; Zhu, L. Engineered LPMO Significantly Boosting Cellulase-Catalyzed Depolymerization of Cellulose. J. Agric. Food Chem. 2020, 68, 15257–15266. [Google Scholar] [CrossRef] [PubMed]
- Jagadeeswaran, G.; Veale, L.; Mort, A.J. Do Lytic Polysaccharide Monooxygenases Aid in Plant Pathogenesis and Herbivory? Trends Plant Sci. 2020, 26, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Loose, J.S.M.; Boudes, M.; Bergoin, M.; Coulibaly, F.; Vaaje-Kolstad, G. The Melolontha Melolontha Entomopoxvirus Fusolin Protein Is a Chitin-active Lytic Polysaccharide Monooxygenase That Displays Extreme Stability. FEBS Lett. 2023, 597, 1375–1383. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).