Abstract
Climate warming is a hot environmental issue of global concern. As one of the major methane sinks, the process of methane oxidation coupled with denitrification (MOD) reduces the environmental impact brought by the greenhouse effect and water eutrophication. In addition, as an energy substance, methane can also improve its economic value by transforming into other liquid chemicals. Previous studies on the mechanism of the process have mainly focused on the extracellular electron transfer between species. However, in recent years, the production of intermediates influenced by different factors, and the existence of a large number of acid-producing bacteria and methanogens under anaerobic conditions, has led some researchers to pursue research into a new mechanism of the process. Moreover, the discovery of CO2 as a potential electron acceptor in products is certainly exciting, being a big opportunity under the ‘carbon neutral’ policy. This review looks back at the development of the MOD process and describes its functional microorganism and mechanism in detail when summarizing the types of microorganisms and intermediates at different oxygen levels, and introduces some traditional and novel biotechnologies, such as metagenomics, meta-transcriptomics, and meta-proteomics, etc., to help explore the novel mechanism of the process of MOD mediated by intermediates.
1. Introduction
Methane is the third largest greenhouse gas, second only to carbon dioxide and water vapor [1], whose global warming potential (GWP) will be 28 times that of CO2 in 100 years [2]. The global emissions of methane are about 600–900 Tg annually [3], among which, wetlands [4] are the most significant natural source of methane emissions; however, due to the widespread existence of methane-oxidizing bacteria in natural habitats, the amount of methane eventually entering the atmosphere is far less than the emissions. The main man-made sources of methane emissions [5], such as fossil fuel extraction and burning, biomass burning, animal husbandry, rice farming, landfill and composting, and anaerobic wastewater treatment, are not effectively treated, so most of the methane is released into the environment. To date, the concentration of methane in the atmosphere has increased 2.6 times over the pre-industrial level (1.8 ppm) [6].
Studies have shown that the methane sink intensity increases in a positive proportion to the increase in methane concentration in the atmosphere. Among them, biomethane oxidation, as one of the major methane sinks, can not only greatly reduce methane from gas hydrates [5], but also be coupled with other organisms to remove some oxidizing pollutants. According to the oxygen content in the environment, the MOD process can be divided into aerobic methane oxidation coupled with denitrification (AME-D) and anaerobic methane oxidation coupled with denitrification (ANME-D). The AME-D process was first confirmed by Rhee and Fuhs in 1978 [7] in the reclamation of methane from sewage plants and landfills. However, it was not until 2006 that Raghoebarsing [8] proved the existence of ANME-D in experimental conditions for the first time. Since then, methane oxidation-coupled denitrification has attracted wide attention and a series of in-depth studies have been carried out [2,9,10]. At present, the main functional strains and related metabolic pathways have been described in detail, laying a foundation for future engineering applications; this has important significance in environmental protection.
In addition, it is worth noting that methane itself, as an abundant and cheap energy substance, can be of higher value as a fuel or when converted into other liquid chemicals if properly utilized [11]. The existing gas-to-liquid (GTL) technologies, such as the Fischer-Tropsch process, are technically complex, costly, and carbon or energy inefficient [12]. The methanotrophs can activate C–H bonds of methane through methane monooxygenase (MMO) under aerobic conditions, which requires the participation of electrons. Due to the loss of electrons in O2 and CO2, aerobic methane oxidation has relatively low energy and carbon efficiency [13]. Anaerobic methanotrophs (ANMEs) oxidize methane through the “reverse methanogenesis pathway”, in which methane is activated by Mcr without the need for electrons or oxygen, thus improving carbon efficiency and energy efficiency. However, practical engineering applications face challenges such as the low mass transfer of methane and oxygen, sluggish biological growth, and other factors that contribute to high economic costs and the low volume productivity of methane conversion. To address these drawbacks, new separation technologies can be utilized to optimize strains, metabolic engineering can be employed to modify the carbon assimilation pathway, the growth conditions of the strains can be optimized, new reactors can be designed, and hollow fiber membrane or nano diffusion technology can be applied [14,15,16]. To date, some SCFAs production has been found in ANME-D [17,18]. These small molecular acids can be used as precursors for chemical fuel production [19,20], which can also be utilized as readily available carbon sources [21] to improve the removal of nitrogen from the environment, which undoubtedly provides another possible metabolic pathway for the ANME-D process.
Based on the collection and analysis of the literature, this review summarizes the main functional microorganisms and metabolic pathways of the traditional process of MOD, the effect of different O2/CH4 ratios and inorganic nitrogen types on AME-D, and the newly discovered intermediates and other functional microorganisms present in the process of nitrite/nitrate-dependent AOM(N-DAMO). Furthermore, several current and potential technologies and future research concerns for understanding these processes are also presented.
2. Microbial Mechanism of the AME-D Process
As early as the early 20th century, methanotrophs were isolated and purified, but the classification work did not begin until Whittenbury isolated and identified more than one hundred methanotrophs [22]. In 1978, the AME-D process was confirmed to further promote the application of aerobic methane oxidation in practical processes, which plays a significant role in the biogeochemical cycle of C and N in environmental protection [10].
2.1. Microorganisms Involved in the AME-D Process
Aerobic methanotrophs and the relevant denitrifying bacteria are the main microorganisms involved in the AME-D process. The extensive and inefficient use of pesticides and fertilizers in modern agriculture has resulted in the loss of a large number of nitrogen sources to the ecological environment, such as lakes, wetlands, groundwater, etc. [23], the oxic–anoxic environments that provide a livable place for methanotrophs and denitrifiers [4].
The aerobic methanotrophs, belonging to Gram-negative bacteria, are a unique subgroup of methylotrophs [24], which can utilize CH4 as the only carbon source and energy source [25]. According to their physiological characteristics, such as their morphological characteristics, membrane structure, guanine and cytosine (GC) content, and phospholipid fatty acids (PLFAs) composition, they are divided into type I, II and III [10], among which type I can be further subdivided into type Ia and Type Ib, which is also known as the type X methanotroph [24]. Moreover, Methylacidiphilum and Methylomirabilis, which belong to NC10, should be divided into a new category because of their unique physiological and ecological characteristics and due to differences in their key steps of methane oxidation [26]. Type I aerobic methanotrophs are mainly distributed in the Methylococcaceae of γ-Proteobacteria. In addition, Crenotrichaceae [27] and Methylothermaceae [28] are two branches of type I. The internal cytoplasmic membranes (ICM) of Type Ia aerobic methanotrophs, which use a ribulose monophosphate pathway (Rump) of carbon assimilation, are bundles of vesicle discs, with 14C PLFAs and 16C PLFAs predominating. The X aerobic methanotrophs use Rump as the main pathway for formaldehyde assimilation, which can also utilize the serine cycle (SC) and Calvin–Benson–Bassham cycle (CBB) for carbon assimilation. Compared with type I, type X can grow at higher temperatures, have a higher molar percentage of G-C, and possess the nitrogen-fixing capacity that most type I bacteria lack [24]. Type II aerobic methanotrophs are distributed in the order Rhizobia of the α-Proteobacteria, including two families. Its incomplete ICMs are arranged along the cell perimeter, similar to cell membranes, and are dominated by 18C PLFAs, which are often used as a marker for the presence of type II in the environment [29]. Moreover, the bacteria employ the SC pathway for carbon assimilation [30]. Type III aerobic methanotrophs are scattered in the Verrucomicrobia, which currently contains only the Methylacidiphilum isolated from extremely acidophilic and thermophilic environments. It does not have a typical inner membrane, while the tubular membrane of the inward membrane can perform the same function, containing C14, C15, and C18 fatty acids, and assimilating CO2 via the CBB pathway [31].
The oxidation process of methane driven by aerobic methanotrophs is shown in Figure 1. Firstly, methane is oxidized to methanol by MMO, which is unique to aerobic methanotrophs and divided into two categories. The soluble MMO (sMMO) has broad substrate specificity, which is particularly effective in co-metabolizing halogenated aliphatic compounds; however, it is produced by only a few methanotrophs (mainly type II methanotrophs) [24], whose expression is inhibited in high copper environments (>2.5 μmol·g−1) [32]. Thus, it is more meaningful to study the application of particulate MMO (pMMO) in methane oxidation [33]. The pMMO consists of α, β, and γ subunits, in which the pmoA gene encoding α subunit can be used as a functional biomarker of proteobacteria in aerobic methanotrophs [34]; this is except for Verrucomicrobia and Crenotrichaceae, because of the phylogenetic differences in pMMO [27,35]. Methanol is then calcined to formaldehyde by the pyrroloquinolinequinone (PQQ)-dependent methanol dehydrogenase (MDH), which is also classified into two types. The MxaF-MDH contains the cofactors PQQ and calcium requiring cytochrome c as an electron acceptor [36]; the other is the Xoxf-type MDH, found in the phylum Verrucomicrobia [37] and in the NC10 bacteria [38], which requires lanthanum to maintain activity; notably, it can directly oxidize methanol to formate via a four-electron transport pathway [39]. Subsequently, a part of formaldehyde was assimilated via the Rump or SC pathway, releasing organic intermediates such as acetate and citrate. The other portion is dissimilated and continues to be oxidized to produce CO2, which is released to the outside of the cell, and the other part is fixed by the CBB cycle to provide the necessary carbon via formaldehyde dehydrogenase and formate dehydrogenase. Importantly, the process in turn provides a reduction equivalent (NAD(P)H) for the initial oxidation step of methane oxidation [40]. In terms of electron transfer, MMO utilizes electrons to convert methane into methanol. Among the electron donors of particulate pMMO, three possibilities exist: (1) Ubiquinol (Q8H2) serves as the electron donor for methane oxidation while MDH directly supplies electrons to complex IV via cytochrome c; (2) the composite structure of pMMO and MDH directly facilitates electron transfer from MDH to pMMO, in which MDH acts as the electron donor for methane oxidation; (3) reverse electron flow occurs from cytochrome c to ubiquinone via the ubiquinol–cytochrome c reductase [41,42,43]. The produced methanol is subsequently oxidized to formaldehyde, formate, and CO2, with electron transfer coupled to cyt cox/cyt cred or NAD/NADH, respectively. This enables the completion of the electron transfer process. The NADH generated during this process can return to the initial step, acting as an electron donor for sMMO [44]. Additionally, the small organic molecules produced during this process and the carbon assimilation pathway can serve as electron donors for the denitrifiers involved in nitrogen removal. Furthermore, when microorganisms exhibit a high energy demand or when the nitrate concentration in the environment is relatively high, direct electron transfer via methane–nitrate/nitrite coupling is often employed [45].
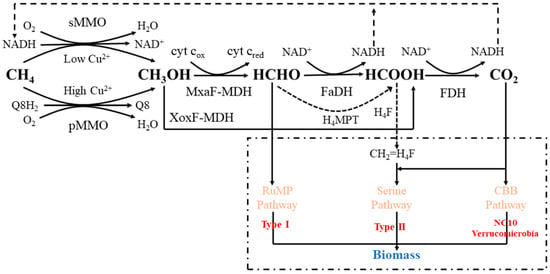
Figure 1.
The metabolic pathway of aerobic methanotrophs. Abbreviations for the cofactors and enzymes in the figure are as follows: NAD, nicotinamide adenine dinucleotide; PQQ, pyrroloquinoline quinone; H4MPT, tetrahydromethanopterin; H4F, tetrahydrofolate; sMMO, soluble methane monooxygenase; pMMO, particulate methane monooxygenase; MDH, methanol dehydrogenase; FaDH, formaldehyde dehydrogenase; FDH, formate dehydrogenase.
Compared with aerobic methanotrophs, denitrifiers have a wider category and distribution. At present, more than 50 genera and 130 species of denitrification microorganisms have been isolated, among which the strains often isolated are different from the ones that actually dominate in the wastewater denitrification treatment system; most of them belong to Proteobacteria (59%) and Bacteroides (16%) [46].
Complete denitrification is a process in which NO3− is successively reduced to NO2−, NO, N2O, and finally N2. Based on the physiological function, intracellular location, and structure of the molybdenum active site, nitrate reductase (Nar), which performs nitrate reduction, can be divided into membrane-bound nitrate reductase (Nar), periplasmic nitrate reductase (Nap), and assimilative nitrate reductase (Nas) [47], in which the formation of Nap is not dependent on nitrite induction or anaerobic conditions, nor is it sensitive to ammonia inhibition [48], whose role in distinct bacteria is suspected to be different but is still unclear. The nitrite reductase (Nir) can be divided into two groups according to the structure and prosthesis metal in the cell: the Cu-Nir and the cytochrome cd1-Nir encoded by nirK and nirS, respectively. Approximately 3/4 of the strains worldwide possess the latter, which is particularly common in Pseudomonas and sensitive to Cyanide and azide, but not to CO in the process of cd1-Nir-catalyzed nitrite reduction [49]. The nitric oxide reductase (Nor) is encoded by the norCB gene in anoxic/anaerobic conditions, performing unstably. Nitrous oxide reductase (Nos) is a poly copper enzyme encoded by nosZ. In addition to Nap, other reductases, including Nar, Nir, Nor, and Nos, are sensitive to oxygen and their activity can be inhibited by specific concentrations of oxygen [50].
2.2. Metabolic Mechanism of the AME-D Process
The Ammonia monooxygenase (AMO) produced by ammonia-oxidizing bacteria (AOB) is a cognate of PMMO that oxidizes methane [51]. Moreover, some aerobic methanotrophs oxidize ammonium or metabolites in the process via pMMO, expressing and translating the homolog gene of hydroxylamine oxidase (HAO) or cytochrome P460 [52] to complete the whole nitrification process [26,53]. Simultaneously, the enzyme-encoding genes required for complete denitrification can be found in aerobic methanotrophs [54,55,56], which do not rely on these pathways for growth. The Methylomonas denitrificans strain FJG1T has been found to possess genes not only for the whole process of methane oxidation, but also for the process of NO3− to N2O [54,57]. However, no aerobic methanotrophs holding all the genes needed for complete denitrification have been found, while most of the remaining bacteria need to cooperate to complete the whole course. Moreover, since most bacteria do not have the nosZ gene required to encode N2O reductase, the process of denitrification generally produces a large amount of N2O due to the nosZ gene in the largest proportion of them [55]. Nevertheless, Cheng et al. [58] found that the aerobic methanotrophs alone or coupled with denitrifiers use N2O as the electron acceptor for methane in anoxic wetland environments, which is of great significance for slowing down the greenhouse effect and reducing nitrogen pollution. Additionally, Ren et al. [59] found that the Methylobacter strain T20, which does not possess genes associated with denitrification, could oxidize CH4 when NO3− was used as the nitrogen source under oxygen-limited conditions, which may be due to a variety of enzyme-encoding genes required for denitrification.
In addition to the independent coupling process of methane oxidation and denitrification, another conjecture is that it uses methane as an energy and carbon source in which the soluble organic intermediates generated will serve as electron donors for the denitrifiers to promote denitrifying. The concentration of oxygen is the main factor that determines the types and quantities of aerobic methanotrophs and denitrifiers presently, deciding the generation of intermediates; this will be described in detail in the third part.
3. Microbial Mechanism of the ANME-D Process
For the past 40 years, anaerobic methane oxidation (AOM) has been considered a prominent link for carbon balance in marine and freshwater systems, which is inferred from the concentration profile of CH4 and SO42− in marine sediments [60]. The AOM process in different habitats will employ different electron acceptors, such as sulfate, nitrate, perchlorate, bromate, heavy metal ions, humus, and even CO2. From the view of thermodynamics, the N-DAMO process will release more energy, which is conducive to the vital activity of microorganisms.
3.1. Microorganisms Involved in the ANME-D Process
For a long time, it has been difficult to demonstrate the slow growth of microorganisms in the N-DAMO process. In 2009, Ettwig et al. [61] demonstrated that NC10 bacteria were responsible for the coupling of AMO and nitrite reduction by enriching an NC10 bacterial subgroup, which they named [62] Candidatus ‘Methylomirabilis oxyfera’ in 2010. Cells of Ca. ‘M. oxyfera’ fission in the form of binary fission with a star type of short rod, 0.3–1.1 μm in length and 0.06–0.26 μm in width, which is surrounded by a special layer of protein outside the cell, usually occur as single-cell or multicellular aggregates. Except for a few obvious longitudinal ridges, the cell surface is relatively smooth; however, theICMs generally found in proteobacteria are not found [63]. Therefore, it and Verrucomicrobia represent the only non-proteobacteria methanotrophs. Moreover, Ca. ‘M. oxyfera’ can accomplish the N-DAMO process when the concentration of dissolved methane is lower than 0.6 μm, manifesting extreme affinity for CH4 [8]. In addition, an alien from the traditional pathway of N-DAMO, O2 is produced by denitrification, prompting aerobic methane oxidation under anaerobic conditions [62].
In 2013, Haroon et al. [64] enriched ANME-2d archaea and Anammox bacteria by employing CH4, NO3−, and NH4+ as substrates under anaerobic conditions, naming the former as Candidatus ‘Methanoperedens nitroreducens’. The cells of Ca. ‘M. nitroreducens’ are irregular polygonal spheres with a diameter of around 1.5 μm, which generally exist in the form of aggregate extruding tubular structures that possess putative cell-to-cell contacts among each other. Additionally, the cytochrome c proteins were localized in the cytoplasm and periplasm [65]. The ANME-2d archaea are easily inhibited by NO2− during the culture process so that co-enrichment culture is indispensably performed by microorganisms using NO2− as the substrate [61]. In contrast to the complete DAMO process, the coupling of ANME-2d archaea with Anammox enables the complete denitrification of wastewater [66].
3.2. Metabolic Mechanism of the ANME-D Process
Initially, it was suggested that N-DAMO was accomplished via the reverse methanogenesis pathway under the synergistic action of N-DAMO archaea and bacteria. However, Ettwig et al. [67] first demonstrated that NC10 bacteria could independently perform nitrite-dependent AOM; subsequently, Haroon et al. [64] confirmed that ANME-2d archaea could independently perform nitrate-dependent AOM.
3.2.1. Methane Oxidation Coupled with Nitrate Reduction by ANME-2d Archaea
As shown in Figure 2, the CH4 is first converted to Methyl coenzyme M (methyl-S-COM) by the action of Mcr during AOM, which is the rate-limiting step of the reverse methanogenesis pathway [68], so that the rate of AOM can be enhanced by elevating the partial pressure of methane [69] and the concentration of Mcr [68]; thereafter, the methyl of Methyl-S-CoM is transferred to H4MPT to form Methyl-H4MPT under the function of Mtr [70]; subsequently, Methyl-H4MPT is converted to CO2 via a series of enzymatic reactions. Apart from all the genes involved in the AOM process, there is also the gene narGH that encodes nitrate reductase in Ca. ‘M. nitroreducens’, which may be obtained via lateral gene transfer via Protein Proteobacteria [64]. Nevertheless, Ca. ‘M. nitroreducens’ can only accomplish part of the denitrification process when the genes encoding the subsequent steps of denitrification are not detected.
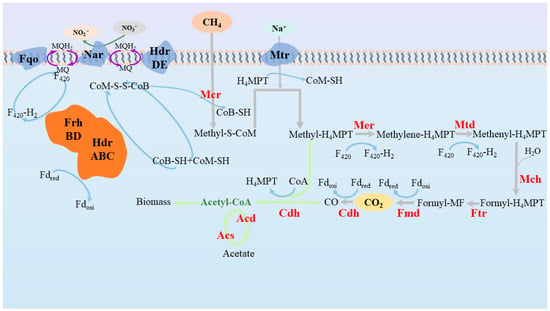
Figure 2.
Reverse methanogenesis pathway of Ca. ‘M. nitroreducens’. Abbreviations for the cofactors and enzymes in the figure are as follows: CoB-SH, coenzyme B; CoM-SH, coenzyme M; F420, coenzyme F420; Fd, ferredoxin; Fqo, F420-H2: quinone oxidoreductase; Hdr, heterodisulfide reductase; Frh, F420-reducing hydrogenase; MQ, menaquinone; MQH2, menaquinol; Nar, nitrate reductase; Mcr, methyl-coenzyme M reductase; Mtr, Na+-translocating methyl-H4MPT coenzyme M methyltransferase; Mer, F420-dependent methylene-H4MPT reductase; Mtd, F420-dependent methylene H4MPT dehydrogenase; Mch, methentl-H4MPT cyclohydrolase; Ftr, formylmethanofuran-H4MPT formyltransferase; Fmd, formylmethanofuran dehydrogenase; Cdh, CO dehydrogenase; Acd, acetyl-CoA synthetase; Acs, Acyl-CoA synthetase.
In terms of electron transfer, some researchers believe that Ca. ‘M. nitroreducens’ make use of membrane-bound, quinone-dependent electron transport proteins that couple nitrate reduction with AOM [2]. First of all, CoM-S-S-CoB (the heterodisulfide of coenzyme M and coenzyme B) is a heterodisulfide reductase that releases CoB-SH (coenzyme B) and CoM-SH (coenzyme M), respectively, via two reactions, accompanied by the transfer of two electrons. Coenzyme B and coenzyme M resynthesize CoM-S-S-CoB via the REDOX loop, releasing electrons transferred directly to the in-membrane electron carrier menaquinone (MQ) via the membrane-bound HdrDE. Subsequently, NO3− is reduced by accepting electrons from the quinone loop. Additionally, an HdrABC–FrhB complex located in the cytoplasm can be coupled to the oxidation of CoB-SH and CoM-SH, to oxidized ferredoxin, and to the reduction of F420, in which electrons released by F420H2 oxidization via the membrane binding of Fqo and quinone loops to Nar are later applied to the reduction of NO3− [71].
3.2.2. Methane Oxidation Coupled with Nitrite Reduction by NC10 Phylum Bacteria
Ettwig et al. examined the genome, transcriptome, and proteome of NC10-enriched cultures, finding that Ca. ‘M. oxyfera’ comprised genes involved in complete denitrification, such as nar, nap, nir, and nor, in which the last three are all expressed as proteins. Nevertheless, the transcriptional expression of nap is quite low, showing that the role of Ca. ‘M. oxyfera’ in nitrate reduction can be ignored. However, an n-DAMO bacterial genome recovered by Li et al. [72] via metagenomic and macro-transcriptome techniques can highly express a complete denitrification genome, indicating that it has the ability to independently reduce nitrate to nitrogen. In addition, the gene encoding N2O reductase has not been found, but for the phenomenon of the presence of N2 in the product with little N2O, it is proposed that the existence of Nor may be for detoxification. There is another unknown NO dismutase (NOD) involved in the conversion of NO into N2 and O2, which is more difficult in the kinetic process but thermodynamically possible. Wu et al. [73] found that there is a genome of Ca. ‘M. oxyfera’ that harbors four sets of genes encoding terminal respiratory oxidases: two cytochrome c oxidases, a third putative bo-type ubiquinol oxidase, and a cyanide-insensitive alternative oxidase, which all are transcribed; however, this is in a low level, further proving the intracellular oxygen production function of Ca. ‘M. oxyfera’.
Subsequently, Wu et al. [74] further refined the energy metabolic pathways and central metabolic processes of Ca. ‘M. oxyfera’ through genomics. First, NO2− is converted to NO by cd1-Nir, which is later converted to N2 and O2 by NOD, in which 25% of the O2 enters the terminal respiratory chain for productivity, and the remainder is used for aerobic methane oxidation [73]. Later, the part comprising formaldehyde is oxidized to CO2 via the H4MPT pathway under the action of FaDH and FDH. In terms of electron transport, NAD(P)H dehydrogenase can be coupled with the NAD(P)H oxidation process and to the quinone reduction process to achieve proton pumping. In addition, the cytochrome bc1 complex can oxidize quinone, realizing the diffusion transfer of protons. Finally, the electrons carried by cytochrome c transfer to cd1-Nir to complete the reduction of NO2− [74] (Figure 3).
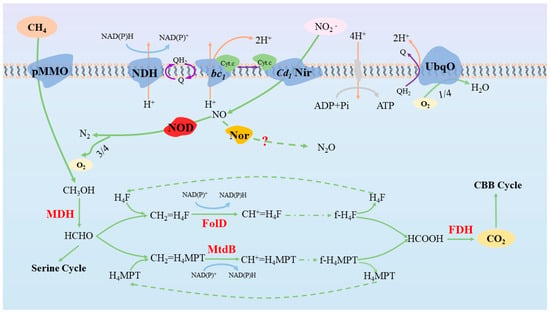
Figure 3.
Metabolic mechanism of Ca. ‘M. oxyfera’. Abbreviations for the cofactors and enzymes in the figure are as follows: NDH, NAD(P)H dehydrogenase complex; UbqO, ubiquinol oxidase; Cyt.c, cytochrome c; NOD, NO dismutase (unknown); Nor, NO reductase; FolD, methylene-H4F dehydrogenase/methenyl-H4F cyclohydrolase; MtdB, methylene-H4MPT dehydrogenase.
4. Microbial Community Structure and Intermediates of the MOD Process
Different types of methanotrophs have a distinct tolerance for diverse circumstances. For aerobic methanotrophs, type I tends to dominate under the condition of eutrophication and a high O2/CH4 ratio. In contrast, type II is prone to being the majority under low O2 and nitrogen conditions. However, Xingkun Xu et al. [75] proposed that type I (especially Methylosarcina and Methyloparacoccus) is constantly the main aerobic methanotroph regardless of the O2/CH4 ratio due to the RuMP pathway (65–80%) being more effective than the SC pathway (40–60%) in formaldehyde conversion. However, the relative abundance of Methylosarcina decreased significantly when the ratio of O2/CH4 was reduced [76]. Zhu et al. raised that the increase in oxygen concentration promoted the production of intermediate metabolites compared to the O2/CH4 ratio changing from 0–0.25, which means that there were enough carbon sources to facilitate complete denitrification. The denitrifiers containing the nirK gene and those tolerable to oxygen account for the majority, which are minimally affected by the exaltation of O2 as a result of exposure seldom caused by the agglomeration or granulation of sludge under a micro-oxygen environment. However, O2 will affect the process of denitrification when O2/CH4 changes from 0.25–1 due to the competition between electron acceptors [77]. Moreover, Cao et al. [78] found that Methylovorus, methyloversalis, Methylotenera, and other methylotrophs were abundant when the concentration of O2 was 21%; this would be changed when the density of O2 was reduced to 10% while denitrifiers such as Azoarcus, Thauera, and Thiobacillus dominated.
In brief, depending on the O2 content, methanotrophs will produce different intermediates, such as methanol, formaldehyde, formate [79], acetate, propionate [80], lactate [81], butyrate, succinate, PHB [82], citrate, iso-citrate [7], carbohydrate [83] and protein [84]; even in some specific circumstances, methanogens can release nucleic acid [85] by cracking, as shown in Table 1, differing in the metabolic mechanism and the type of denitrifier. Moreover, oxygen can also affect the rate of denitrification by influencing the oxidation rate of methane. Therefore, the coupling process of methane oxidation and denitrification can be divided into four types according to the concentration of oxygen.

Table 1.
The main functional microorganisms and intermediates of the MOD process.
4.1. Aerobic Methane Oxidation Coupled to Denitrification (AME-D)
Under aerobic conditions, the methanotrophs promote denitrification by releasing intermediates and consuming O2 to create the anoxic environment required for denitrification [106]. However, an O2 content that is too high will affect the selection of electron acceptors by denitrifiers. Furthermore, the oxidation of methanol to formaldehyde is much faster than its subsequent conversion to formate and CO2, leading to the accumulation of formaldehyde, which may cause cell poisoning under high oxygen conditions [107]. Thalasso et al. [108] proposed that a higher partial pressure of O2 in the gas supply would lead to a higher total nitrogen removal rate, while a higher denitrification ratio will happen in a lower oxygen partial pressure, indicating that the AME-D process occurs frequently under the condition of low O2.
4.2. Microaerobic Methane Oxidation Coupled to Denitrification (MAME-D)
The main functional microorganisms in aerobic and microaerobic conditions are Methylococcaceae (Methylobacte, Methylomonas), Methylophilaceae (Methylotenera), Hyphomicrobium and Thermomonas. First, Methylococcaceae oxidizes CH4, in which formaldehyde is converted into pyruvate via the Embden–Meyerhof–Parnas (EMP) or Entner–Doudoroff (EDD) pathways, which is aerobically decomposed into AcCoA by pyruvate dehydrogenase. Formate converted to CH2=H4F can also be converted to AcCoA via the SC pathway. These two substances act as intermediates for the production of other organics from methane, initiating the biological transformation of methanotrophs into 1-, 2-, 3- and 4-carbon intermediates [41]; this includes pyruvate oxidation decarboxylation combined with phosphoric acid, which can produce acetate, butyrate and other substances. On the other hand, AcCoA associated with the EMP, SC, citric acid cycle (TCA) and ethylmalonyl-CoA (EMC) pathways can generate acetate, 3-hydroxypropionic acid, butyrate, succinate, etc. [16]. Later, midbodies with methyls, such as acetate and butyrate, are taken as electron donors by Methylotenera, Hyphomicrobium, and other Methylotrophic denitrifiers that are generally aerobic for denitrification in order to produce a large amount of N2O [109]. Meanwhile, non-methyl intermediates like formate and citrate are utilized by Pseudomonas sp., Thermomonas, etc. to convert NO3− to N2O or N2 [78,110] (Figure 4).
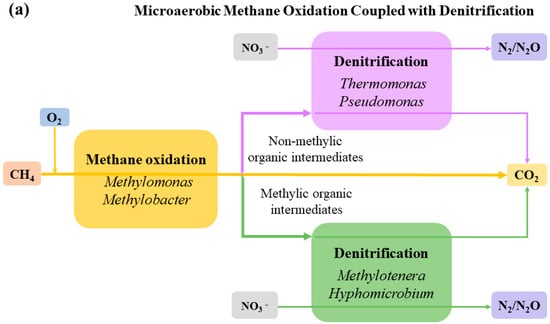
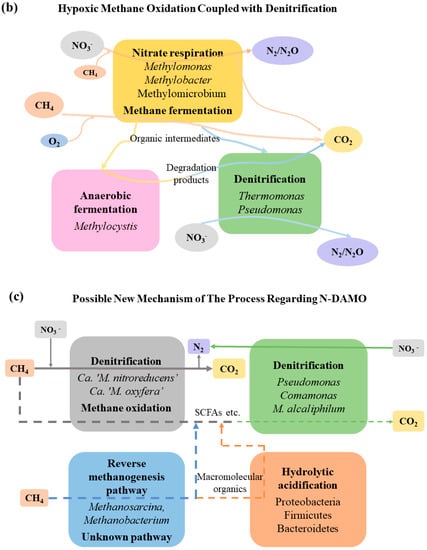
Figure 4.
The MOD process mediated by intermediates.
It is beneficial for aerobic methanotrophs to produce methanol, formate, and acetate under oxygen-limited conditions. With the increase in the O2 concentration varying from 0 to 0.25, the number and type of organic intermediates are continuously enriched, the content of formate, acetate, and citrate is continuously increased, and trace amounts of methanol and butyrate emerge (<1.20 μg·L−1) [77]. Although there was no significant difference in the concentration of organic compounds under the micro-aerobic and aerobic conditions, Liu et al. [94] have speculated that the denitrification efficiency was better when O2 and NO3− are both used as electron acceptors. Moreover, some research has shown that high levels of NO2− and NO3− stimulate the growth of type I even under low O2 conditions; in particular, type X dominated after the addition of nitrate [79,91,94]. The Methanotrophs with nitrogen-fixation ability, such as Methylocystis, Methylosinus, and Methylocella, are more likely to survive in low-oxygen conditions under the nitrogen-limited condition since nitrogenase is sensitive to O2 (4–10%) [111].
Aerobic denitrifiers utilize diverse carbon sources for aerobic respiration and denitrification, but are partial to one-carbon compounds, such as methanol and formate [109]. Thermodynamically, methanol may be the most ideal single electron donor that can maximize the efficiency of nitrate removal [10]. However, it is physiologically difficult for methanol to be discharged into the external environment as a metabolite. C. Costa et al. studied the methane oxidation process through 13C-NMR with 5% O2, finding a large number of acetate-utilizing denitrifiers, but only a few methanol-degrading denitrifying bacteria [93]. One of the reasons for this phenomenon is that the composite structure of MDH and pMMO enables the methanol produced to be immediately captured by MDH and converted into formaldehyde. Myronova et al. [112] found that pMMO hydroxylase (pMMO-H) formed a cap-structure with MDH, on the one hand increasing the stability of purified pMMO-H and thus its activity; on the other hand, the complex structure enables the electrons existing in the process of methanol oxidation to be recycled into the methane oxidation [113], which reduces the consumption of NADH (limiting growth factor) and thus improves the growth efficiency [114]. Secondly, from the perspective of kinetics, aerobic methanotrophs establish oxidation kinetics that are faster for methanol than for methane [93]. Furthermore, methanol is a key substrate for the reducing agent required for the initial reaction involved in methane oxidation [10,115]. How to improve the methanol yield is a problem worthy of attention and, therefore, the following solutions are given in this paper:
- (1)
- Inhibitors: the productivity of methanol can be improved by adding some inhibitors of MDH, such as CO2, phosphate, cyclopropyl alcohol, sodium chloride, ammonium chloride or EDTA, and other selective inhibitors [11]. However, when summarizing the productivity of methanol under different inhibitors, Wang et al. [116] suggested that the maximum accumulated concentration of methanol was lower than 12 mmol·L−1, which may be triggered by the fact that MMO requires the participation of NADH, which is mainly derived from the subsequent process of methanol oxidation. Therefore, exogenous electron donors such as sodium formate [117] or NADH must be added to promote the continuation of the reaction, which inevitably adds to the cost.
- (2)
- Low ratio of C/N: the aerobic methanotrophs will adjust the metabolic pathway to suit the environment by releasing methanol under certain conditions. Wolfe et al. [118] studied the ‘acetate switch’ of E. coli under different conditions, taking the shake bottle experiment of Bacto Tryptone broth as an example. E. coli first consumed L-aspartic acid in a strict order while catabolizing acetic acid during exponential growth; when it reached the fixed period, namely the decelerated growth period, it began to use L-tryptophan and assimilate acetic acid; the ammonia produced by the utilization of amino acids and the consumption of acetate in the early stage led to an alkaline environment, causing the arrival of the stage of acetate generation. Based on this, Zhu et al. [10] proposed that acetate-degrading denitrifiers were major players in the growth stage, while methanol-degrading denitrifying bacteria were more dominant in the stable stage. Moreover, via thermodynamic calculation, it was concluded that the lower the C/N ratio, the smaller the proportion of methanol utilized by aerobic methanotrophs is, which is theoretically no less than 40% of the methanol produced.
- (3)
- The circumstance of low oxygen: Krause et al. [95] found that non-methanotrophic partners induce an alteration in the MDH of aerobic methanotrophs under low O2/CH4 conditions, which prompts the lanthanide-dependent MDH (XoxF-MDH) with high methanol affinity to transform into calcium-dependent MDH (MxaF-MDH) with a low methanol affinity, thus allowing more methanol to be freed into the external environment.
- (4)
- Optimization of culture condition: It is possible to increase the methanol yield by optimizing the pH, temperature [119], culture time, culture method, methane concentration, bacterial solution concentration [120], the concentration of phosphoric acid buffer, and even the concentration of Cu2+ [121]. In addition, cell immobilization is an effective means of methanol accumulation.
- (5)
- Type of bacteria: The methanol production of Verrucomicrobia surviving at extremely low pH levels or high temperatures deserves further study due to the fact that type I and II methanotrophs cannot adapt to the extreme environment [122].
- (6)
- AOB-mediated oxidation of methane: AOB-mediated bio-methanol production can be performed via the continuous flow process due to the similar characteristics of AMO and pMMO. First, a high NH4+/CH4 inlet ratio should be maintained to prevent the growth of methanotrophs; next, it is better to select NH3 as the electron donor in water resource recovery facilities (WRRFs) because it is a nearly ideal electron donor for methanol production, is cheap and easy to obtain, and does not increase the exogenous load of nitrogen from the perspective of practical application, although NH2OH can avoid the competitive inhibition of CH4 and NH3 on AMO and any potential inhibition of a reducing equivalent [123].
4.3. Hypoxic Methane Oxidation Coupled to Denitrification (HYME-D)
Methylobacter is one of the main methanotrophs in low-oxygen habitats and has the ability to oxidize methane at low oxygen levels at the nanoscale due to having a high-affinity cytochrome (cytochrome bd ubiquinol-oxidoreductase) [99]. In addition, pMMO-containing aerobic methanotrophs generally contain Hemerythrin (Hmrs), a non-heme oxygen-binding protein that transports O2 from the cytoplasm to the cytoplasmic membrane for use by pMMO [124], showing that Hmrs can improve the activity of pMMO [125]. In addition, the increased expression of Hmrs is stimulated under oxygen-restricted conditions [97,126]. Methylomonas show three times the expression of the pmoA gene compared with NC10 bacteria in anoxic (5%) conditions but those rich in methane and nitrate [127].
The metabolic mechanism of HYME-D is different from the previous two (Figure 4). Methylobacter and Methylomonas, etc. [57], use O2/NO3− to co-oxidize methane, while methanotrophic cultures switch to a fermentation mode under low oxygen conditions. Kalyuzhnaya et al. [97] found that Methylomicrobium, etc., produce organic acids such as formate, acetate, succinate, and lactate, etc., via a method of methane fermentation, namely, the pyrophosphate-mediated Embden–Meyerhof–Parnas pathway, by studying the C1 assimilation pathway of type I [128], of which the production of formate is three times that of the aerobic condition; this provides denitrifiers (Thermomonas, Hyphomicrobium) for denitrification that are used for the anaerobic fermentation of Methylocystis at a low O2/CH4 ratio. Vecherskaya et al. [129] found that Methylocystis can degrade PHB under the anaerobic condition to produce ß-hydroxybutyric acid, butyrate, acetate, acetone, isopropanol, 2,3-butanediol, and succinate, etc., which may be utilized as intermediates by denitrifying bacteria [78,110].
When exploring mixed microbial culture under CH4/O2-limited conditions in a continuous flow fermenter, Wilkinson T. G. et al. [130] showed that Hyphomicrobium sp. employed the methanol accumulated in the process of methane oxidation to denitrify, eliminating the inhibition of the low concentration of methanol on Pseudomonas sp. Additionally, Rahalkar et al. [98] found that the optimal concentration of O2 is 2% for Methylosoma difficile sp. nov. The oxidation rate of methane is low due to the weak O2 content, which leads to a low denitrification rate. When conducting a leach-bed bioreactor semi-continuous experiment under different oxygen conditions, Cao et al. [78] found that the denitrification efficiency of MAME-D was 70.25%, and that that of HYME-D was 34.80%; they also discovered that Methylobacter and Methylomonas performed methane oxidation under micro-aerobic conditions and that Methylophilaceae completed denitrification, the efficiency of which was 20.36 mg·(L·d)−1. However, under hypoxic conditions, Methylomonas and Methylobacter perform methane oxidation in an intermediate-mediated manner, a phenomenon first observed by Cuba et al. [131]. Later, intermediate organisms are used by Methylophilaceae to further complete denitrification, the productivity of which is 8.09 mg·(L·d)−1 [110]. The relative abundance levels of Methylococcaceae (7.52%) and Methylophilaceae (3.51%) are higher, the reason for which may be that the extremely anoxic environment stimulates the nitrate respiration of Methylococcaceae, while facultative Methylophilaceae can not only survive in an anoxic environment, but also use intermediates for denitrification.
Nutrients and O2 are the predisposing factors affecting PHB synthesis and degradation; for instance, hypotrophic conditions will induce the synthesis of PHB. In addition, O2, as an electron acceptor in the process of methane oxidation and ATP synthesis, preferentially assimilates methane into cellular material under the condition of low oxygen, blocking the synthesis of ATP so that PHB will be degraded to provide energy and a reduction equivalent (NADH) for cells [132]. Type II has advantages in the production of biopolymers such as PHA/PHB, entering the PHA/PHB cycle and producing CO2 after passing via the SC pathway under the condition of lacking nutrients [133]. Among them, the PHB synthesis process takes AcCoA as the starting point, while the three necessary enzymes of the catalytic pathway are β-ketothiolase, acetoacetyl-CoA reductase, and PHA synthase [16]. Fereshteh Rahnama et al. [86] found that Methylocystis hirsuta could use biogas to produce the most PHB when CH4/O2 was one in the bubble tower bioreactor. Kalyuzhnaya et al. [97] found a small amount of PHB accumulated in a hypoxic sealed vial experiment. Vecherskaya et al. [82] isolated Methylocystis from a micro-aerobic denitrification reactor, finding that Methylocystis synthesized PHB, accompanied by the production of succinate, acetate, and 2, 3-butanediol, etc., under micro-oxygen conditions (5–10%); the degradation of intracellular PHB occurs under anoxic/anaerobic conditions without external carbon sources, producing ß-hydroxybutyric acid, butyrate, acetate, acetone, isopropanol, 2,3-butanediol, and succinate, etc., which can be used as an energy source by methanotrophs or a substrate by denitrifiers [129]. It was beneficial to maintain the cellular MMO oxidation activity when enough PHBs were accumulated (1–5%), thus being conducive to PHB synthesis [134].
4.4. Anaerobic Methane Oxidation Coupled to Denitrification (ANME-D)
The ANME-D process is jointly completed by Ca. ‘M. nitroreducens’ and Ca. ‘M. oxyfera’. However, the presence of acetyl-CoA synthetase and the complete reductive acetyl-CoA pathway suggest that Ca. ‘M. nitroreducens’ is possibly able to produce acetate [64]. In order to verify this possibility, Cai et al. [18] utilized NO2−/NO3− as an electron receptor for the culture of Ca. ‘M. nitroreducens’, and identified that the microorganism may produce acetate (1620 μmol·L−1) by oxidizing the PHA [18,104] produced intracellularly under the restriction of NO2−/NO3−; meanwhile, other relatively low concentrations (150 μmol·L−1) of SCFAs, such as propionate, iso-butyrate, butyrate, isovalerate, valerate, and caproate, were found. In addition, the storage of PHA could be heightened by increasing the ratio of C/N [135]. However, Zhao et al. [103] found that the production rate of SCFAs was higher at a high electron accepter, up to 2425.7 mg·(L·d)−1; this was determined via the study of feeding nitrate, nitrite, and nitrate/nitrite-independent membrane biofilm reactors (MBfRs). Ca. ‘M. oxyfera’ is also a dominant performer in the ANME-D process. Luo et al. [80] found that it is involved in the synthesis of SCFAs, which do good to the heterotrophic denitrifiers (Comamonas, Azoarcus) of the system via the internal oxygen production pathway.
However, Chen et al. [17] found that Ca. ‘M. nitroreducens’ and Ca. ‘M. oxyfera’, as inoculants, gradually disappeared as the culture process went on. The microbial community in the reactor was dominated by Bacteroidetes, Firmicutes, and Actinobacteria at 129 d, with a higher abundance at 345 d. In addition, there was Methylomonas, an aerobic methanotroph, which disappeared at 345 d; the abundance of Methanosarcina and Methanobacterium was higher at 345 d. There were no anaerobic/aerobic methanotrophs detected during the rapid generation period of the short-chain fatty acids (SCFAs) while methanogen had a high abundance, which may be involved in the activation of methane. In brief, the relatively high abundance of methanogens (14.7%) and acid-producing bacteria (25.1%) suggests that both of them have the potential to co-convert methane to SCFAs [104,105]. Methane is converted by methanogens to some organic intermediates, which are further degraded by acid-producing bacteria to SCFAs [136], as shown in Figure 4.
In addition, another possibility is that the methanogen produces SCFAs directly via AOM. The methanogens can only oxidize methane during net methane production while producing trace amounts of CO2, a process known as trace methane oxidation (TMO). In Zehnder et al.’s experiments, the amount of oxidized methane only varies between 0.001% to 0.3% compared to the production of methane, but theoretically, the oxidation of methane into CO2 and hydrogen is exergonic under extreme conditions, namely a high partial pressure of methane (pCH4 ≥ 100 atm), a very low concentration of hydrogen (pH2 ≤ 10−5 atm), and a moderate bicarbonate buffered system (HCO3−~10−2 M) [137]. However, Valentine et al. [138] found that continuous H2 production and methane oxidation did not occur at a partial pressure of low H2 and that high CH4 might result from some methanogens preferring to use other electron donors. Methanosarcina, which is frequently discovered in the system, can oxidize methane to produce acetate or methanol [137] and form a progressive coupling with a variety of electron acceptors, such as nitrate [17], bromate [139], antimonat [140], and perchlorate [141,142], etc. Soo et al. [143] transferred the Mcr in ANME-1 to Methanosarcina acetivorans, which used methane to produce acetate via “the reverse methanogenesis pathway”, realizing the first pure culture of reverse methanogen. In addition to the reverse methanogenesis pathway, Caldewll et al. [144] summarized acetogenesis and methylogenesis as two AOM biochemical pathways; the former has two hypotheses: methanogen oxidizes two molecules of CH4 to produce H2 and acetate [138]; methanogen utilizes CO2 and CH4 to generate acetate [145]. The latter is the coupling between methanogen related to ANME-2d couples with SBR via the production of methyl-sulfides [146].
In addition to a large number of methanogens and acid-producing bacteria, some researchers have shown that the process also includes some aerobic methanotrophs, which oxidize methane as intermediates via the infiltration of oxygen within the system [17], and a large number of heterotrophic denitrifying bacteria that produce SCFAs and are used as electron donors of heterotrophic denitrifying bacteria for denitrification [101,103]. However, the enrichment of heterotrophic bacteria and the complete oxidation of methane in MBfR are harmful to the overall production of SCFAs. In addition, Chen et al. [147] found that the Chloroflexi phylum [148], which is able to oxidize CH4, may explain why CH4 is oxidized into intermediates for further anaerobic fermentation by acid-producing bacteria when methanogen (Methanosarcina, Methanobacterium, and Methanospirillum) are seldom.
Additionally, due to the infiltration of oxygen into the tube system, it is assumed that the nitrate and O2 of the system are both electron acceptors inverting CH4 to SCFAs whose rate of production is only 11% of the actual measurement. Therefore, it is speculated that CO2, as the potential electron acceptor, co-utilizes methane and bicarbonate with NO2−/NO3−, thus ensuring the thermodynamic feasibility of Equation (1).
Hadi et al. [149] proved that the process can be carried out in the direction of inverse thermodynamics with appropriate electron acceptors like Fe, identifying the range of the co-utilization of M. acetivorans in the presence of Fe in order to convert CH4, CO, and CO2 into liquid chemicals via metabolic modeling. Chen et al. [101] studied membrane biofilm reactors (MBfRs) containing nitrates, nitrites, low-rate O2 supply, and high-rate O2 supply, understanding that all the electron acceptors used could promote the conversion of methane to SCFAs, with carbon efficiency ranging from 7.9 ± 1.4% to 148.5 ± 1.3%, among which a carbon efficiency of more than 100% also indicates that CO2 may act as potential electron acceptor. In addition, the carbon efficiency and SCFA productivity are highest when O2 is supplied at a low rate. Moreover, Averesch [150] also proposed that methanogens could fix CO2 via the Wood–Ljungdahl pathway to form reductive acetyl CoA under anaerobic conditions, which is another parallel pathway of AOM besides Mcr-AOM; this realizes the co-utilization of CO2 and CH4 [143] and thus improves the yield of acetate [151]. In addition to co-utilization, the homoacetogens can reduce CO2 to acetate via autotrophic (H2) or heterotrophic (mainly sugar) means [152]. For autotrophy, the homoacetogens accomplish the process by passing via the carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) pathway, also referred to as the Wood/Ljungdhal pathway [153]. The hydrogen required for this process derives from the stripping of H+ when NAD(P)H is converted to NAD(P) [154]. In contrast to most hydrogen-producing microorganisms in anaerobic environments [155,156], Methylomonas sp. DH-1 can produce hydrogen from methane under low oxygen conditions, which is a reversible process, possibly due to the presence of hydrogenase and the effect of oxygen concentration, the production of hydrogen under low oxygen conditions, the absorption of hydrogen when oxygen levels increase, and the recovery of the hydrogen production capacity when conditions become slightly aerobic again. In addition, cell density affects the amount of hydrogen production [157]. Methylomicrobium alcaliphilum 20Z produces a certain amount of hydrogen via methane fermentation under micro-oxygen conditions, especially in vial incubations, with a hydrogen yield of up to 2237 ± 38 μmol·g·DCW−1. In addition, Alexey et al. [154] also found that Methylomicrobium buryatense 5GB1C has the ability to produce hydrogen from methane. In addition, by fixing CO2 to metabolic intermediates such as pyruvate or acetyl-CoA, and by further connecting other metabolic pathways, formate, acetate, lactate, succinate, H2 [158], and acetone [159], etc., can also be produced. Succinate is generated when CO2 is used as a carbonate source or when CH4 enters the TCA cycle via the Rump pathway. The supply and solubility of CO2 limit the production of succinate, which may lead to the formation of formate, lactate, and acetate, whereas MgCO3 and an exogenous hydrogen supply can increase the yield of succinate [160]. However, according to the study of Chen [147], the number of methanogens is low, but acid-producing bacteria and heterotrophic denitrifying bacteria occupy a dominant position, which is responsible for some carbon efficiency exceeding 100%; in particular, denitrifiers such as Pseudomonas, Azospira and Comamonas are involved in the production of SCFAs as they generate intracellular (glycogen, PHA, etc.) or extracellular compounds (EPS, etc.) [161,162,163].
5. Current and Potential Technologies for Understanding the MOD Process
The identification of functional groups and metabolic pathways are the two basic directions for understanding the process of MOD [10], in which using the pure culture is one of the significant methods by which to ascertain the process. On the one hand, methanotrophs can be enriched and cultured in the laboratory under similar living conditions [164]; on the other hand, individual cells, such as microbial fuel cells (MFCs) [165] can be isolated and artificial electron acceptors [166] that promote the decoupling of anaerobic methanotrophs from each other or from reducing bacteria such as SBR can be added. In addition, an optofluidic platform for the automated sorting of stable-isotope-probing-labeled microbial cells that combines microfluidics, optical tweezing [167], and Raman microspectroscopy and rRNA-targeted Magneto-FISH [168] provides a pretty good choice for separating anaerobic methanotrophs. However, the fact that anaerobic methanotrophs have not been purely cultivated might be mainly due to the long time required for incubation [2], but they presently live independently [166,169]. In addition, Ca. ‘M. oxyfera’ cannot synthesize cofactor PQQ, whose production needs to involve other microorganisms [170]. Therefore, it is necessary to explore the pure culture by adding artificial PQQ. Kalyuzhnaya et al. [171] used a new fluorescent dye, combining flow cytometry and cell sorting to isolate aerobic methanotrophs that had previously been uncultured. In addition, MMO, as a key enzyme required in methane activation, has been purified [172,173,174,175], while Mcr, which is involved in the N-DAMO process, has only been isolated and purified from thermophilic bacteria [68,176], and most studies focus on its redox morphology [176] and assembly [177]. Therefore, the purification and structural characterization of Mcr should be the direction of future research. However, the reliance on cultivation has always been detrimental to the full mastery of the process [10].
At present, PCR based on a clone library of 16S rRNA [61,94,178,179,180], pmoA [27,94,181], Mcr [182], niK/nirS [57,183], etc., and some combined techniques such as DNA-SIP [184], RNA-SIP [185], and PLFA-SIP [185], has been developed to a fairly mature level, and is regularly used to identify the main functional microorganisms of MOD. RNA-SIP has a higher sensitivity since RNA can be labeled by 13CH4 faster than DNA [186], but the sensitiveness of PLFA-SIP is highest and thus often utilized to confirm the presence of methanotrophs in an environment containing a high concentration of CH4 [187]. In addition, some functional genes (mxaF, nifH, fhcD) that are unique to non-methanotrophs are also employed for identification [188]. However, PCR based on pmoA ignores the existence of Verrucomicrobia due to the lack of some specific primers; this means the functional microorganisms of the community can only be ensured according to known specific functional genes in the bargain, while some dynamic processes, such as the other functions and metabolic pathways of microorganisms, cannot be determined.
Genome sequencing technology independent of PCR not only reveals all the genetic information of a certain organism, but also predicts many important functions of the organism, such as the potential capacity of some methanotrophs for denitrification [39,54] and that of methanogens for methane oxidation via the reverse methanogenesis pathway [149], etc. However, the process of MOD is usually completed by a biological community, part of which cannot however be separated and purified presently. Therefore, the use of metagenomic sequencing technology could obtain the whole genome of cultured/uncultured microorganisms in the community, which is favorable in terms of grasping the interaction between the species of a community and between species and the environment, as well as genetic characteristics and metabolic pathways [189,190]. However, the large and complete information acquisition method of metagenomics brings difficulties in subsequent analysis. Although the combination of DNA–SIP and metagenomics can significantly diminish the community complexity of whole-genome shotgun sequencing, the great cost of sequencing [191] and its inability to determine whether genes are expressed under actual conditions have been the bottleneck problems of metagenomic sequencing [10].
Meta-transcriptomics involves the random sequencing of the mRNA of microbial communities without primers, probes, and predicting key genes in advance; it encounters fewer errors when sequencing transcription products, which makes it widely applicable [192]. Dumont et al. [100] identified the major aerobic methanotrophs in lake sediments by obtaining a targeted transcriptome using SIP. In addition to the identification of strains, the main feature is meta-transcriptomics, which is used to produce an immediate regulatory response to the alteration of environmental conditions. However, mRNA molecules are not stable and the complete recovery of them from the environment is almost impossible [186]. To minimize the changes to the transcriptional profile that may result from sampling, it is indispensable to rapidly freeze the samples in liquid nitrogen or transfer them to an RNA preservation solution [193].
Although meta-proteomics has some technical shortcomings, for instance, it is impossible to achieve 100% efficiency regarding the extraction of protein from environmental samples, LC technology cannot completely separate protein fragments from the complex mixture when a false or missing assignment of MS/MS spectra to bioinformatic databases occurs [194]; meanwhile, it provides the most direct functional insight and previously unavailable information at a limited cost [195], making it a powerful tool for studies of the process [194,196,197,198]. However, at present, it is mainly focused on the traditional MOD process. Consequently, it should pay close attention to the research of methanogens, acid-producing bacteria, and novel metabolic mechanisms concerning intermediates in the future.
Metabolomics is an emerging discipline developed in the late 1990s that discusses the type, quantity, and change rule of endogenous metabolites after organisms are disturbed due to alterations in genes or the environment. Metabolomics is downstream of gene and protein network regulation, with fewer metabolites that can thus more directly reflect the state of the organism. The target or non-target analysis of metabolites, using methods such as NMR [199,200]/GC–MS [201,202]/LC–MS [203,204], can be used for the qualitative and quantitative detection of amino acids and SCFAs. Combining SIP or staining techniques with metagenomics, meta-transcriptomics, metabolomics, and other-omics approaches, as well as with advanced imaging technologies such as SEER-FISH [205] for the spatial distribution analysis of microbial communities, and with MALDI MSI [206] for visualizing changes in metabolite distribution, could be a highly effective research strategy for elucidating the metabolic regulatory networks of organisms. Moreover, Wang et al. [207] have developed a theoretical framework and mathematical model for analyzing the quantitative rules of microbial community assembly in terms of the metabolic division of labor. This framework may prove valuable in the quantitative analysis of the metabolic mechanisms and community structure within the context of methane oxidation dynamics.
6. Conclusions
The coupling process of methane oxidation and denitrification is a significant methane sink, with its importance lying in the biogeochemical significance of the methane and nitrogen cycles. At present, the understanding of this process is generally divided into two kinds: (1) methanotrophs act alone or cooperate with denitrifiers via electron transfer to complete the coupling process; (2) the process of MOD, mediated by intermediates including methanol, formaldehyde, formate, acetate, propionate, lactate, butyrate, succinate, PHB, citrate, iso-citrate, carbohydrate, protein, and nucleic acid, etc., emerges in the process of methane oxidation or fermentation. The intermediate-mediated AME-D process has been confirmed, but the generation of midbodies in the N-DAMO process is still in the initial stage of research. The combination of SIP and emerging biotechnology, advanced imaging techniques, and the establishment of related metabolic division models may be helpful in the exploration of this hidden mechanism.
7. Future Prospects
Regarding the current limitations and future key research directions of the MOD process, the authors put forward the following points:
- (1)
- The slow growth rate of anaerobic methanotrophs and the lack of a PQQ synthesis mechanism of Ca. ‘M. oxyfera’ are the reasons why it is difficult to conduct pure culture. Therefore, ways in which to improve environmental conditions or develop new biotechnology to promote their enrichment or even pure culture are the focus of future research. Moreover, Mcr, as a key enzyme in the catalytic anaerobic activation of methane, has only been purified in thermophilic bacteria, meaning that its purification and structural characterization in ANME-2d should be paid attention to.
- (2)
- The ratio of C/N, O2/CH4, the concentration of phosphate, and the pH, T, and other factors will have an impact on the production of intermediates in the process, among which O2/CH4 has a considerable effect on the AME-D. Although there is quite a lot of research on the AME-D mechanism under different O2/CH4 conditions, there is no clear and unified conclusion regarding the metabolic mechanism of AME-D under the influence of O2/CH4 due to the ambiguous O2 range and the differences in experimental conditions. Furthermore, with the decrease in the O2 concentration, methane oxidation gradually transitioned into a fermentation process. However, there is little knowledge about when and how to switch methane oxidation from the respiratory mode to the fermentation mode presently.
- (3)
- The acetyl-coA pathway of Ca. ‘M. nitroreducens’ or the endogenous oxygen production pathway of Ca. ‘M. oxyfera’ makes it possible to produce acetate under low concentrations of NO2− or NO3−. However, under some anaerobic/micro-aerobic conditions, with the proceeding of the anaerobic coupling process, some N-DAMO archaea and bacteria gradually disappeared; in contrast, a large number of methanogens, acid-producing bacteria and heterotrophic denitrifiers took their place when numerous acetates, propionates and other SCFAs accumulated. In addition, CO2, as a potential electron acceptor, may also be involved in the synthesis of SCFAs. All of this indicates that, apart from the previous mode of direct electron transfer, the N-DAMO process may be mediated by intermediate products such as SCFAs and thatCO2 might especially partake in the process, which makes the discussion of a new mechanism of great significance with regard to the global carbon cycle, the global climate and environmental change. Therefore, the identification of related strains and their roles, and whether CO2 is a potential electron acceptor involved in the production of intermediates, both deserve further investigation.
Author Contributions
Conceptualization, L.Z.; resources, L.Z.; writing—original draft preparation, X.-C.Z.; writing—review and editing, H.-S.L., Z.-H.W. and Z.-F.S.; supervision, L.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (No. U22A20443); the Fundamental Research Funds for the Central Universities (Grant No. HIT. OCEF. 2021031); the China Postdoctoral Science Foundation (No. 2018M640299); the Heilongjiang Postdoctoral Foundation (No. LBHZ18091), China; the State Key Laboratory of Urban Water Resource and Environment, Harbin Institute of Technology (No. 2021TS21), China; Open Project of Key Laboratory of Environmental Biotechnology, CAS (Grant No. kf2020009); Foundation from National Engineering Research Center for Bioenergy (Grant No. 2021B008), and Heilongjiang Touyan Innovation Team Program.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Van Amstel, A. Methane. A review. J. Integr. Environ. Sci. 2012, 9, 5–30. [Google Scholar] [CrossRef]
- Cai, C.; Zhang, X.; Wu, M.; Liu, T.; Lai, C.-Y.; Frank, J.; He, B.; Marcellin, E.; Guo, J.; Hu, S.; et al. Roles and opportunities for microbial anaerobic oxidation of methane in natural and engineered systems. Energy Environ. Sci. 2021, 14, 4803–4830. [Google Scholar] [CrossRef]
- Haque, M.F.U.; Crombie, A.T.; Murrell, J.C. Novel facultative Methylocella strains are active methane consumers at terrestrial natural gas seeps. Microbiome 2019, 7, 134. [Google Scholar] [CrossRef] [PubMed]
- Conrad, R. The global methane cycle: Recent advances in understanding the microbial processes involved: Global Methane Cycle. Environ. Microbiol. Rep. 2009, 1, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Bačėninaitė, D.; Džermeikaitė, K.; Antanaitis, R. Global Warming and Dairy Cattle: How to Control and Reduce Methane Emission. Animals 2022, 12, 2687. [Google Scholar] [CrossRef]
- Patel, S.K.; Shanmugam, R.; Kalia, V.C.; Lee, J.-K. Methanol production by polymer-encapsulated methanotrophs from simulated biogas in the presence of methane vector. Bioresour. Technol. 2020, 304, 123022. [Google Scholar] [CrossRef]
- Rhee, G.-Y.; Fuhs, G.W. Wastewater Denitrification with One-Carbon Compounds as Energy Source. J. WPCF 1978, 50, 2111–2119. [Google Scholar]
- Raghoebarsing, A.A.; Pol, A.; van de Pas-Schoonen, K.T.; Smolders, A.J.P.; Ettwig, K.F.; Rijpstra, W.I.C.; Schouten, S.; Damsté, J.S.S.; Camp, H.J.M.O.D.; Jetten, M.S.M.; et al. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 2006, 440, 918–921. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Knief, C. Diversity and Phylogeny of Described Aerobic Methanotrophs. In Methane Biocatalysis: Paving the Way to Sustainability; Kalyuzhnaya, M.G., Xing, X.-H., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 17–42. ISBN 978-3-319-74865-8. [Google Scholar]
- Zhu, J.; Wang, Q.; Yuan, M.; Tan, G.-Y.A.; Sun, F.; Wang, C.; Wu, W.; Lee, P.-H. Microbiology and potential applications of aerobic methane oxidation coupled to denitrification (AME-D) process: A review. Water Res. 2016, 90, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Strong, P.J.; Xie, S.; Clarke, W.P. Methane as a Resource: Can the Methanotrophs Add Value? Environ. Sci. Technol. 2015, 49, 4001–4018. [Google Scholar] [CrossRef]
- Haynes, C.A.; Gonzalez, R. Rethinking biological activation of methane and conversion to liquid fuels. Nat. Chem. Biol. 2014, 10, 331–339. [Google Scholar] [CrossRef]
- Strong, P.; Kalyuzhnaya, M.; Silverman, J.; Clarke, W. A methanotroph-based biorefinery: Potential scenarios for generating multiple products from a single fermentation. Bioresour. Technol. 2016, 215, 314–323. [Google Scholar] [CrossRef]
- Lee, O.K.; Hur, D.H.; Nguyen, D.T.N.; Lee, E.Y. Metabolic engineering of methanotrophs and its application to production of chemicals and biofuels from methane. Biofuels, Bioprod. Biorefining 2016, 10, 848–863. [Google Scholar] [CrossRef]
- Hwang, I.Y.; Nguyen, A.D.; Nguyen, T.T.; Nguyen, L.T.; Lee, O.K.; Lee, E.Y. Biological conversion of methane to chemicals and fuels: Technical challenges and issues. Appl. Microbiol. Biotechnol. 2018, 102, 3071–3080. [Google Scholar] [CrossRef]
- Gęsicka, A.; Oleskowicz-Popiel, P.; Łężyk, M. Recent trends in methane to bioproduct conversion by methanotrophs. Biotechnol. Adv. 2021, 53, 107861. [Google Scholar] [CrossRef]
- Chen, H.; Zhao, L.; Hu, S.; Yuan, Z.; Guo, J. High-Rate Production of Short-Chain Fatty Acids from Methane in a Mixed-Culture Membrane Biofilm Reactor. Environ. Sci. Technol. Lett. 2018, 5, 662–667. [Google Scholar] [CrossRef]
- Cai, C.; Shi, Y.; Guo, J.; Tyson, G.W.; Hu, S.; Yuan, Z. Acetate Production from Anaerobic Oxidation of Methane via Intracellular Storage Compounds. Environ. Sci. Technol. 2019, 53, 7371–7379. [Google Scholar] [CrossRef] [PubMed]
- Agler, M.T.; Spirito, C.M.; Usack, J.G.; Werner, J.J.; Angenent, L.T. Chain elongation with reactor microbiomes: Upgrading dilute ethanol to medium-chain carboxylates. Energy Environ. Sci. 2012, 5, 8189–8192. [Google Scholar] [CrossRef]
- Steinbusch, K.J.; Hamelers, H.V.; Buisman, C.J. Alcohol production through volatile fatty acids reduction with hydrogen as electron donor by mixed cultures. Water Res. 2008, 42, 4059–4066. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Shen, Y.; An, D.; Voordouw, G. Use of Acetate, Propionate, and Butyrate for Reduction of Nitrate and Sulfate and Methanogenesis in Microcosms and Bioreactors Simulating an Oil Reservoir. Appl. Environ. Microbiol. 2017, 83, e02983-16. [Google Scholar] [CrossRef]
- Whittenbury, R.; Phillips, K.C.; Wilkinson, J.F. Enrichment, Isolation and Some Properties of Methane-utilizing Bacteria. J. Gen. Microbiol. 1970, 61, 205–218. [Google Scholar] [CrossRef]
- Galloway, J.N.; Townsend, A.R.; Erisman, J.W.; Bekunda, M.; Cai, Z.; Freney, J.R.; Martinelli, L.A.; Seitzinger, S.P.; Sutton, M.A. Transformation of the Nitrogen Cycle: Recent Trends, Questions, and Potential Solutions. Science 2008, 320, 889–892. [Google Scholar] [CrossRef] [PubMed]
- Hanson, R.S.; Hanson, T. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar] [CrossRef] [PubMed]
- Fei, Q.; Guarnieri, M.T.; Tao, L.; Laurens, L.M.; Dowe, N.; Pienkos, P.T. Bioconversion of natural gas to liquid fuel: Opportunities and challenges. Biotechnol. Adv. 2014, 32, 596–614. [Google Scholar] [CrossRef]
- Stein, L.Y.; Roy, R.; Dunfield, P.F. Aerobic Methanotrophy and Nitrification: Processes and Connections. In eLS; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2012; ISBN 978-0-470-01617-6. [Google Scholar]
- Stoecker, K.; Bendinger, B.; Schöning, B.; Nielsen, P.H.; Nielsen, J.L.; Baranyi, C.; Toenshoff, E.R.; Daims, H.; Wagner, M. Cohn’s Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. Proc. Natl. Acad. Sci. USA 2006, 103, 2363–2367. [Google Scholar] [CrossRef]
- Hirayama, H.; Abe, M.; Miyazaki, M.; Nunoura, T.; Furushima, Y.; Yamamoto, H.; Takai, K. Methylomarinovum caldicuralii gen. nov., sp. nov., a moderately thermophilic methanotroph isolated from a shallow submarine hydrothermal system, and proposal of the family Methylothermaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.P.; Skerratt, J.H.; Nichols, P.D.; Sly, L.I. Phospholipid fatty acid and lipopolysaccharide fatty acid signature lipids in methane-utilizing bacteria. FEMS Microbiol. Lett. 1991, 85, 15–22. [Google Scholar] [CrossRef]
- Semrau, J.D.; DiSpirito, A.A.; Yoon, S. Methanotrophs and copper. FEMS Microbiol. Rev. 2010, 34, 496–531. [Google Scholar] [CrossRef]
- Pol, A.; Heijmans, K.; Harhangi, H.R.; Tedesco, D.; Jetten, M.S.M.; Op Den Camp, H.J.M. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 2007, 450, 874–878. [Google Scholar] [CrossRef] [PubMed]
- Stanley, S.H.; Prior, S.D.; Leak, D.J.; Dalton, H. Copper stress underlies the fundamental change in intracellular location of methane mono-oxygenase in methane-oxidizing organisms: Studies in batch and continuous cultures. Biotechnol. Lett. 1983, 5, 487–492. [Google Scholar] [CrossRef]
- Phelps, P.A.; Agarwal, S.K.; Speitel, G.E.; Georgiou, G. Methylosinus trichosporium OB3b Mutants Having Constitutive Expression of Soluble Methane Monooxygenase in the Presence of High Levels of Copper. Appl. Environ. Microbiol. 1992, 58, 3701–3708. [Google Scholar] [CrossRef] [PubMed]
- Bourne, D.G.; McDonald, I.R.; Murrell, J.C. Comparison of pmoA PCR Primer Sets as Tools for Investigating Methanotroph Diversity in Three Danish Soils. Appl. Environ. Microbiol. 2001, 67, 3802–3809. [Google Scholar] [CrossRef] [PubMed]
- Dunfield, P.F.; Yuryev, A.; Senin, P.; Smirnova, A.V.; Stott, M.B.; Hou, S.; Ly, B.; Saw, J.H.; Zhou, Z.; Ren, Y.; et al. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 2007, 450, 879–882. [Google Scholar] [CrossRef] [PubMed]
- Blake, C.; Ghosh, M.; Harlos, K.; Avezoux, A.; Anthony, C. The Active-Site of Methanol Dehydrogenase Contains a Disulfide Bridge Between Adjacent Cysteine Residues. Nat. Struct. Biol. 1994, 1, 557. [Google Scholar]
- Pol, A.; Barends, T.R.M.; Dietl, A.; Khadem, A.F.; Eygensteyn, J.; Jetten, M.S.M.; Op den Camp, H.J.M. Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ. Microbiol. 2014, 16, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Versantvoort, W.; Guerrero-Cruz, S.; Speth, D.R.; Frank, J.; Gambelli, L.; Cremers, G.; Van Alen, T.; Jetten, M.S.M.; Kartal, B.; Camp, H.O.D.; et al. Comparative Genomics of Candidatus Methylomirabilis Species and Description of Ca. Methylomirabilis Lanthanidiphila. Front. Microbiol. 2018, 9, 1672. [Google Scholar] [CrossRef]
- Khadem, A.F.; Wieczorek, A.S.; Pol, A.; Vuilleumier, S.; Harhangi, H.R.; Dunfield, P.F.; Kalyuzhnaya, M.G.; Murrell, J.C.; Francoijs, K.-J.; Stunnenberg, H.G.; et al. Draft Genome Sequence of the Volcano-Inhabiting Thermoacidophilic Methanotroph Methylacidiphilum fumariolicum Strain SolV. J. Bacteriol. 2012, 194, 3729–3730. [Google Scholar] [CrossRef] [PubMed]
- Lieberman, R.L.; Rosenzweig, A.C. Biological Methane Oxidation: Regulation, Biochemistry, and Active Site Structure of Particulate Methane Monooxygenase. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 147–164. [Google Scholar] [CrossRef] [PubMed]
- Kalyuzhnaya, M.G.; Puri, A.W.; Lidstrom, M.E. Metabolic engineering in methanotrophic bacteria. Metab. Eng. 2015, 29, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Naizabekov, S.; Lee, E.Y. Genome-Scale Metabolic Model Reconstruction and in Silico Investigations of Methane Metabolism in Methylosinus trichosporium OB3b. Microorganisms 2020, 8, 437. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.D.; Lee, E.Y. Engineered Methanotrophy: A Sustainable Solution for Methane-Based Industrial Biomanufacturing. Trends Biotechnol. 2021, 39, 381–396. [Google Scholar] [CrossRef]
- Fox, B.G.; Froland, W.A.; Dege, J.E.; Lipscomb, J. Methane monooxygenase from Methylosinus trichosporium OB3b. Purification and properties of a three-component system with high specific activity from a type II methanotroph. J. Biol. Chem. 1989, 264, 10023–10033. [Google Scholar] [CrossRef] [PubMed]
- Robertson, L.A.; van Niel, E.W.J.; Torremans, R.A.M.; Kuenen, J.G. Simultaneous Nitrification and Denitrification in Aerobic Chemostat Cultures of Thiosphaera pantotropha. Appl. Environ. Microbiol. 1988, 54, 2812–2818. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Chandran, K.; Stensel, D. Microbial ecology of denitrification in biological wastewater treatment. Water Res. 2014, 64, 237–254. [Google Scholar] [CrossRef] [PubMed]
- Sparacino-Watkins, C.; Stolz, J.F.; Basu, P. Nitrate and periplasmic nitrate reductases. Chem. Soc. Rev. 2014, 43, 676–706. [Google Scholar] [CrossRef]
- Warnecke-Eberz, U.; Friedrich, B. Three nitrate reductase activities in Alcaligenes eutrophus. Arch. Microbiol. 1993, 159, 405–409. [Google Scholar] [CrossRef]
- Zumft, W.G. Cell Biology and Molecular Basis of Denitrification. Microbiol. Mol. Biol. Rev. 1997, 61, 533–616. [Google Scholar]
- Yamanaka, T.; Ota, A.; Okunuki, K. A nitrite reducing system reconstructed with purified cytochrome components of Pseudomonas aeruginosa. Biochim. et Biophys. Acta 1961, 53, 294–308. [Google Scholar] [CrossRef]
- Holmes, A.J.; Costello, A.; Lidstrom, M.E.; Murrell, J.C. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 1995, 132, 203–208. [Google Scholar] [CrossRef]
- Elmore, B.O.; Bergmann, D.J.; Klotz, M.G.; Hooper, A.B. Cytochromes P460 and c′-beta; A new family of high-spin cytochromes c. FEBS Lett. 2007, 581, 911–916. [Google Scholar] [CrossRef]
- Tamas, I.; Smirnova, A.V.; He, Z.; Dunfield, P.F. The (d)evolution of methanotrophy in the Beijerinckiaceae—A comparative genomics analysis. ISME J. 2013, 8, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Orata, F.D.; Kits, K.D.; Stein, L.Y. Complete Genome Sequence of Methylomonas denitrificans Strain FJG1, an Obligate Aerobic Methanotroph That Can Couple Methane Oxidation with Denitrification. Genome Announc. 2018, 6, e00276-18. [Google Scholar] [CrossRef] [PubMed]
- Stein, L.Y.; Klotz, M.G. Nitrifying and denitrifying pathways of methanotrophic bacteria. Biochem. Soc. Trans. 2011, 39, 1826–1831. [Google Scholar] [CrossRef]
- Dam, B.; Dam, S.; Blom, J.; Liesack, W. Genome Analysis Coupled with Physiological Studies Reveals a Diverse Nitrogen Metabolism in Methylocystis sp. Strain SC2. PLoS ONE 2013, 8, e74767. [Google Scholar] [CrossRef]
- Kits, K.D.; Klotz, M.G.; Stein, L.Y. Methane oxidation coupled to nitrate reduction under hypoxia by the Gammaproteobacterium Methylomonas denitrificans, sp. nov. type strain FJG1. Environ. Microbiol. 2015, 17, 3219–3232. [Google Scholar] [CrossRef]
- Cheng, C.; Zhang, J.; He, Q.; Wu, H.; Chen, Y.; Xie, H.; Pavlostathis, S.G. Exploring simultaneous nitrous oxide and methane sink in wetland sediments under anoxic conditions. Water Res. 2021, 194, 116958. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.; Roy, R.; Knowles, R. Production and Consumption of Nitric Oxide by Three Methanotrophic Bacteria. Appl. Environ. Microbiol. 2000, 66, 3891–3897. [Google Scholar] [CrossRef]
- Iversen, N.; Jorgensen, B.B. Anaerobic methane oxidation rates at the sulfate-methane transition in marine sediments from Kattegat and Skagerrak (Denmark)1: Anaerobic Methane Oxidation. Limnol. Oceanogr. 1985, 30, 944–955. [Google Scholar] [CrossRef]
- Ettwig, K.F.; van Alen, T.; van de Pas-Schoonen, K.T.; Jetten, M.S.M.; Strous, M. Enrichment and Molecular Detection of Denitrifying Methanotrophic Bacteria of the NC10 Phylum. Appl. Environ. Microbiol. 2009, 75, 3656–3662. [Google Scholar] [CrossRef]
- Ettwig, K.F.; Butler, M.K.; Le Paslier, D.; Pelletier, E.; Mangenot, S.; Kuypers, M.M.; Schreiber, F.; Dutilh, B.E.; Zedelius, J.; de Beer, D.; et al. Nitrite-driven anaerobic me-thane oxidation by oxygenic bacteria. Nature 2010, 464, 543–548. [Google Scholar] [CrossRef]
- Wu, M.L.; van Teeseling, M.C.F.; Willems, M.J.R.; van Donselaar, E.G.; Klingl, A.; Rachel, R.; Geerts, W.J.C.; Jetten, M.S.M.; Strous, M.; van Niftrik, L. Ultrastructure of the Denitrifying Methanotroph “Candidatus Methylomirabilis oxyfera,” a Novel Polygon-Shaped Bacterium. J. Bacteriol. 2012, 194, 284–291. [Google Scholar] [CrossRef]
- Haroon, M.F.; Hu, S.; Shi, Y.; Imelfort, M.; Keller, J.; Hugenholtz, P.; Yuan, Z.; Tyson, G.W. Erratum: Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 2013, 501, 578. [Google Scholar] [CrossRef]
- Gambelli, L.; Guerrero-Cruz, S.; Mesman, R.J.; Cremers, G.; Jetten, M.S.M.; Camp, H.J.M.O.D.; Kartal, B.; Lueke, C.; van Niftrik, L. Community Composition and Ultrastructure of a Nitrate-Dependent Anaerobic Methane-Oxidizing Enrichment Culture. Appl. Environ. Microbiol. 2018, 84, e02186-17. [Google Scholar] [CrossRef]
- Liu, T.; Hu, S.; Yuan, Z.; Guo, J. Microbial Stratification Affects Conversions of Nitrogen and Methane in Biofilms Coupling Anammox and n-DAMO Processes. Environ. Sci. Technol. 2023, 57, 4608–4618. [Google Scholar] [CrossRef]
- Ettwig, K.F.; Shima, S.; van de Pas-Schoonen, K.T.; Kahnt, J.; Medema, M.H.; Camp, H.J.M.O.D.; Jetten, M.S.M.; Strous, M. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ. Microbiol. 2008, 10, 3164–3173. [Google Scholar] [CrossRef] [PubMed]
- Scheller, S.; Goenrich, M.; Boecher, R.; Thauer, R.K.; Jaun, B. The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 2010, 465, 606–608. [Google Scholar] [CrossRef] [PubMed]
- Nauhaus, K.; Boetius, A.; Kruger, M.; Widdel, F. In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ. Microbiol. 2002, 4, 296–305. [Google Scholar] [CrossRef]
- Timmers, P.H.A.; Welte, C.U.; Koehorst, J.J.; Plugge, C.M.; Jetten, M.S.M.; Stams, A.J.M. Reverse Methanogenesis and Respiration in Methanotrophic Archaea. Archaea 2017, 2017, 1654237. [Google Scholar] [CrossRef]
- He, B.; Cai, C.; McCubbin, T.; Muriel, J.C.; Sonnenschein, N.; Hu, S.; Yuan, Z.; Marcellin, E. A Genome-Scale Metabolic Model of Methanoperedens nitroreducens: Assessing Bioenergetics and Thermodynamic Feasibility. Metabolites 2022, 12, 314. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, T.; McIlroy, S.J.; Tyson, G.W.; Guo, J. Phylogenetic and metabolic diversity of microbial communities performing anaerobic ammonium and methane oxidations under different nitrogen loadings. ISME Commun. 2023, 3, 39. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.L.; de Vries, S.; van Alen, T.A.; Butler, M.K.; Camp, H.J.M.O.D.; Keltjens, J.T.; Jetten, M.S.M.; Strous, M. Physiological role of the respiratory quinol oxidase in the anaerobic nitrite-reducing methanotroph ‘Candidatus Methylomirabilis oxyfera’. Microbiology 2011, 157, 890–898. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.L.; Ettwig, K.F.; Jetten, M.S.; Strous, M.; Keltjens, J.T.; van Niftrik, L. A new intra-aerobic metabolism in the nitrite-dependent anaerobic methane-oxidizing bacterium Candidatus ‘Methylomirabilis oxyfera’. Biochem. Soc. Trans. 2011, 39, 243–248. [Google Scholar] [CrossRef]
- Fineberg, S.J.; Nandyala, S.V.; Marquez-Lara, A.; Oglesby, M.W.; Pelton, M.A.; Patel, A.A.; Singh, K. The Biochemistry of Methylotrophs. Comp. Biochem. Phys. A 1983, 75, 497. [Google Scholar]
- Xu, X.; Qin, Y.; Li, X.; Ma, Z.; Wu, W. Heterogeneity of CH4-derived carbon induced by O2:CH4 mediates the bacterial community assembly processes. Sci. Total. Environ. 2022, 829, 154442. [Google Scholar] [CrossRef]
- Zhu, J.; Xu, X.; Yuan, M.; Wu, H.; Ma, Z.; Wu, W. Optimum O2:CH4 Ratio Promotes the Synergy between Aerobic Methanotrophs and Denitrifiers to Enhance Nitrogen Removal. Front. Microbiol. 2017, 8, 1112. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Liu, X.; Li, N.; Xie, Z.; Li, Z.; Li, D. Stable-isotopic analysis and high-throughput pyrosequencing reveal the coupling process and bacteria in microaerobic and hypoxic methane oxidation coupled to denitrification. Environ. Pollut. 2019, 250, 863–872. [Google Scholar] [CrossRef]
- Osaka, T.; Ebie, Y.; Tsuneda, S.; Inamori, Y. Identification of the bacterial community involved in methane-dependent denitrification in activated sludge using DNA stable-isotope probing: Characterization of a MDD Ecosystem by SIP. FEMS Microbiol. Ecol. 2008, 64, 494–506. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.-H.; Chen, H.; Yuan, Z.; Guo, J. Methane-supported nitrate removal from groundwater in a membrane biofilm reactor. Water Res. 2018, 132, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Henard, C.A.; Smith, H.; Dowe, N.; Kalyuzhnaya, M.G.; Pienkos, P.T.; Guarnieri, M.T. Bioconversion of methane to lactate by an obligate methanotrophic bacterium. Sci. Rep. 2016, 6, 21585. [Google Scholar] [CrossRef]
- Vecherskaya, M.; Dijkema, C.; Stams, A.J.M. Intracellular PHB conversion in a Type II methanotroph studied by 13 C NMR. J. Ind. Microbiol. Biotechnol. 2001, 26, 15–21. [Google Scholar] [CrossRef]
- Nesterov, A.I.; Ivanova, T.I.; Il’Chenko, V.Y.; Gayazov, R.R.; Mshenskii, Y.N. Protein and Polysaccharide Composition during Growth of Methylomonas Methanica under Chemostat Conditions. Sov. Biotechnol. 1988, 5, 18–22. [Google Scholar]
- Eisentraeger, A.; Klag, P.; Vansbotter, B.; Heymann, E.; Dott, W. Denitrification of groundwater with methane as sole hydrogen donor. Water Res. 2001, 35, 2261–2267. [Google Scholar] [CrossRef] [PubMed]
- Linton, J.D.; Buckee, J.C. Interactions in a Methane-utilizing Mixed Bacterial Culture in a Chemostat. J. Gen. Microbiol. 1977, 101, 219–225. [Google Scholar] [CrossRef]
- Rahnama, F.; Vasheghani-Farahani, E.; Yazdian, F.; Shojaosadati, S.A. PHB production by Methylocystis hirsuta from natural gas in a bubble column and a vertical loop bioreactor. Biochem. Eng. J. 2012, 65, 51–56. [Google Scholar] [CrossRef]
- Liu, T.; Lu, Y.; Zheng, M.; Hu, S.; Yuan, Z.; Guo, J. Efficient nitrogen removal from mainstream wastewater through coupling Partial Nitritation, Anammox and Methane-dependent nitrite/nitrate reduction (PNAM). Water Res. 2021, 206, 117723. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.-J.; Zhang, H.; Li, W.; Yi, J.-B.; Sun, F.-Y.; Zhao, Y.-W.; Feng, L.; Li, Z.; Dong, W.-Y. Biofilm stratification in counter-diffused membrane biofilm bioreactors (MBfRs) for aerobic methane oxidation coupled to aerobic/anoxic denitrification: Effect of oxygen pressure. Water Res. 2022, 226, 119243. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.-J.; Shen, Q.; Li, X.-Y.; Sun, F.-Y.; Yi, J.-B.; Dong, W.-Y.; Yan, W.-J.; Du, H.; Mu, J.-L. Surface-manipulated membranes to accelerate biofilm formation and to resist bacterial detachment in MBfR for aerobic methane oxidation coupled to denitrification. Chem. Eng. J. 2022, 430, 132629. [Google Scholar] [CrossRef]
- Singh, A.K.; Nakhate, S.P.; Gupta, R.K.; Chavan, A.R.; Poddar, B.J.; Prakash, O.; Shouche, Y.S.; Purohit, H.J.; Khardenavis, A.A. Mining the landfill soil metagenome for denitrifying methanotrophic taxa and validation of methane oxidation in microcosm. Environ. Res. 2022, 215, 114199. [Google Scholar] [CrossRef]
- Beck, D.A.; Kalyuzhnaya, M.G.; Malfatti, S.; Tringe, S.G.; del Rio, T.G.; Ivanova, N.; Lidstrom, M.E.; Chistoserdova, L. A metagenomic insight into freshwater methane-utilizing communities and evidence for cooperation between the Methylococcaceae and the Methylophilaceae. PeerJ 2013, 1, e23. [Google Scholar] [CrossRef]
- Mechsner, K.L.; Hamer, G.; Paulis, A.D. Denitrification with Methanotrophic/Methylotrophic Bacterial Associations in the Presence of Oxygen. Site 1985, 1, 135–213. [Google Scholar]
- Costa, C.; Dijkema, C.; Friedrich, M.W.; Garcia-Encina, P.A.; Fernández-Polanco, F.; Stams, A.J.M. Denitrification with methane as electron donor in oxygen-limited bioreactors. Appl. Microbiol. Biotechnol. 2000, 53, 754–762. [Google Scholar] [CrossRef]
- Liu, J.; Sun, F.; Wang, L.; Ju, X.; Wu, W.; Chen, Y. Molecular characterization of a microbial consortium involved in methane oxidation coupled to denitrification under micro-aerobic conditions. Microb. Biotechnol. 2014, 7, 64–76. [Google Scholar] [CrossRef]
- Krause, S.M.B.; Johnson, T.; Karunaratne, Y.S.; Fu, Y.; Beck, D.A.C.; Chistoserdova, L.; Lidstrom, M.E. Lanthanide-dependent cross-feeding of methane-derived carbon is linked by microbial community interactions. Proc. Natl. Acad. Sci. USA 2017, 114, 358–363. [Google Scholar] [CrossRef]
- Nyerges, G.; Han, S.-K.; Stein, L.Y. Effects of Ammonium and Nitrite on Growth and Competitive Fitness of Cultivated Methanotrophic Bacteria. Appl. Environ. Microbiol. 2010, 76, 5648–5651. [Google Scholar] [CrossRef]
- Kalyuzhnaya, M.G.; Yang, S.; Rozova, O.N.; Smalley, N.E.; Clubb, J.; Lamb, A.; Gowda, G.A.N.; Raftery, D.; Fu, Y.; Bringel, F.; et al. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat. Commun. 2013, 4, 2785. [Google Scholar] [CrossRef]
- Rahalkar, M.; Bussmann, I.; Schink, B. Methylosoma difficile gen. nov., sp. nov., a novel methanotroph enriched by gradient cultivation from littoral sediment of Lake Constance. Int. J. Syst. Evol. Microbiol. 2007, 57, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.; Amaral, J.A.; Knowles, R. The response of methane consumption by pure cultures of methanotrophic bacteria to oxygen. Can. J. Microbiol. 1997, 43, 925–928. [Google Scholar] [CrossRef]
- Dumont, M.G.; Pommerenke, B.; Casper, P. Using stable isotope probing to obtain a targeted metatranscriptome of aerobic methanotrophs in lake sediment: SIP-Metatranscriptomics of Methanotrophs. Environ. Microbiol. Rep. 2013, 5, 757–764. [Google Scholar] [CrossRef]
- Chen, H.; Luo, J.; Liu, S.; Yuan, Z.; Guo, J. Microbial Methane Conversion to Short-Chain Fatty Acids Using Various Electron Acceptors in Membrane Biofilm Reactors. Environ. Sci. Technol. 2019, 53, 12846–12855. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Zheng, Y.; Hou, L.; Zhou, J.; Yin, G.; Liu, M. Denitrifying anaerobic methane oxidation in marsh sediments of Chongming eastern intertidal flat. Mar. Pollut. Bull. 2020, 150, 110681. [Google Scholar] [CrossRef]
- Zhao, L.; Chen, H.; Yuan, Z.; Guo, J. Interactions of functional microorganisms and their contributions to methane bioconversion to short-chain fatty acids. Water Res. 2021, 199, 117184. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.-N.; Zhang, F.; Yu, L.-P.; Zhang, Y.-L.; Wu, Y.; Lau, T.-C.; Zhao, H.-P.; Zeng, R.J. Acetate and electricity generation from methane in conductive fiber membrane- microbial fuel cells. Sci. Total. Environ. 2022, 804, 150147. [Google Scholar] [CrossRef]
- Guo, X.; Lai, C.-Y.; Hartmann, E.M.; Zhao, H.-P. Heterotrophic denitrification: An overlooked factor that contributes to nitrogen removal in n-DAMO mixed culture. Environ. Res. 2023, 216, 114802. [Google Scholar] [CrossRef]
- Modin, O.; Fukushi, K.; Yamamoto, K. Denitrification with methane as external carbon source. Water Res. 2007, 41, 2726–2738. [Google Scholar] [CrossRef]
- Costa, C.; Vecherskaya, M.; Dijkema, C.; Stams, A.J.M. The effect of oxygen on methanol oxidation by an obligate methanotrophic bacterium studied by in vivo 13 C nuclear magnetic resonance spectroscopy. J. Ind. Microbiol. Biotechnol. 2001, 26, 9–14. [Google Scholar] [CrossRef]
- Thalasso, F.; Vallecillo, A.; García-Encina, P.; Fdz-Polanco, F. The use of methane as a sole carbon source for wastewater denitrification. Water Res. 1997, 31, 55–60. [Google Scholar] [CrossRef]
- Takaya, N.; Catalan-Sakairi, M.A.B.; Sakaguchi, Y.; Kato, I.; Zhou, Z.; Shoun, H. Aerobic Denitrifying Bacteria That Produce Low Levels of Nitrous Oxide. Appl. Environ. Microbiol. 2003, 69, 3152–3157. [Google Scholar] [CrossRef]
- Cao, Q.; Liu, X.; Ran, Y.; Li, Z.; Li, D. Methane oxidation coupled to denitrification under microaerobic and hypoxic conditions in leach bed bioreactors. Sci. Total. Environ. 2019, 649, 1–11. [Google Scholar] [CrossRef]
- Chu, K.-H.; Alvarez-Cohen, L. Treatment of Chlorinated Solvents by Nitrogen-Fixing and Nitrate-Supplied Methane Oxidizers in Columns Packed with Unsaturated Porous Media. Environ. Sci. Technol. 2000, 34, 1784–1793. [Google Scholar] [CrossRef]
- Myronova, N.; Kitmitto, A.; Collins, R.F.; Miyaji, A.; Dalton, H. Three-Dimensional Structure Determination of a Protein Supercomplex That Oxidizes Methane to Formaldehyde in Methylococcus capsulatus (Bath). Biochemistry 2006, 45, 11905–11914. [Google Scholar] [CrossRef] [PubMed]
- Tonge, G.; Harrison, D.; Knowles, C.; Higgins, I. Properties and partial purification of the methane-oxidising enzyme system from Methylosinus trichosporium. FEBS Lett. 1975, 58, 293–299. [Google Scholar] [CrossRef]
- Leak, D.J.; Dalton, H. Growth Yields of Methanotrophs. Appl. Microbiol. Biotechnol. 1986, 23, 470–476. [Google Scholar] [CrossRef]
- Trotsenko, Y.A.; Murrell, J.C. Metabolic Aspects of Aerobic Obligate Methanotrophy⋆. In Advances in Applied Microbiology; Elsevier: Amsterdam, The Netherlands, 2008; Volume 63, pp. 183–229. ISBN 978-0-444-53191-9. [Google Scholar]
- Wang, S.; An, Z.; Wang, Z.-W. Bioconversion of methane to chemicals and fuels by methane-oxidizing bacteria. In Advances in Bioenergy; Elsevier: Amsterdam, The Netherlands, 2020; Volume 5, pp. 169–247. ISBN 978-0-12-820744-4. [Google Scholar]
- Sheets, J.P.; Ge, X.; Li, Y.-F.; Yu, Z.; Li, Y. Biological conversion of biogas to methanol using methanotrophs isolated from solid-state anaerobic digestate. Bioresour. Technol. 2016, 201, 50–57. [Google Scholar] [CrossRef]
- Wolfe, A.J. The Acetate Switch. Microbiol. Mol. Biol. Rev. 2005, 69, 12–50. [Google Scholar] [CrossRef] [PubMed]
- Furuto, T.; Takeguchi, M.; Okura, I. Semicontinuous methanol biosynthesis by Methylosinus trichosporium OB3b. J. Mol. Catal. A Chem. 1999, 144, 257–261. [Google Scholar] [CrossRef]
- Lee, S.G.; Goo, J.H.; Kim, H.G.; Oh, J.-I.; Kim, Y.M.; Kim, S.W. Optimization of methanol biosynthesis from methane using Methylosinus trichosporium OB3b. Biotechnol. Lett. 2004, 26, 947–950. [Google Scholar] [CrossRef]
- Markowska, A.; Michalkiewicz, B. Biosynthesis of methanol from methane by Methylosinus trichosporium OB3b. Chem. Pap. 2009, 63, 105–110. [Google Scholar] [CrossRef]
- Guerrero-Cruz, S.; Vaksmaa, A.; Horn, M.A.; Niemann, H.; Pijuan, M.; Ho, A. Methanotrophs: Discoveries, Environmental Relevance, and a Perspective on Current and Future Applications. Front. Microbiol. 2021, 12, 678057. [Google Scholar] [CrossRef]
- Su, Y.-C.; Sathyamoorthy, S.; Chandran, K. Bioaugmented methanol production using ammonia oxidizing bacteria in a continuous flow process. Bioresour. Technol. 2019, 279, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Rahalkar, M.C.; Bahulikar, R.A. Hemerythrins are widespread and conserved for methanotrophic guilds. Gene Rep. 2018, 11, 250–254. [Google Scholar] [CrossRef]
- Chen, K.H.-C.; Wu, H.-H.; Ke, S.-F.; Rao, Y.-T.; Tu, C.-M.; Chen, Y.-P.; Kuei, K.-H.; Chen, Y.-S.; Wang, V.C.-C.; Kao, W.-C.; et al. Bacteriohemerythrin bolsters the activity of the particulate methane monooxygenase (pMMO) in Methylococcus capsulatus (Bath). J. Inorg. Biochem. 2012, 111, 10–17. [Google Scholar] [CrossRef]
- Yu, Z.; Pesesky, M.; Zhang, L.; Huang, J.; Winkler, M.; Chistoserdova, L. A Complex Interplay between Nitric Oxide, Quorum Sensing, and the Unique Secondary Metabolite Tundrenone Constitutes the Hypoxia Response in Methylobacter. Msystems 2020, 5, e00770-19. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Cruz, S.; Cremers, G.; van Alen, T.A.; Op den Camp, H.J.M.; Jetten, M.S.M.; Rasigraf, O.; Vaksmaa, A. Response of the Anaerobic Methanotroph “Candidatus Methanoperedens nitroreducens” to Oxygen Stress. Appl. Environ. Microbiol. 2018, 84, e01832-18. [Google Scholar] [CrossRef] [PubMed]
- Strom, T.; Ferenci, T.; Quayle, J.R. The Carbon Assimilation Pathways of Methylococcus Capsulatus, Pseudomonas Methanica and Methylosinus Trichosporium (OB3B) during Growth on Methane. Biochem. J. 1974, 144, 76–465. [Google Scholar] [CrossRef]
- Vecherskaya, M.; Dijkema, C.; Saad, H.R.; Stams, A.J.M. Microaerobic and anaerobic metabolism of a Methylocystis parvus strain isolated from a denitrifying bioreactor: Microaerobic and Anaerobic Metabolism of a Methanotroph. Environ. Microbiol. Rep. 2009, 1, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, T.G.; Topiwala, H.H.; Hamer, G. Interactions in a mixed bacterial population growing on methane in continuous culture. Biotechnol. Bioeng. 1974, 16, 41–59. [Google Scholar] [CrossRef]
- Cuba, R.M.F.; Duarte, I.C.; Saavedra, N.K.; Varesche, M.B.A.; Foresti, E. Denitrification coupled with methane anoxic oxidation and microbial community involved identification. Braz. Arch. Biol. Technol. 2011, 54, 173–182. [Google Scholar] [CrossRef]
- Pieja, A.J.; Sundstrom, E.R.; Criddle, C.S. Cyclic, alternating methane and nitrogen limitation increases PHB production in a methanotrophic community. Bioresour. Technol. 2012, 107, 385–392. [Google Scholar] [CrossRef]
- Yan, C.; Mei, J.; Zhao, Y.-C. Aerobic Methanotrophs and Its Application in Engineering. Chin. J. Biotechnol. 2022, 38, 1322–1338. [Google Scholar]
- Zhang, Y.; Xin, J.; Chen, L.; Xia, C. The Methane Monooxygenase Intrinsic Activity of Kinds of Methanotrophs. Appl. Biochem. Biotechnol. 2009, 157, 431–441. [Google Scholar] [CrossRef]
- Zhu, G.-C.; Lu, Y.-Z.; Xu, L.-R. Effects of the carbon/nitrogen (C/N) ratio on a system coupling simultaneous nitrification and denitrification (SND) and denitrifying phosphorus removal (DPR). Environ. Technol. 2021, 42, 3048–3054. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Xia, Y.; Nielsen, P.H. Activity and identity of fermenting microorganisms in full-scale biological nutrient removing wastewater treatment plants. Environ. Microbiol. 2008, 10, 2008–2019. [Google Scholar] [CrossRef] [PubMed]
- Zehnder, A.J.; Brock, T.D. Methane formation and methane oxidation by methanogenic bacteria. J. Bacteriol. 1979, 137, 420–432. [Google Scholar] [CrossRef]
- Valentine, D.; Reeburgh, W.S. New perspectives on anaerobic methane oxidation. Environ. Microbiol. 2000, 2, 477–484. [Google Scholar] [CrossRef]
- Luo, J.-H.; Wu, M.; Yuan, Z.; Guo, J. Biological Bromate Reduction Driven by Methane in a Membrane Biofilm Reactor. Environ. Sci. Technol. Lett. 2017, 4, 562–566. [Google Scholar] [CrossRef]
- Lai, C.-Y.; Dong, Q.-Y.; Rittmann, B.E.; Zhao, H.-P. Bioreduction of Antimonate by Anaerobic Methane Oxidation in a Membrane Biofilm Batch Reactor. Environ. Sci. Technol. 2018, 52, 8693–8700. [Google Scholar] [CrossRef]
- Lv, P.-L.; Shi, L.-D.; Wang, Z.; Rittmann, B.; Zhao, H.-P. Methane oxidation coupled to perchlorate reduction in a membrane biofilm batch reactor. Sci. Total. Environ. 2019, 667, 9–15. [Google Scholar] [CrossRef]
- Xie, T.; Yang, Q.; Winkler, M.K.; Wang, D.; Zhong, Y.; An, H.; Chen, F.; Yao, F.; Wang, X.; Wu, J.; et al. Perchlorate bioreduction linked to methane oxidation in a membrane biofilm reactor: Performance and microbial community structure. J. Hazard. Mater. 2018, 357, 244–252. [Google Scholar] [CrossRef]
- Averesch, N.J.H.; Kracke, F. Metabolic Network Analysis of Microbial Methane Utilization for Biomass Formation and Upgrading to Bio-Fuels. Front. Energy Res. 2018, 6, 106. [Google Scholar] [CrossRef]
- Caldwell, S.L.; Laidler, J.R.; Brewer, E.A.; Eberly, J.O.; Sandborgh, S.C.; Colwell, F.S. Correction to “Anaerobic Oxidation of Methane: Mechanisms, Bioenergetics, And the Ecology of Associated Microorganisms”. Environ. Sci. Technol. 2010, 44, 3200. [Google Scholar] [CrossRef]
- Hoehler, T.M.; Alperin, M.J.; Albert, D.B.; Martens, C.S. Field and laboratory studies of methane oxidation in an anoxic marine sediment: Evidence for a methanogen-sulfate reducer consortium. Glob. Biogeochem. Cycles 1994, 8, 451–463. [Google Scholar] [CrossRef]
- Moran, J.J.; Beal, E.J.; Vrentas, J.M.; Orphan, V.J.; Freeman, K.H.; House, C.H. Methyl sulfides as intermediates in the anaerobic oxidation of methane. Environ. Microbiol. 2008, 10, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, S.; Liu, T.; Yuan, Z.; Guo, J. Efficient nitrate removal from synthetic groundwater via in situ utilization of short-chain fatty acids from methane bioconversion. Chem. Eng. J. 2020, 393, 124594. [Google Scholar] [CrossRef]
- Ward, L.M.; Shih, P.M.; Hemp, J.; Kakegawa, T.; Fischer, W.W.; McGlynn, S.E. Phototrophic Methane Oxidation in a Member of the Chloroflexi Phylum. Microbiology 2019, 26, 531582. [Google Scholar] [CrossRef]
- Nazem-Bokaee, H.; Maranas, C.D. A Prospective Study on the Fermentation Landscape of Gaseous Substrates to Biorenewables Using Methanosarcina acetivorans Metabolic Model. Front. Microbiol. 2018, 9, 1855. [Google Scholar] [CrossRef]
- De Souza, Y.P.A.; Rosado, A.S. Opening the Black Box of Thermophilic Autotrophic Bacterial Diversity. In Microbial Diversity in the Genomic Era; Elsevier: Amsterdam, The Netherlands, 2019; pp. 333–343. ISBN 978-0-12-814849-5. [Google Scholar]
- Nazem-Bokaee, H.; Gopalakrishnan, S.; Ferry, J.G.; Wood, T.K.; Maranas, C.D. Assessing methanotrophy and carbon fixation for biofuel production by Methanosarcina acetivorans. Microb. Cell Factories 2016, 15, 10. [Google Scholar] [CrossRef]
- Liu, C.; Ren, L.; Yan, B.; Luo, L.; Zhang, J.; Awasthi, M.K. Electron transfer and mechanism of energy production among syntrophic bacteria during acidogenic fermentation: A review. Bioresour. Technol. 2021, 323, 124637. [Google Scholar] [CrossRef]
- Hattori, S. Syntrophic Acetate-Oxidizing Microbes in Methanogenic Environments. Microbes Environ. 2008, 23, 118–127. [Google Scholar] [CrossRef]
- Gilman, A.; Fu, Y.; Hendershott, M.; Chu, F.; Puri, A.W.; Smith, A.L.; Pesesky, M.; Lieberman, R.; Beck, D.A.; Lidstrom, M.E. Oxygen-limited metabolism in the methanotroph Methylomicrobium buryatense 5GB1C. PeerJ 2017, 5, e3945. [Google Scholar] [CrossRef]
- Maintinguer, S.I.; Lazaro, C.Z.; Pachiega, R.; Varesche, M.B.A.; Sequinel, R.; de Oliveira, J.E. Hydrogen bioproduction with Enterobacter sp. isolated from brewery wastewater. Int. J. Hydrogen Energy 2017, 42, 152–160. [Google Scholar] [CrossRef]
- Kim, M.S.; Kim, H.-H.; Lee, K.-M.; Lee, H.-J.; Lee, C. Oxidation of microcystin-LR by ferrous-tetrapolyphosphate in the presence of oxygen and hydrogen peroxide. Water Res. 2017, 114, 277–285. [Google Scholar] [CrossRef]
- Jo, S.Y.; Na Rhie, M.; Jung, S.M.; Sohn, Y.J.; Yeon, Y.J.; Kim, M.-S.; Park, C.; Lee, J.; Park, S.J.; Na, J.-G. Hydrogen Production from Methane by Methylomonas sp. DH-1 under Micro-aerobic Conditions. Biotechnol. Bioprocess Eng. 2020, 25, 71–77. [Google Scholar] [CrossRef]
- Nisar, A.; Khan, S.; Hameed, M.; Nisar, A.; Ahmad, H.; Mehmood, S.A. Bio-conversion of CO2 into biofuels and other value-added chemicals via metabolic engineering. Microbiol. Res. 2021, 251, 126813. [Google Scholar] [CrossRef]
- Kremp, F.; Müller, V. Methanol and methyl group conversion in acetogenic bacteria: Biochemistry, physiology and application. FEMS Microbiol. Rev. 2021, 45, fuaa040. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.N.; Sarkar, O.; Chandrasekhar, K.; Raj, T.; Narisetty, V.; Mohan, S.V.; Pandey, A.; Varjani, S.; Kumar, S.; Sharma, P.; et al. Upgrading the value of anaerobic fermentation via renewable chemicals production: A sustainable integration for circular bioeconomy. Sci. Total. Environ. 2022, 806, 150312. [Google Scholar] [CrossRef] [PubMed]
- Myszka, K.; Czaczyk, K. Characterization of Adhesive Exopolysaccharide (EPS) Produced by Pseudomonas aeruginosa under Starvation Conditions. Curr. Microbiol. 2009, 58, 541–546. [Google Scholar] [CrossRef]
- Ye, F.; Ye, Y.; Li, Y. Effect of C/N ratio on extracellular polymeric substances (EPS) and physicochemical properties of activated sludge flocs. J. Hazard. Mater. 2011, 188, 37–43. [Google Scholar] [CrossRef]
- Su, J.F.; Yang, S.; Huang, T.L.; Li, M.; Liu, J.R.; Yao, Y.X. Enhancement of the denitrification in low C/N condition and its mechanism by a novel isolated Comamonas sp. YSF15. Environ. Pollut. 2020, 256, 113294. [Google Scholar] [CrossRef] [PubMed]
- Girguis, P.R.; Orphan, V.J.; Hallam, S.J.; DeLong, E.F. Growth and Methane Oxidation Rates of Anaerobic Methanotrophic Archaea in a Continuous-Flow Bioreactor. Appl. Environ. Microbiol. 2003, 69, 5472–5482. [Google Scholar] [CrossRef]
- Ding, J.; Lu, Y.-Z.; Fu, L.; Ding, Z.-W.; Mu, Y.; Cheng, S.H.; Zeng, R.J. Decoupling of DAMO archaea from DAMO bacteria in a methane-driven microbial fuel cell. Water Res. 2017, 110, 112–119. [Google Scholar] [CrossRef]
- Scheller, S.; Yu, H.; Chadwick, G.L.; McGlynn, S.E.; Orphan, V.J. Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science 2016, 351, 703–707. [Google Scholar] [CrossRef]
- Lee, K.S.; Palatinszky, M.; Pereira, F.C.; Nguyen, J.; Fernandez, V.I.; Mueller, A.J.; Menolascina, F.; Daims, H.; Berry, D.; Wagner, M.; et al. An automated Raman-based platform for the sorting of live cells by functional properties. Nat. Microbiol. 2019, 4, 902–903. [Google Scholar] [CrossRef]
- Trembath-Reichert, E.; Green-Saxena, A.; Orphan, V.J. Whole Cell Immunomagnetic Enrichment of Environmental Microbial Consortia Using RRNA-Targeted Magneto-FISH. In Microbial Metagenomics, Metatranscriptomics, and Metaproteomics; DeLong, E.F., Ed.; Elsevier Academic Press Inc.: San Diego, CA, USA, 2013; Volume 531, pp. 21–44. ISBN 978-0-12-407863-5. [Google Scholar]
- McGlynn, S.E.; Chadwick, G.L.; Kempes, C.P.; Orphan, V.J. Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 2015, 526, 531–535. [Google Scholar] [CrossRef]
- Welte, C.U.; Rasigraf, O.; Vaksmaa, A.; Versantvoort, W.; Arshad, A.; Camp, H.J.O.D.; Jetten, M.S.; Lüke, C.; Reimann, J. Nitrate- and nitrite-dependent anaerobic oxidation of methane. Environ. Microbiol. Rep. 2016, 8, 941–955. [Google Scholar] [CrossRef]
- Kalyuzhnaya, M.G.; Lidstrom, M.E.; Chistoserdova, L. Real-time detection of actively metabolizing microbes by redox sensing as applied to methylotroph populations in Lake Washington. ISME J. 2008, 2, 696–706. [Google Scholar] [CrossRef]
- Kitmitto, A.; Myronova, N.; Basu, P.; Dalton, H. Characterization and Structural Analysis of an Active Particulate Methane Monooxygenase Trimer from Methylococcus capsulatus (Bath). Biochemistry 2005, 44, 10954–10965. [Google Scholar] [CrossRef] [PubMed]
- Fuller, M.E.; Rezes, R.T.; Hedman, P.C.; Jones, J.C.; Sturchio, N.C.; Hatzinger, P.B. Biotransformation of the insensitive munition constituents 3-nitro-1,2,4-triazol-5-one (NTO) and 2,4-dinitroanisole (DNAN) by aerobic methane-oxidizing consortia and pure cultures. J. Hazard. Mater. 2021, 407, 124341. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.-N.; Yu, C.-L.; Ma, Q.-Q.; Li, S.-B. Direct Evidence for a Soluble Methane Monooxygenase from Type I Methanotrophic Bacteria: Purification and Properties of a Soluble Methane Monooxygenase fromMethylomonassp. GYJ3. Arch. Biochem. Biophys. 1997, 345, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Ukaegbu, U.E.; Henery, S.; Rosenzweig, A.C. Biochemical Characterization of MmoS, a Sensor Protein Involved in Copper-Dependent Regulation of Soluble Methane Monooxygenase. Biochemistry 2006, 45, 10191–10198. [Google Scholar] [CrossRef] [PubMed]
- Mahlert, F.; Bauer, C.; Jaun, B.; Thauer, R.K.; Duin, E.C. The nickel enzyme methyl-coenzyme M reductase from methanogenic archaea: In vitro induction of the nickel-based MCR-ox EPR signals from MCR-red2. JBIC J. Biol. Inorg. Chem. 2002, 7, 500–513. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Z.; Chou, C.-W.; Shi, H.; Wang, L.; Ghebreab, R.; Phillips, D.; Yan, Y.; Duin, E.C.; Whitman, W.B. Assembly of Methyl Coenzyme M Reductase in the Methanogenic Archaeon Methanococcus maripaludis. J. Bacteriol. 2018, 200, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Ding, Z.-W.; Fu, L.; Lu, Y.-Z.; Cheng, S.H.; Zeng, R.J. New primers for detecting and quantifying denitrifying anaerobic methane oxidation archaea in different ecological niches. Appl. Microbiol. Biotechnol. 2015, 99, 9805–9812. [Google Scholar] [CrossRef] [PubMed]
- Goevert, D.; Conrad, R. Effect of Substrate Concentration on Carbon Isotope Fractionation during Acetoclastic Methanogenesis by Methanosarcina barkeri and M. acetivorans and in Rice Field Soil. Appl. Environ. Microbiol. 2009, 75, 2605–2612. [Google Scholar] [CrossRef]
- Abou-Zeid, D.M.; Biebl, H.; Spröer, C.; Müller, R.-J. Propionispora hippei sp. nov., a novel Gram-negative, spore-forming anaerobe that produces propionic acid. Int. J. Syst. Evol. Microbiol. 2004, 54, 951–954. [Google Scholar] [CrossRef]
- Wang, S.; Liu, Y.; Liu, G.; Huang, Y.; Zhou, Y. A New Primer to Amplify pmoA Gene From NC10 Bacteria in the Sediments of Dongchang Lake and Dongping Lake. Curr. Microbiol. 2017, 74, 908–914. [Google Scholar] [CrossRef] [PubMed]
- Vaksmaa, A.; Jetten, M.S.M.; Ettwig, K.F.; Lüke, C. McrA primers for the detection and quantification of the anaerobic archaeal methanotroph ‘Candidatus Methanoperedens nitroreducens’. Appl. Microbiol. Biotechnol. 2017, 101, 1631–1641. [Google Scholar] [CrossRef]
- Priemé, A.; Braker, G.; Tiedje, J.M. Diversity of Nitrite Reductase (nirK and nirS) Gene Fragments in Forested Upland and Wetland Soils. Appl. Environ. Microbiol. 2002, 68, 1893–1900. [Google Scholar] [CrossRef] [PubMed]
- Bing, H.; Yin, C.; Guy, A.; Hao, J.; Levente, B.; Zhao, J.; Colin, M.J.; Xing, X.H. Diversity and Activity of Methanotrophs in Alkaline Soil from a Chinese Coal Mine. FEMS Microbiol. Ecol. 2010, 70, 196–207. [Google Scholar]
- He, R.; Wooller, M.J.; Pohlman, J.W.; Tiedje, J.M.; Leigh, M.B. Methane-derived carbon flow through microbial communities in arctic lake sediments. Environ. Microbiol. 2015, 17, 3233–3250. [Google Scholar] [CrossRef]
- Dumont, M.G.; Pommerenke, B.; Casper, P.; Conrad, R. DNA-, rRNA- and mRNA-based stable isotope probing of aerobic methanotrophs in lake sediment. Environ. Microbiol. 2011, 13, 1153–1167. [Google Scholar] [CrossRef]
- Neufeld, J.D.; Dumont, M.G.; Vohra, J.; Murrell, J.C. Methodological Considerations for the Use of Stable Isotope Probing in Microbial Ecology. Microb. Ecol. 2007, 53, 435–442. [Google Scholar] [CrossRef]
- McDonald, I.R.; Bodrossy, L.; Chen, Y.; Murrell, J.C. Molecular Ecology Techniques for the Study of Aerobic Methanotrophs. Appl. Environ. Microbiol. 2008, 74, 1305–1315. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, Z.; Xia, S.; Ding, N.; Long, X.; Wang, J.; Chen, M.; Ye, C.; Chen, S. Combined genome-centric metagenomics and stable isotope probing unveils the microbial pathways of aerobic methane oxidation coupled to denitrification process under hypoxic conditions. Bioresour. Technol. 2020, 318, 124043. [Google Scholar] [CrossRef]
- Lee, J.; Alrashed, W.; Engel, K.; Yoo, K.; Neufeld, J.D.; Lee, H.-S. Methane-based denitrification kinetics and syntrophy in a membrane biofilm reactor at low methane pressure. Sci. Total. Environ. 2019, 695, 133818. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Murrell, J.C. When metagenomics meets stable-isotope probing: Progress and perspectives. Trends Microbiol. 2010, 18, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Moran, M.A. Metatranscriptomics: Eavesdropping on Complex Microbial Communities. Microbe 2009, 4, 329–335. [Google Scholar] [CrossRef]
- Carvalhais, L.C.; Dennis, P.G.; Tyson, G.W.; Schenk, P.M. Application of metatranscriptomics to soil environments. J. Microbiol. Methods 2012, 91, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Hanson, B.T.; Madsen, E.L. In situ expression of nitrite-dependent anaerobic methane oxidation proteins by Candidatus Methylomirabilis oxyfera co-occurring with expressed anammox proteins in a contaminated aquifer. Environ. Microbiol. Rep. 2015, 7, 252–264. [Google Scholar] [CrossRef]
- Lue, F.; Bize, A.; Guillot, A.; Monnet, V.; Madigou, C.; Chapleur, O.; Mazéas, L.; He, P.; Bouchez, T. Metaproteomics of cellulose methanisation under thermophilic conditions reveals a surprisingly high proteolytic activity. ISME J. 2014, 8, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Skennerton, C.T.; Chourey, K.; Iyer, R.; Hettich, R.L.; Tyson, G.W.; Orphan, V.J. Methane-Fueled Syntrophy through Extracellular Electron Transfer: Uncovering the Genomic Traits Conserved within Diverse Bacterial Partners of Anaerobic Methanotrophic Archaea. mBio 2017, 8, e01561-17. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Okubo, T.; Kubota, K.; Kasahara, Y.; Tsurumaru, H.; Anda, M.; Ikeda, S.; Minamisawa, K. Metaproteomic Identification of Diazotrophic Methanotrophs and Their Localization in Root Tissues of Field-Grown Rice Plants. Appl. Environ. Microbiol. 2014, 80, 5043–5052. [Google Scholar] [CrossRef] [PubMed]
- Taubert, M.; Grob, C.; Crombie, A.; Howat, A.M.; Burns, O.J.; Weber, M.; Lott, C.; Kaster, A.-K.; Vollmers, J.; Jehmlich, N.; et al. Communal metabolism byMethylococcaceaeandMethylophilaceaeis driving rapid aerobic methane oxidation in sediments of a shallow seep near Elba, Italy. Environ. Microbiol. 2019, 21, 3780–3795. [Google Scholar] [CrossRef] [PubMed]
- Hindmarsh, J.P.; Awati, A.; Edwards, P.J.; Moughan, P. NMR-based metabonomics detection of differences in the metabolism of hydrolysed versus intact protein of similar amino acid profile. J. Sci. Food Agric. 2012, 92, 2013–2016. [Google Scholar] [CrossRef]
- Cui, M.; Trimigno, A.; Aru, V.; Khakimov, B.; Engelsen, S.B. Human Faecal 1H NMR Metabolomics: Evaluation of Solvent and Sample Processing on Coverage and Reproducibility of Signature Metabolites. Anal. Chem. 2020, 92, 9546–9555. [Google Scholar] [CrossRef] [PubMed]
- Putri, S.P.; Ikram, M.M.M.; Sato, A.; Dahlan, H.A.; Rahmawati, D.; Ohto, Y.; Fukusaki, E. Application of gas chromatography-mass spectrometry-based metabolomics in food science and technology. J. Biosci. Bioeng. 2022, 133, 425–435. [Google Scholar] [CrossRef]
- Gu, H.; Jasbi, P.; Patterson, J.; Jin, Y. Enhanced Detection of Short-Chain Fatty Acids Using Gas Chromatography Mass Spectrometry. Curr. Protoc. 2021, 1, e177. [Google Scholar] [CrossRef]
- Miyano, H.; Nakayama, A. Development of Precolumn Derivatization–LC/MS for Amino-Acid-Focused Metabolomics. Chromatography 2021, 42, 17–27. [Google Scholar] [CrossRef]
- McKay, M.J.; Castaneda, M.; Catania, S.; Charles, K.A.; Shanahan, E.; Clarke, S.J.; Engel, A.; Varelis, P.; Molloy, M.P. Quantification of short-chain fatty acids in human stool samples by LC-MS/MS following derivatization with aniline analogues. J. Chromatogr. B 2023, 1217, 123618. [Google Scholar] [CrossRef]
- Cao, Z.; Zuo, W.; Wang, L.; Chen, J.; Qu, Z.; Jin, F.; Dai, L. Spatial profiling of microbial communities by sequential FISH with error-robust encoding. Nat. Commun. 2023, 14, 1477. [Google Scholar] [CrossRef]
- Sgobba, E.; Daguerre, Y.; Giampà, M. Unravel the Local Complexity of Biological Environments by MALDI Mass Spectrometry Imaging. Int. J. Mol. Sci. 2021, 22, 12393. [Google Scholar] [CrossRef]
- Wang, M.; Chen, X.; Liu, X.; Fang, Y.; Zheng, X.; Huang, T.; Tang, Y.-Q.; Ackermann, M.; Nie, Y.; Wu, X.-L. Even allocation of benefits stabilizes microbial community engaged in metabolic division of labor. Cell Rep. 2022, 40, 111410. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).