Feasibility Study of Biohydrogen Production from Acid Cheese Whey via Lactate-Driven Dark Fermentation
Abstract
1. Introduction
2. Materials and Methods
2.1. Feedstock
2.2. Inoculum
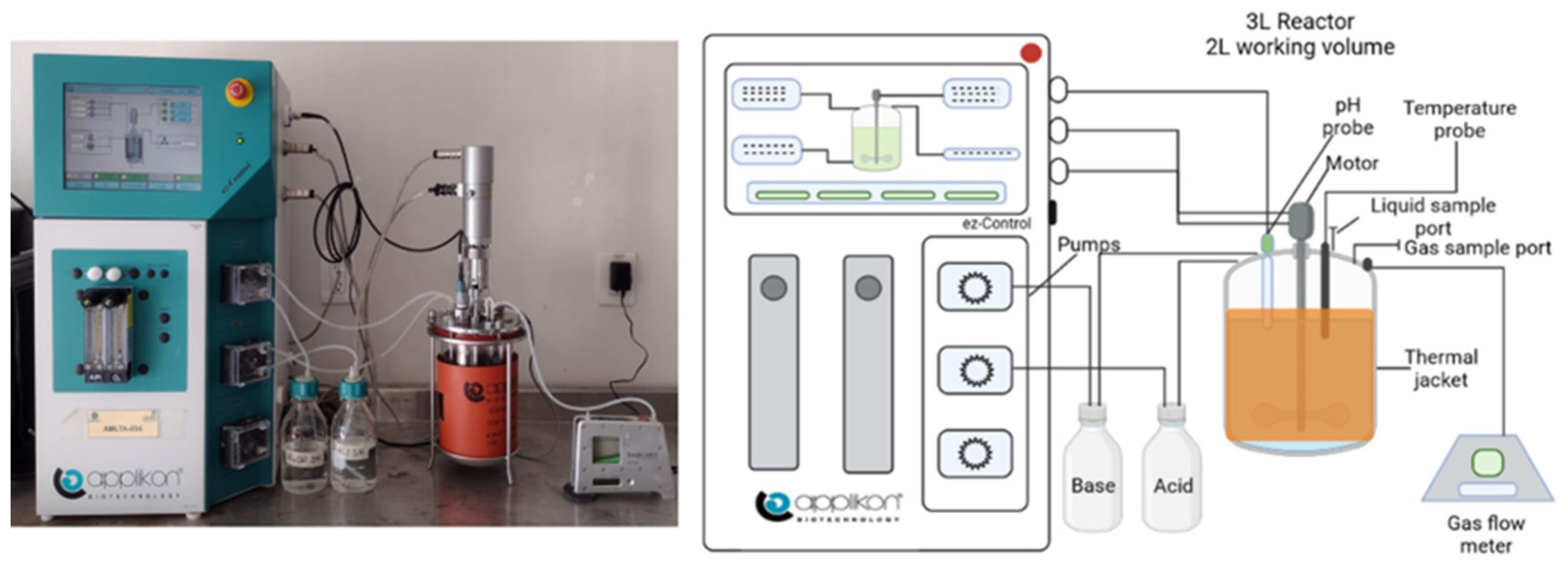
2.3. Experimental Setup and Fermentation Conditions
2.4. Analytical Methods
2.5. Microbial Community Analysis
3. Results and Discussion
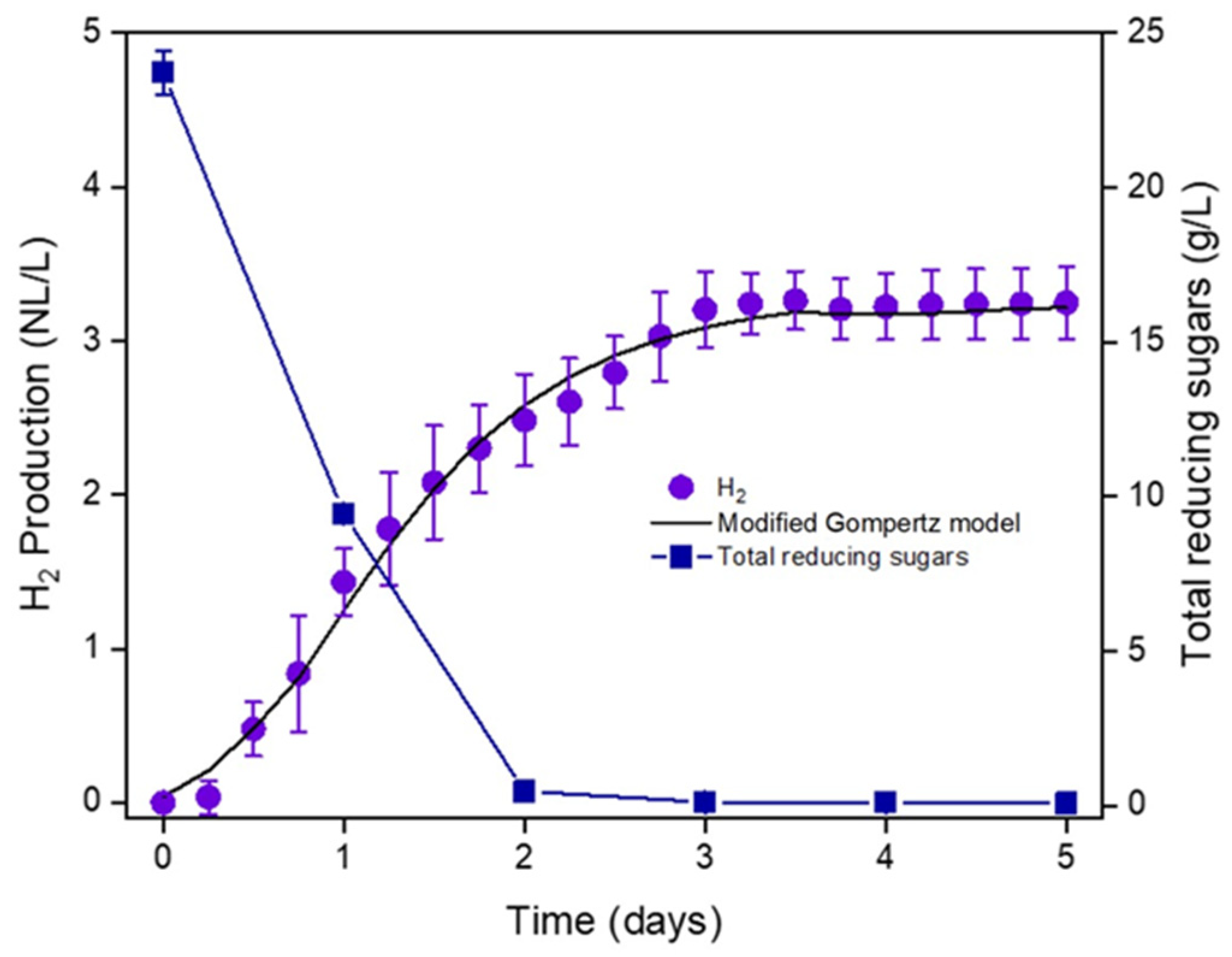
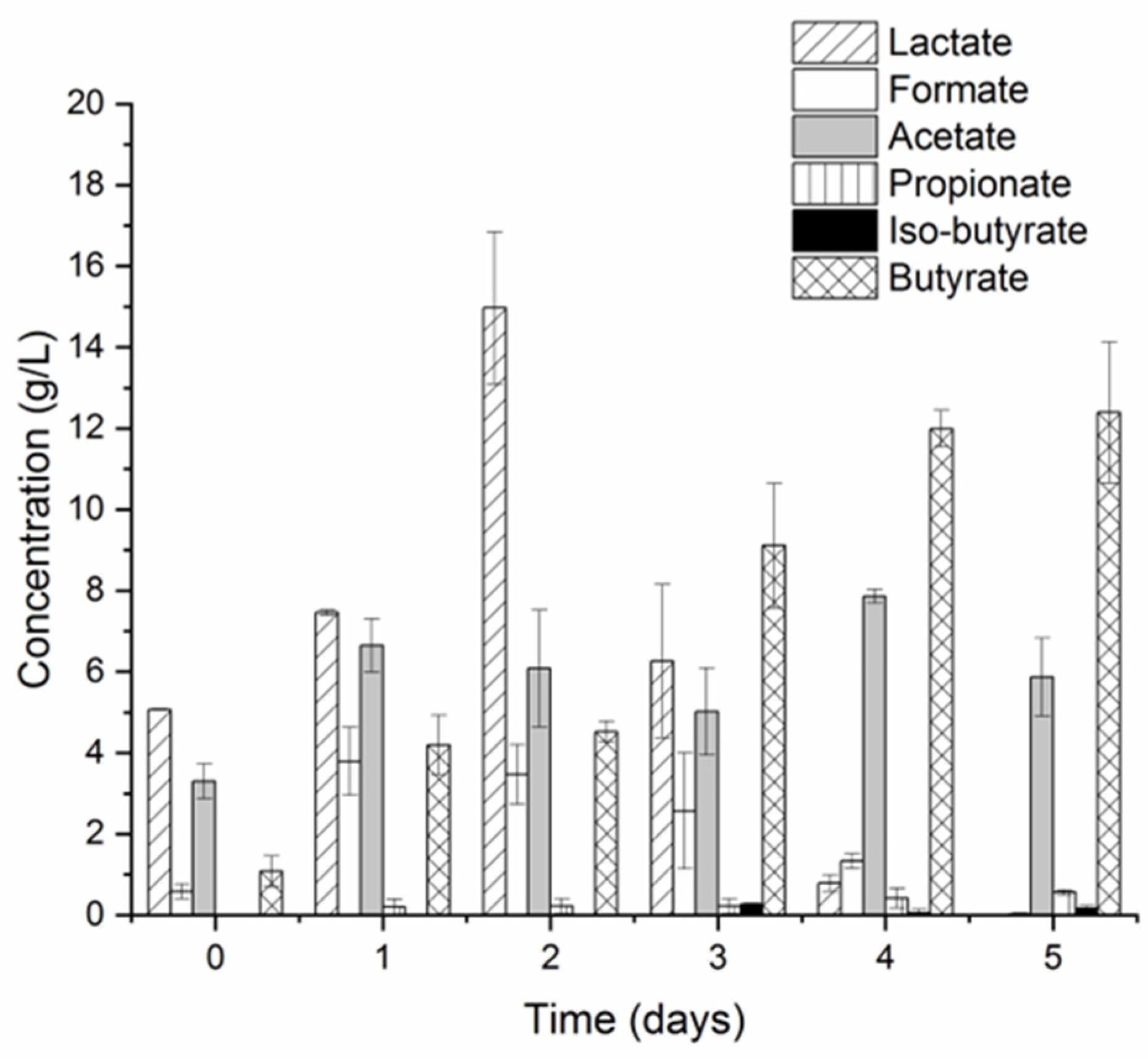
3.1. Undiluted CW Can Support Hydrogen Production via LD-DF
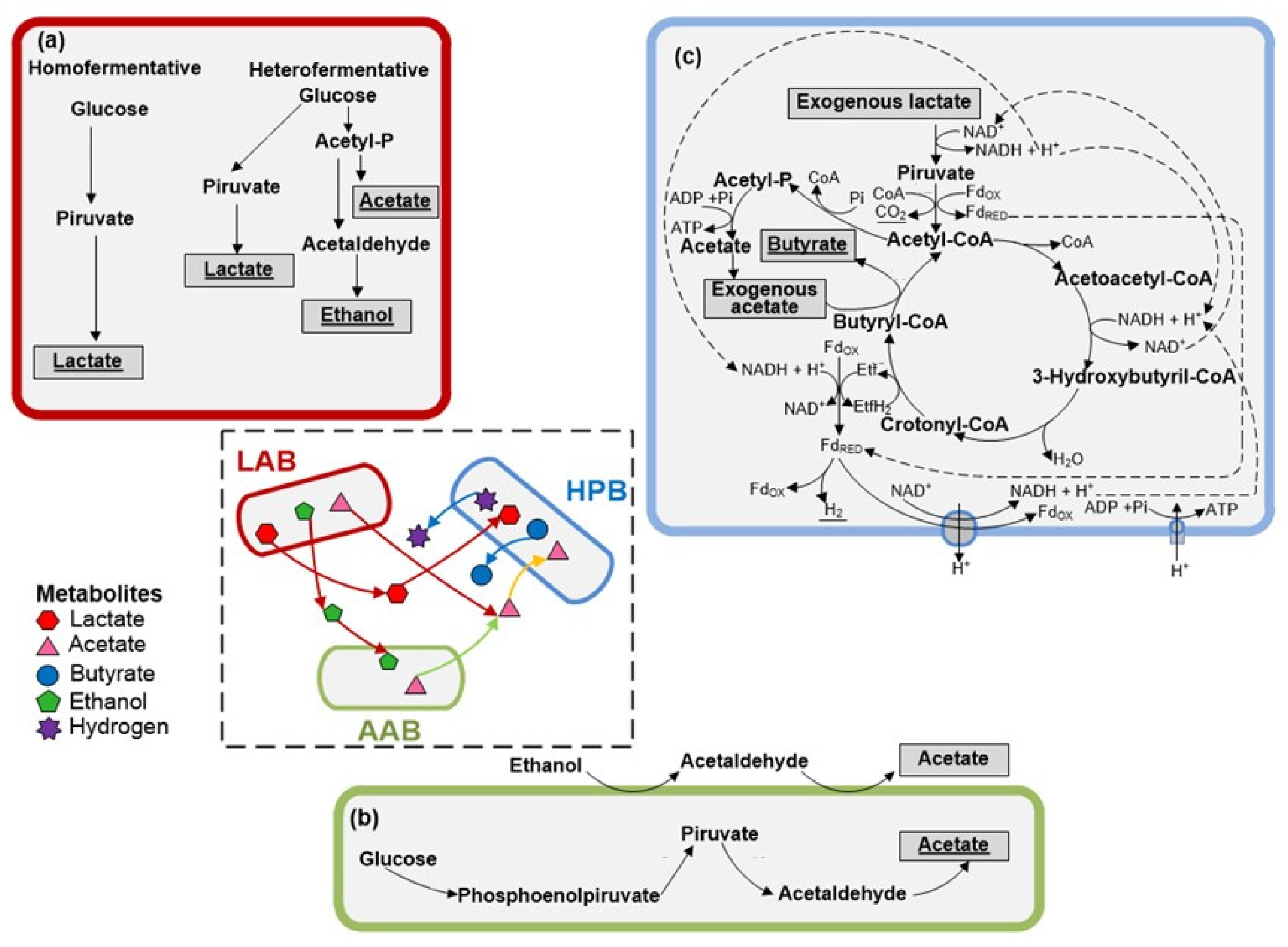
3.2. Microbial Communities and Metabolic Pathways
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prazeres, A.R.; Carvalho, F.; Rivas, J. Cheese whey management: A review. J. Environ. Manag. 2012, 110, 48–68. [Google Scholar] [CrossRef] [PubMed]
- Zotta, T.; Solieri, L.; Iacumin, L.; Picozzi, C.; Gullo, M. Valorization of cheese whey using microbial fermentations. Appl. Microbiol. Biotechnol. 2020, 104, 2749–2764. [Google Scholar] [CrossRef] [PubMed]
- Goyal, C.; Dhyani, P.; Rai, D.C.; Tyagi, S.; Dhull, S.B.; Sadh, P.K.; Duhan, J.S.; Saharan, B.S. Emerging Trends and Advancements in the Processing of Dairy Whey for Sustainable Biorefining. J. Food Process. Preserv. 2023, 2023, 6626513. [Google Scholar] [CrossRef]
- Osorio-González, C.S.; Gómez-Falcon, N.; Brar, S.K.; Ramírez, A.A. Cheese whey as a potential feedstock for producing renewable biofuels: A Review. Energies 2022, 15, 6828. [Google Scholar] [CrossRef]
- Monsalve-Atencio, R.; Sanchez-Soto, K.; Chica, J.; Echavarría, J.A.C.; Vega-Castro, O. Interaction between phospholipase and transglutaminase in the production of semi-soft fresh cheese and its effect on the yield, composition, microstructure and textural properties. LWT 2022, 154, 112722. [Google Scholar] [CrossRef]
- Asunis, F.; De Gioannis, G.; Dessì, P.; Isipato, M.; Lens, P.N.; Muntoni, A.; Polettini, A.; Pomi, R.; Rossi, A.; Spiga, D. The dairy biorefinery: Integrating treatment processes for cheese whey valorisation. J. Environ. Manag. 2020, 276, 111240. [Google Scholar] [CrossRef]
- Karadag, D.; Köroğlu, O.E.; Özkaya, B.; Cakmakci, M.; Heaven, S.; Banks, C.; Koroglu, E.O. A review on fermentative hydrogen production from dairy industry wastewater. J. Chem. Technol. Biotechnol. 2014, 89, 1627–1636. [Google Scholar] [CrossRef]
- Moreno, R.; Escapa, A.; Cara, J.; Carracedo, B.; Gómez, X. A two-stage process for hydrogen production from cheese whey: Integration of dark fermentation and biocatalyzed electrolysis. Int. J. Hydrogen Energy 2015, 40, 168–175. [Google Scholar] [CrossRef]
- Ahmad, T.; Aadil, R.M.; Ahmed, H.; Rahman, U.U.; Soares, B.C.; Souza, S.L.; Pimentel, T.C.; Scudino, H.; Guimarães, J.T.; Esmerino, E.A.; et al. Treatment and utilization of dairy industrial waste: A review. Trends Food Sci. Technol. 2019, 88, 361–372. [Google Scholar] [CrossRef]
- Dessì, P.; Asunis, F.; Ravishankar, H.; Cocco, F.G.; De Gioannis, G.; Muntoni, A.; Lens, P.N. Fermentative hydrogen production from cheese whey with in-line, concentration gradient-driven butyric acid extraction. Int. J. Hydrogen Energy 2020, 45, 24453–24466. [Google Scholar] [CrossRef]
- Ubando, A.T.; Chen, W.-H.; Hurt, D.A.; Conversion, A.; Rajendran, S.; Lin, S.-L. Biohydrogen in a circular bioeconomy: A critical review. Bioresour. Technol. 2022, 366, 128168. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, R.; Menon, R.K. An overview of industrial uses of hydrogen. Int. J. Hydrogen Energy 1998, 23, 593–598. [Google Scholar] [CrossRef]
- Chezeau, B.; Danican, A.; Fontaine, J.-P.; Vial, C. Characterization of the local hydromechanical stress through experimental and numerical analysis of hydrodynamics under dark fermentation operating conditions. Chem. Eng. J. 2020, 382, 122748. [Google Scholar] [CrossRef]
- Hallenbeck, P.C.; Ghosh, D. Advances in fermentative biohydrogen production: The way forward? Trends Biotechnol. 2009, 27, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Show, K.-Y.; Lee, D.-J.; Chang, J.-S. Bioreactor and process design for biohydrogen production. Bioresour. Technol. 2011, 102, 8524–8533. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.M.; Trably, E.; Latrille, E.; Carrère, H.; Steyer, J.-P. Hydrogen production from agricultural waste by dark fermentation: A review. Int. J. Hydrogen Energy 2010, 35, 10660–10673. [Google Scholar] [CrossRef]
- Ntaikou, I.; Antonopoulou, G.; Lyberatos, G. Biohydrogen Production from Biomass and Wastes via Dark Fermentation: A Review. Waste Biomass-Valorization 2010, 1, 21–39. [Google Scholar] [CrossRef]
- Martinez-Burgos, W.J.; do Nascimento Junior, J.R.; Medeiros, A.B.P.; Herrmann, L.W.; Sydney, E.B.; Soccol, C.R. Biohydrogen Production from Agro-industrial Wastes Using Clostridium beijerinckii and Isolated Bacteria as Inoculum. BioEnergy Res. 2021, 15, 987–997. [Google Scholar] [CrossRef]
- Asunis, F.; Carucci, A.; De Gioannis, G.; Farru, G.; Muntoni, A.; Polettini, A.; Pomi, R.; Rossi, A.; Spiga, D. Combined biohydrogen and polyhydroxyalkanoates production from sheep cheese whey by a mixed microbial culture. J. Environ. Manag. 2022, 322, 116149. [Google Scholar] [CrossRef]
- Polettini, A.; Pomi, R.; Rossi, A.; Zonfa, T.; De Gioannis, G.; Muntoni, A. Continuous fermentative hydrogen production from cheese whey—New insights into process stability. Int. J. Hydrogen Energy 2022, 47, 21044–21059. [Google Scholar] [CrossRef]
- Polettini, A.; Pomi, R.; Rossi, A.; Zonfa, T.; De Gioannis, G.; Muntoni, A. Factor-based assessment of continuous bio-H2 production from cheese whey. Chemosphere 2022, 308, 136174. [Google Scholar] [CrossRef]
- Litti, Y.; Khuraseva, N.; Vishnyakova, A.; Zhuravleva, E.; Kovalev, A.; Kovalev, D.; Panchenko, V.; Parshina, S. Comparative study on biohydrogen production by newly isolated Clostridium butyricum SP4 and Clostridium beijerinckii SP6. Int. J. Hydrogen Energy, 2023; in press. [Google Scholar] [CrossRef]
- Castelló, E.; Perna, V.; Wenzel, J.; Borzacconi, L.; Etchebehere, C. Microbial community composition and reactor performance during hydrogen production in a UASB reactor fed with raw cheese whey inoculated with compost. Water Sci. Technol. 2011, 64, 2265–2273. [Google Scholar] [CrossRef]
- Castelló, E.; Braga, L.; Fuentes, L.; Etchebehere, C. Possible causes for the instability in the H2 production from cheese whey in a CSTR. Int. J. Hydrogen Energy 2018, 43, 2654–2665. [Google Scholar] [CrossRef]
- Gomes, B.C.; Rosa, P.R.F.; Etchebehere, C.; Silva, E.L.; Varesche, M.B.A. Role of homo-and heterofermentative lactic acid bacteria on hydrogen-producing reactors operated with cheese whey wastewater. Int. J. Hydrogen Energy 2015, 40, 8650–8660. [Google Scholar] [CrossRef]
- Ottaviano, L.M.; Ramos, L.R.; Botta, L.S.; Varesche, M.B.A.; Silva, E.L. Continuous thermophilic hydrogen production from cheese whey powder solution in an anaerobic fluidized bed reactor: Effect of hydraulic retention time and initial substrate concentration. Int. J. Hydrogen Energy 2017, 42, 4848–4860. [Google Scholar] [CrossRef]
- Nascimento, T.R.D.; Cavalcante, W.A.; de Oliveira, G.H.D.; Zaiat, M.; Ribeiro, R. Modeling dark fermentation of cheese whey for H2 and n-butyrate production considering the chain elongation perspective. Bioresour. Technol. Rep. 2022, 17, 100940. [Google Scholar] [CrossRef]
- Ribeiro, J.C.; Mota, V.T.; de Oliveira, V.M.; Zaiat, M. Hydrogen and organic acid production from dark fermentation of cheese whey without buffers under mesophilic condition. J. Environ. Manag. 2022, 304, 114253. [Google Scholar] [CrossRef]
- García-Depraect, O.; Mirzazada, I.; Martínez-Mendoza, L.J.; Regueira-Marcos, L.; Muñoz, R. Biotic and abiotic insights into the storage of food waste and its effect on biohydrogen and methane production potential. J. Water Process. Eng. 2023, 53, 103840. [Google Scholar] [CrossRef]
- Noike, T.; Takabatake, H.; Mizuno, O.; Ohba, M. Inhibition of hydrogen fermentation of organic wastes by lactic acid bacteria. Int. J. Hydrogen Energy 2002, 27, 1367–1371. [Google Scholar] [CrossRef]
- Sikora, A.; Błaszczyk, M.; Jurkowski, M.; Zielenkiewicz, U. Lactic Acid Bacteria in Hydrogen-Producing Consortia: On Purpose or by Coincidence? INTECH Open Science Open Minds: London, UK, 2013; pp. 488–514. [Google Scholar]
- Azbar, N.; Dokgöz, F.T.Ç.; Keskin, T.; Korkmaz, K.S.; Syed, H.M. Continuous fermentative hydrogen production from cheese whey wastewater under thermophilic anaerobic conditions. Int. J. Hydrogen Energy 2009, 34, 7441–7447. [Google Scholar] [CrossRef]
- García-Depraect, O.; Muñoz, R.; Rodríguez, E.; Rene, E.R.; León-Becerril, E. Microbial ecology of a lactate-driven dark fermentation process producing hydrogen under carbohydrate-limiting conditions. Int. J. Hydrogen Energy 2021, 46, 11284–11296. [Google Scholar] [CrossRef]
- Pérez-Rangel, M.; Corona, J.E.B.; Díaz, M.N.; Escalante, A.E.; Valdez-Vazquez, I. The duo Clostridium and Lactobacillus linked to hydrogen production from a lignocellulosic substrate. Water Sci. Technol. 2021, 83, 3033–3040. [Google Scholar] [CrossRef] [PubMed]
- Detman, A.; Mielecki, D.; Chojnacka, A.; Salamon, A.; Błaszczyk, M.K.; Sikora, A. Cell factories converting lactate and acetate to butyrate: Clostridium butyricum and microbial communities from dark fermentation bioreactors. Microb. Cell Factories 2019, 18, 36. [Google Scholar] [CrossRef] [PubMed]
- Belenguer, A.; Holtrop, G.; Duncan, S.H.; Anderson, S.E.; Calder, A.G.; Flint, H.J.; Lobley, G.E. Rates of production and utilization of lactate by microbial communities from the human colon. FEMS Microbiol. Ecol. 2011, 77, 107–119. [Google Scholar] [CrossRef]
- Blanco, V.; Oliveira, G.; Zaiat, M. Dark fermentative biohydrogen production from synthetic cheese whey in an anaerobic structured-bed reactor: Performance evaluation and kinetic modeling. Renew. Energy 2019, 139, 1310–1319. [Google Scholar] [CrossRef]
- De Gioannis, G.; Friargiu, M.; Massi, E.; Muntoni, A.; Polettini, A.; Pomi, R.; Spiga, D. Biohydrogen production from dark fermentation of cheese whey: Influence of pH. Int. J. Hydrogen Energy 2014, 39, 20930–20941. [Google Scholar] [CrossRef]
- Muñoz-Páez, K.M.; Vargas, A.; Buitrón, G. Feedback control-based strategy applied for biohydrogen production from acid cheese whey. Waste Biomass-Valorization 2023, 14, 447–460. [Google Scholar] [CrossRef]
- García-Depraect, O.; León-Becerril, E. Fermentative biohydrogen production from tequila vinasse via the lactate-acetate pathway: Operational performance, kinetic analysis and microbial ecology. Fuel 2018, 234, 151–160. [Google Scholar] [CrossRef]
- García-Depraect, O.; Rene, E.R.; Diaz-Cruces, V.F.; León-Becerril, E. Effect of process parameters on enhanced biohydrogen production from tequila vinasse via the lactate-acetate pathway. Bioresour. Technol. 2019, 273, 618–626. [Google Scholar] [CrossRef]
- García-Depraect, O.; Rene, E.R.; Gómez-Romero, J.; López-López, A.; León-Becerril, E. Enhanced biohydrogen production from the dark co-fermentation of tequila vinasse and nixtamalization wastewater: Novel insights into ecological regulation by pH. Fuel 2019, 253, 159–166. [Google Scholar] [CrossRef]
- García-Depraect, O.; Valdez-Vázquez, I.; Rene, E.R.; Gómez-Romero, J.; López-López, A.; León-Becerril, E. Lactate- and acetate-based biohydrogen production through dark co-fermentation of tequila vinasse and nixtamalization wastewater: Metabolic and microbial community dynamics. Bioresour. Technol. 2019, 282, 236–244. [Google Scholar] [CrossRef] [PubMed]
- APHA. Standard Methods for the Examination of Water and Wastewater, 21st ed.; American Public Health Association: Washington, DC, USA; American Water Works Association: Washington, DC, USA; Water Environmental Federation: Washington, DC, USA, 2005. [Google Scholar]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- RTL Genomics. Data Analysis Methodology. 2019. Available online: http://www.rtlgenomics.com/docs/Data_Analysis_Methodology.pdf (accessed on 23 September 2022).
- Asunis, F.; De Gioannis, G.; Isipato, M.; Muntoni, A.; Polettini, A.; Pomi, R.; Rossi, A.; Spiga, D. Control of fermentation duration and pH to orient biochemicals and biofuels production from cheese whey. Bioresour. Technol. 2019, 289, 121722. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.-H.; Whang, L.-M.; Chung, M.-C.; Chan, K.-C. Biological hydrogen and methane production from bagasse bioethanol fermentation residues using a two-stage bioprocess. Bioresour. Technol. 2016, 210, 49–55. [Google Scholar] [CrossRef] [PubMed]
- König, H.; Fröhlich, J. Lactic acid bacteria. In Biology of Microorganisms on Grapes, in Must and in Wine; Springer: Berlin/Heidelberg, Germany, 2017; pp. 3–41. [Google Scholar]
- Martinez FA, C.; Balciunas, E.M.; Salgado, J.M.; González, J.M.D.; Converti, A.; de Souza Oliveira, R.P. Lactic acid properties, applications and production: A review. Trends Food Sci. Technol. 2013, 30, 70–83. [Google Scholar] [CrossRef]
- Dzulkarnain, E.L.N.; Audu, J.O.; Wan Dagang, W.R.Z.; Abdul-Wahab, M.F. Microbiomes of biohydrogen production from dark fermentation of industrial wastes: Current trends, advanced tools and future outlook. Bioresour. Bioprocess. 2022, 9, 16. [Google Scholar] [CrossRef]
- Lee, M.-J.; Kim, T.-H.; Min, B.; Hwang, S.-J. Sodium (Na+) concentration effects on metabolic pathway and estimation of ATP use in dark fermentation hydrogen production through stoichiometric analysis. J. Environ. Manag. 2012, 108, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Ghimire, A.; Frunzo, L.; Pirozzi, F.; Trably, E.; Escudie, R.; Lens, P.N.; Esposito, G. A review on dark fermentative biohydrogen production from organic biomass: Process parameters and use of by-products. Appl. Energy 2015, 144, 73–95. [Google Scholar] [CrossRef]
- Chen, X.; Sun, Y.; Xiu, Z.; Li, X.; Zhang, D. Stoichiometric analysis of biological hydrogen production by fermentative bacteria. Int. J. Hydrogen Energy 2006, 31, 539–549. [Google Scholar] [CrossRef]
- Tholozan, J.L.; Touzel, J.P.; Samain, E.; Grivet, J.P.; Prensier, G.; Albagnac, G. Clostridium neopropionicum sp. nov., a strict anaerobic bacterium fermenting ethanol to propionate through acrylate pathway. Arch. Microbiol. 1992, 157, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Kandler, O.; Schillinger, U.; Weiss, N. Lactobacillus bifermentans sp. nov., nom. rev., an Organism forming CO2 and H2 from lactic acid. Syst. Appl. Microbiol. 1983, 4, 408–412. [Google Scholar] [CrossRef] [PubMed]
- Diez-Gonzalez, F.; Russell, J.B.; Hunter, J.B. The role of an NAD-independent lactate dehydrogenase and acetate in the utilization of lactate by Clostridium acetobutylicum strain P262. Arch. Microbiol. 1995, 164, 36–42. [Google Scholar] [CrossRef]
- García-Depraect, O.; Castro-Muñoz, R.; Muñoz, R.; Rene, E.R.; León-Becerril, E.; Valdez-Vazquez, I.; Kumar, G.; Reyes-Alvarado, L.C.; Martínez-Mendoza, L.J.; Carrillo-Reyes, J.; et al. A review on the factors influencing biohydrogen production from lactate: The key to unlocking enhanced dark fermentative processes. Bioresour. Technol. 2021, 324, 124595. [Google Scholar] [CrossRef]
- Tao, Y.; Hu, X.; Zhu, X.; Jin, H.; Xu, Z.; Tang, Q.; Li, X. Production of butyrate from lactate by a newly isolated Clostridium sp. BPY5. Appl. Biochem. Biotechnol. 2016, 179, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef]
- Rivière, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and butyrate-producing colon bacteria: Importance and strategies for their stimulation in the human gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef]
| Parameter | Raw CW | Preconditioned CW |
|---|---|---|
| pH | 5.4 ± 0.02 | 5.4 ± 0.04 |
| Total alkalinity (g CaCO3/L) | 3.8 ± 0.1 | 1.0 ± 0.02 |
| Total COD (g/L) | 118.1 ± 1.3 | 83.3 ± 0.5 |
| Soluble COD (g/L) | 95.4 ± 0.64 | 66.3 ± 0.2 |
| Total nitrogen (g/L) | 1.7 ± 0.3 | 0.4 ± 0.09 |
| Ammonia nitrogen (mg/L) | ND | 67.4 ± 0.3 |
| Protein (g/L) | 5.4 ± 0.03 | 3.6 ± 0.03 |
| Total phosphorous (g/L) | 2.9 ± 0.2 | 0.3 ± 0.02 |
| Total sugars (g/L) | 51.8 ± 1.3 | 44.9 ± 1.1 |
| Lipids (g/L) | 6.0 ± 0.01 | 0.1 ± 0.01 |
| Total solids (g/L) | 67.5 ± 0.8 | 52.2 ± 0.6 |
| Volatile solids (g/L) | 62.0 ± 0.1 | 46.8 ± 0.3 |
| Lactate (g/L) | 1.0 ± 0.1 | 2.9 ± 0.1 |
| Formate (g/L) | 1.3 ± 0.4 | 0.4 ± 0.04 |
| Acetate (g/L) | 1.0 ± 0.01 | 3.1 ± 0.02 |
| Parameter | Value |
|---|---|
| λ (h) | 4.2 ± 1.8 |
| Hmax (NmL H2/L) | 3313.2 ± 207.4 |
| Rmax (NmL H2/L-h) | 77.9 ± 20.5 |
| R2 | 0.9959 |
| VHPR (NL H2/L-d) | 1.9 ± 0.5 |
| YH2 (NmL H2/g VSfed-L) | 80.0 ± 5.3 |
| YH2 (NmL H2/g CODfed-L) | 44.5 ± 2.9 |
| COD removal (%) | 24.6 ± 3.9 |
| VS removal (%) | 48.7 ± 3.4 |
| Parameter | Value |
|---|---|
| SCOD effluent (%) | 79.8 ± 6.8 |
| Hydrogen (%) | 3.1 ± 0.2 |
| Biomass (%) | 10.0 ± 0.0 |
| Recovered fraction (%) | 92.9 ± 6.6 |
| Non-recovered fraction (%) | 7.1 ± 6.6 |
| Phylum | Class | Family | Genus | Relative Abundance |
|---|---|---|---|---|
| Firmicutes | Lactobacillales | Lactobacillaceae | Lactobacillus | 86.6 ± 4.2% |
| Clostridiales | Clostridiaceae | Clostridium | 12.3 ± 3.7 | |
| Proteobacteria | Enterobacteriales | Enterobacteriaceae | Klebsiella | 1.0 ± 0.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aranda-Jaramillo, B.; León-Becerril, E.; Aguilar-Juárez, O.; Castro-Muñoz, R.; García-Depraect, O. Feasibility Study of Biohydrogen Production from Acid Cheese Whey via Lactate-Driven Dark Fermentation. Fermentation 2023, 9, 644. https://doi.org/10.3390/fermentation9070644
Aranda-Jaramillo B, León-Becerril E, Aguilar-Juárez O, Castro-Muñoz R, García-Depraect O. Feasibility Study of Biohydrogen Production from Acid Cheese Whey via Lactate-Driven Dark Fermentation. Fermentation. 2023; 9(7):644. https://doi.org/10.3390/fermentation9070644
Chicago/Turabian StyleAranda-Jaramillo, Brenda, Elizabeth León-Becerril, Oscar Aguilar-Juárez, Roberto Castro-Muñoz, and Octavio García-Depraect. 2023. "Feasibility Study of Biohydrogen Production from Acid Cheese Whey via Lactate-Driven Dark Fermentation" Fermentation 9, no. 7: 644. https://doi.org/10.3390/fermentation9070644
APA StyleAranda-Jaramillo, B., León-Becerril, E., Aguilar-Juárez, O., Castro-Muñoz, R., & García-Depraect, O. (2023). Feasibility Study of Biohydrogen Production from Acid Cheese Whey via Lactate-Driven Dark Fermentation. Fermentation, 9(7), 644. https://doi.org/10.3390/fermentation9070644








