Abstract
Pulque is a traditional Mexican fermented beverage associated with numerous health benefits. Over time, there has been considerable interest in studying the bacterial diversity of pulque, and microbial characterization has been carried out using traditional microbiological and molecular methods. Therefore, the objective of this research was to characterize the microbiota of artisanal pulque obtained from the Zacatlán region in Puebla, México, by the means of Illumina sequencing, and to compare it with the microbial diversity of aguamiel (sap before fermentation), commercial pulque (supplemented with additives to increase its yield), and its seed (batch of pulque previously fermented). An analysis of the Shannon index showed medium diversity for both aguamiel and pulque samples (score > 2), while the Chao 1 index exhibited a non-significant difference between them. On the other hand, a principal components analysis confirmed the role of the seed as an essential inoculum to define the microbial diversity of pulque, emphasizing the importance of its preservation as a quality standard during the elaboration process. In addition, results showed that the dominant phyla in artisanal and commercial pulque were Firmicutes and Bacteroidetes. As the fermentation process progressed, it was possible to observe an increase in the population of lactic acid bacteria (LAB) in both types of pulque compared to those detected in aguamiel. Of these, the species Lactobacillus, Leuconostoc, and Lactococcus represented almost 95% of the total LAB. Finally, even though the safety of pulque has been in question due to its non-aseptic manufacturing process, the present study confirmed that less than 1% of its microbiota corresponds to the genera with a pathogenic potential such as γ-proteobacteria (Enterobacter and Hafnia), which decreases as the fermentation process advances.
1. Introduction
Fermentation has been recognized since the beginning of human civilization as one of the most important biotechnological techniques used for the production and preservation of food and beverages [1]. According to the International Scientific Association for Probiotics and Prebiotics (ISAPP), fermented foods can be defined as foods and beverages produced through desired microbial growth and enzymatic conversions of its components in a controlled environment [2]. Currently, numerous fermented products are consumed around the world (kombucha and sauerkraut in China, kefir in Russia, miso in Japan, and kimchi in Korea), many of which are produced on a small scale and are exclusively for local consumption. However, other foods such as cheese, yogurt, wine, and beer have evolved on an industrial scale under strictly regulated production parameters as a result of technological advances in the food industry and a greater understanding of the role of microorganisms in the generation of unique textures and flavors during the fermentation process [3].
In Mexico, a broad spectrum of traditional fermented foods and beverages is produced from different raw materials depending on their region of origin. Some of the most prominent traditional fermented products include colonche, tepache, tesgüino, pozol, mezcal, tequila, bacanora, sotol, and pulque [4]. Particularly, pulque is a traditional Mexican alcoholic beverage that dates to pre-Hispanic times [5] and results from the fermentation of aguamiel, a sap excreted from maguey plants, which contains a high percentage of fermentable carbohydrates such as fructose (32.4%), glucose (26.5%), sucrose (8.8%), and fructo-oligosaccharides (10.2%) [4]. Additionally, aguamiel is considered a rich source of minerals (such as potassium, calcium, iron, and zinc), vitamins (C, B6, B2, and B1), γ-aminobutyric acid (GABA), and essential amino acids [1,6].
The elaboration of pulque starts with the castration of mature plants from 6 to 15 years old, relative to the maguey species [7]. A wide diversity of species of maguey can be used for the production of pulque, with Agave salmiana subsp. salmiana, Agave atrovirens, and Agave applanata subsp. ferox being the most frequently reported [8,9]. During this operation, the central leaves are cut to remove the thicker part known as the meyolote [10,11], and the remaining concavity is where the aguamiel is extracted. The next step consists of the addition of a portion of pulque to the freshly collected aguamiel to act as an inoculum, which is commonly referred to as a seed [5]. From this point, the fermentation begins, and it can vary from 12 to 24 h at 25 °C, depending on the quality of aguamiel, the seed maturity, and the producing region [5,10]. The pulque obtained is a white viscous alcoholic beverage (between 4 and 7° GL) [9] that acts as a significant source of probiotics, mainly lactic acid bacteria (LAB) [11].
Understanding the specific health-promoting properties of fermented beverages, such as pulque, provides a basis for evaluating how those properties are influenced by its method of production and its strain composition [12]. For example, Nazhand et al. [13] evaluated the current information on the therapeutic properties of fermented dairy and non-dairy beverages in terms of probiotic, prebiotic, and synbiotic activities, such as cardiovascular system enhancement, health effects on the digestive tract, improvement of the immune defense, antioxidant activity, and cancer prevention. Zhang and collaborators reported antioxidant and gut-microbiota-regulating activities of fermented milk enhanced with a probiotic system containing Lactobacillus plantarum YW11 [14]. Another study by Torres et al. [15] demonstrated the anti-inflammatory effects of L. plantarum LBH1064, L. sanfranciscensis LBH1068, and L. composti LBH1072 isolated from pulque, while Giles and collaborators [16] measured the antibacterial activity of Leuconostoc mesenteroides P45 isolated from this Mexican fermented beverage against Gram-positive and Gram-negative bacteria, attributing its effect to the production of bacteriocins. On the other hand, Jeong et al. [17] evaluated the therapeutic effect of an inactivated solution of L. rhamnosus in infants with atopic dermatitis, which resulted in a decreased expression of different proteins associated with inflammation. Similarly, Oliviero and Spinella [18] reported a mitigating effect of arthritis symptoms after administering a cell wall extract of probiotic microorganisms that can also be found in pulque, including the genera Bacillus, Leuconostoc, Enterococcus, Saccharomyces, and Kluyveromyces, among others.
Twenty years ago, research on the bacterial diversity of pulque was limited. However, in the last ten years, the number of identified bacterial genera has doubled as a result of the expansion of the research to cover aguamiel, the seed, and the tailstock (see Supplementary Table S1). Between 2004 and 2008, investigations by Escalante et al. [7,19] studied the presence of different microorganisms through an amplified rDNA restriction analysis (ARDRA) of 16S rDNA amplicons, which belonged to the genera Microbacterium, Chryseobacterium, Flavobacterium, Bacillus, Lactobacillus, Lactococcus, Leucostoc, Pediococcus, Streptococcus, Acetoacter, Acineto-bacter, Citrobacter, Enterobacter, Erwinia, Gluconoacter, Hafnia, Klyvera, Providencia, Serratia, Sterotrophomonas, and Zymomonas. By 2019, Villareal and collaborators [20] identified metagenomic populations in aguamiel using denaturing gradient gel electrophoresis and sequencing of the 16S rRNA gene and regions of the 26S rRNA gene, detecting the presence of Leuconostoc sp., Leuconostoc gelidum, Lactococcus lactis, Enterococcus casseliflavus, Pediococcus sp., Trichococcus sp. uncultivated, uncultivated Leuconostoc, and uncultivated Lactococcus. Finally, Rocha et al. [21] studied the microbiota present in aguamiel, the seed, the tailstock, and pulque by means of massive sequencing analysis, and identified 2855 operational taxonomic units (OTUs) of bacteria, including bacteria belonging to the genera Sphingomonas and Weisella.
Despite the efforts of the scientific community to emphasize the properties of pulque in recent years, there has been a reduction in the space dedicated to the cultivation of maguey with this purpose [22]. For example, just in the states of Hidalgo, Puebla, and Tlaxcala, there has been a total loss of up to 624.45 hectares dedicated to the cultivation of maguey accompanied by economic and social restrictions [8]. Additionally, the emergence of negative campaigns around the artisanal production of pulque have contributed to the loss of information about the maguey and its products, leaving all knowledge to the older generations [1]. Therefore, in this research, the characterization of the microbiota of two different pulques, one commercial and one artisanal, was carried out using Illumina massive sequencing to identify changes in the microbial diversity of this traditional fermented beverage after processing, and the relative abundance of each bacterial genus compared with that of aguamiel (before fermentation) and the seed (inoculum).
2. Materials and Methods
2.1. Samples Preparation and Collection
Samples of aguamiel, artisanal and commercial pulque were obtained from a local producer in Zacatlán de las Manzanas, Puebla, México (19°56′31.1″ N 98°03′53.7″ W). Aguamiel was harvested during June (summer season), and both commercial and artisanal pulque were produced using the same Agave salmiana plant sieve and the same seed. Pulque producers defined artisanal pulque as the fermented beverage without any additives, while commercial pulque was understood as a beverage supplemented with different additives to increase its yield. Samples of 100 mL of aguamiel, the seed, artisanal pulque, and commercial pulque were collected in triplicate into 15 mL sterile centrifuge tubes (Corning) and flash-frozen in liquid nitrogen before storage at −80 °C.
2.2. Bacterial Genomic DNA Extraction
Bacterial genomic DNA extraction was performed using the UltraClean® Microbial DNA Isolation Kit (Mo Bio Laboratories (San Diego, CA, USA), Catalog No. 12224), following the manufacturer’s instructions. This kit was chosen for its ability to efficiently isolate high-quality genomic DNA from Gram-positive and Gram-negative bacteria (UltraClean® Microbial DNA Isolation Kit Instruction Manual, [23]). For each extraction, 1.8 mL of each sample were used directly. The purity and concentration of the DNA extracts were evaluated by an agarose gel electrophoresis analysis and by its measurement at 260 and 280 nm using a Genova Nano spectrophotometer (Jenway®, Staffordshire, UK).
2.3. Microbiome Analysis
The hypervariable V3 and V4 regions of the 16S rRNA gene for bacteria were amplified from each sample using barcoded 806R primer, 515F primer, and a MyFi Mix (Bioline, MA, USA). A partial database is available in Zenodo (16S Illumina Sequencing of Aguamiel Microbiota, RRID: SCR_004129). The quality of the amplicons and potential contaminants were examined on an agarose gel. The amplicons were quantified using Picogreen (Invitrogen®, Eugene, OR, USA), according to the manufacturer’s protocol. DNA (240 ng) from each sample was pooled and cleaned using UltraClean PCR Clean-Up Kit (MoBio). The pooled library was quantified against a standard curve using the qPCR-based KAPA Library Quantification Kit. The library was diluted to a concentration of 4 nM, denatured with 0.2 N NaOH and diluted to a concentration 20 pM.
Due to limited sequence diversity among 16S rRNA amplicons, 5% of the PhiX (Illumina) control library made from phiX174 was added. A final concentration of 7 pM of the pooled 16S rRNA library was subjected to the paired-end sequencing using 2 × 150 pb V2 (Illumina), using custom primers. Sequencing was performed on the Illumina MiSeq (SN M02149, with the MiSeq Control Software v 2.5.0.5). After demultiplexing and filtering, 3,177,139 sequences remained, with a mean length of 252.8 ± 0.6889 bases. Demultiplexing and filtering were performed using the QIIME 1.9.11 software package. Sequences were assigned to operational taxonomic units (OTUs), with a similarity threshold of 97% using the uclust-based open reference OTU selection protocol of QIIME against the SILVA reference database (version 128), http://www.arb-silva.de/download/archive/qiime/ (accessed on 15 November 2016). Sequences that did not match the reference database were clustered de novo. The average number of sequences per sample was 263,686 ± 71,901 (min 166,257; max 413,526). All samples were analyzed with the sampling depth of 166,257 sequences. Diversity analysis was performed on the OTU tables, including α and β diversity, as well as the taxonomic summary implemented in the QIIME software package. The results were analyzed according to principal components using GraphPad Prism 9 (UPAEP License).
3. Results
3.1. Qualitative and Quantitative Characterization of the Bacterial Communities Present in Aguamiel, Pulque, and Seed Samples at the Genus Level
To characterize the bacterial community present in aguamiel, the seed, artisanal and commercial pulque at the genus level, Illumina sequencing of the V3 and V4 hypervariable regions of the 16S bacterial gene was performed. These results are shown in Figure 1 and Table 1. Qualitatively, Firmicutes was the dominant phylum found, followed by Bacteroidetes, and in lesser proportion, Proteobacteria (Figure 1). These data coincide with what was previously reported in the studies carried out by Escalante et al. [7,19] and Rocha et al. [21] using 16 rRNA amplicon analysis techniques. More specifically, the bacteria belonging to the phylum Firmicutes were mainly lactic acid bacteria from the families Lactobacillaceae, Leuconostocaceae, and Streptococcaceae, as well as bacteria belonging to the Lachnospiraceae and Erysipelotrichacea families. These last two have not been reported in previous studies, which can be attributed to the precision of Illumina sequencing, and local and regional variability in bacterial diversity.
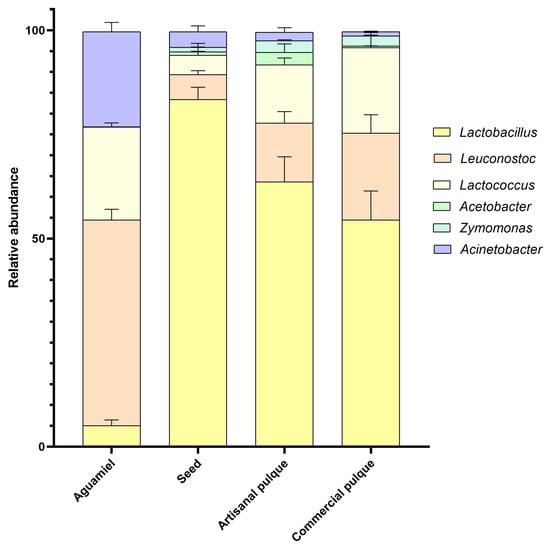
Figure 1.
Relative abundance of 16S amplicons assigned to the bacterial groups found in aguamiel, seed samples, artisanal and commercial pulque from the Zacatlán region. Data points are presented as the percentage of the total generated 16S sequences, and each datum point represents the average of three replicates.

Table 1.
Phylogenetic diversity of bacteria found in aguamiel, seed samples, artisanal pulque and commercial pulque from the Zacatlán.
On the other hand, the Proteobacteria detected in this research belong to the α and γ classes. However, authors such as Rocha et al. [21] and Enriquez et al. [23] have also identified Proteobacteria belonging to classes ϵ, δ, and β. More precisely, the α-Proteobacteria identified in this study include the genera Acetobacter and Zymomonas, while the γ-Proteobacteria comprised the genera Enterobacter, Hafnia, and Acinetobacter (Table 1). It is important to emphasize that some of the γ-Proteobacteria present in aguamiel are widely distributed in different environments, since it is obtained in the field and is exposed to multiple sources of contamination during fermentation. Similarly, the presence of Hafnia can be attributed to its adaptation to different ecological niches such as water, soil, food, and humans [24]. All these processes are considered risk factors, especially human manipulation. Likewise, after the collection of aguamiel, machinery, equipment, containers, domestic and wild animals, workers, dust in the atmosphere, and transport vehicles can influence its microbiota [25].
As the result of the use of next-generation sequencing, the abundance of Acinetobacter in aguamiel was quantified for the first time (22.8%), and was shown to be the second dominant genus, on par with Lactococcus (22.3%), superseded only by Leuconostoc (49.3%). Rocha et al. [21] reported the presence of additional α-Proteobacteria in pulque belonging to the genera Commensalibacter, Gluconacetobacter, Gluconobacter, Kaistobacter, Marivita, Mesorhizobium, Neoasaia, Novosphingobium, Rhodobacter, Rhodovulum, Sphingobium, Sphingomonas, and Sphhaingopyeniaxis. As mentioned before, some of the γ-Proteobacteria present in aguamiel come from different environments, since it is exposed to multiple sources of contamination during extraction and fermentation [26]. In this sense, some of the γ-Proteobacteria that have been reported in both aguamiel and pulque include Citrobacter, Erwinia, Glaciecola, Klebsiella, Kluyvera, Microbulbifer, Providencia, Pseudomonas, Serratia, Sterotrophomonas, and Yokenella [14,19,21].
It is important to emphasize that the presence of pathogenic microorganisms in aguamiel and in both types of pulque is lower than in other bacteria. As shown in Table 1, of the eight identified genera, three are potentially pathogenic (Enterobacter, Hafnia, and Acinetobacter). However, authors such as Aguado-Santacruz and collaborators [27] have reported that Acinetobacter bacteria such as A. baumannii and A. bereziniae are endophytic microorganisms of Agave, responsible for fixing atmospheric nitrogen. Likewise, there are no data on the microbiological safety of pulque, and information on the behavior of pathogenic microorganisms in both aguamiel and pulque is limited. Gómez [28] carried out a test on the behavior of five gastrointestinal pathogenic strains (Salmonella typhimurium, Staphylococcus aureus, Listeria monocytogenes, Shigella flexneri, and Shigella sonnei) during the fermentation of aguamiel from different producers and discarded a potential risk for consumers. Additionally, Huezo [29] analyzed the antibacterial effect of probiotic bacteria isolated from pulque and cultured in aguamiel against five pathogenic strains (S. typhimurium, E. coli, S. flexneri, S. aureus, and E. aerogenes), and concluded that these microorganisms have the ability to inhibit the growth of the pathogens in vitro, which shows that the probability of contracting a pathogen from the consumption of traditional beverages is low thanks to the presence of metabolites with antimicrobial potential produced by LAB [16,30].
Quantitatively, the genera Lactobacillus (5.07%), Leuconostoc (49.36%), and Lactococcus (22.33%) were the dominant LAB present in aguamiel (Supplementary Table S2). According to several studies on the microbiology of pulque, the presence of Lactobacillus spp., Leuconostoc spp., and Zymomonas is considered essential during fermentation [7,19]. As the fermentation process progressed, the presence of LAB increased for the artisan pulque, such that Lactobacillus reached a relative abundance of 63.61%, Leuconostoc 14.06%, and Lactococcus 14.03%. In the case of commercial pulque, its microbial diversity was slightly different from that of artisanal pulque, with 54.43% Lactobacillus, 20.85% Leuconostoc, and 20.52% Lactococcus (Supplementary Table S2). The increase in the relative abundance of LAB in pulque can be attributed to the seed, in which LAB represent 93.67% of its microbiota. Bacteria of the Lactobacillus genus are responsible for the production of lactic acid and acetic acid, which lower the final pH value of pulque, while favoring the formation of aromatic compounds that influence the quality and flavor [31]. Additionally, strains of this genus have been recognized for their ability to function as probiotics by inhibiting pathogens and restoring microbial homeostasis through microbe–microbe interactions, as well as through increased epithelial barrier function and modulation of the immune response [23]. On the other hand, bacteria from the Leuconostoc genera are responsible for the viscous fermentation of pulque [11], which confers additional benefits to the final product. For example, Leuconostoc mesenteroides P45 reported by Escalante [7] can produce antimicrobial compounds, with positive effects at the gastrointestinal level.
Furthermore, the genus Zymomonas was not found in aguamiel but in the seed, highlighting its role in the production of lactic acid, ethanol, and exopolysaccharides during fermentation [19]. Consequently, the presence of these bacteria increased in both types of pulque with no significant differences between the samples. Even though the presence of Zymomonas in the seed and in different pulque samples has been extensively documented [7,19,32], no other studies have been able to quantify its abundance in either substrate. In contrast, the incidence of Hafnia, Acinetobacter, and Enterobacter genera was reduced from 0.07%, 22.83%, and 0.04% in aguamiel, to 0.01%, 2.05%, and 0.01% in artisanal pulque, and to 0.01%, 1.02%, and 0.02% in commercial pulque (Supplementary Table S2). These results reinforce the safety of this traditional beverage, since the bacteria identified as potentially pathogenic are not capable of growing in pulque once it is fermented as a result of the acidification of the medium by lactic acid bacteria and the secretion of antimicrobial compounds by the latter [33].
3.2. Alpha Diversity of Bacterial Communities in Artisanal and Commercial Pulque, Aguamiel, and Seed Samples
To determine the effect that the fermentation process and the supplementation of commercial pulque can have on the alpha diversity of bacterial communities, the diversity (Shannon) and richness (Chao1) indexes were calculated. In particular, the rarefaction curves using the Chao1 index (Figure 2) showed that, within the studied research matrices, the total number of species does not differ in a statistically significant manner, which means that neither the fermentation process nor the use of additives in commercial pulque affects the microbiota abundance. On the contrary, when the alpha diversity of the samples was compared using the Shannon index (Figure 3), only aguamiel, artisanal, and commercial pulque showed similar scores (>2), which referred to a medium diversity count [34]. These Shannon indexes are slightly higher than those estimated by Sepulveda [35], whose values for aguamiel and pulque alpha diversity were less than 1.5 and 2, respectively. However, as it was mentioned before, microbial composition can be influenced by the quality of aguamiel, the species of Agave sieve, and the manipulation during collection and fermentation [5].
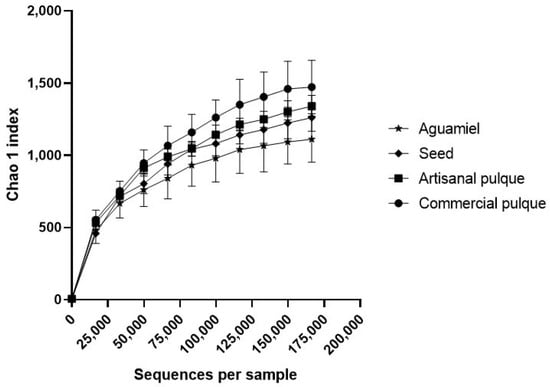
Figure 2.
Alpha diversity (Chao1 index) of bacteria in aguamiel, the seed, artisanal and commercial pulque. Rarefaction plot of alpha diversity using the Chao1 index in aguamiel (cross), seed (diamond), artisanal (square) and adulterated (circle) pulque. Each datum point represents the average of three independent replicates.
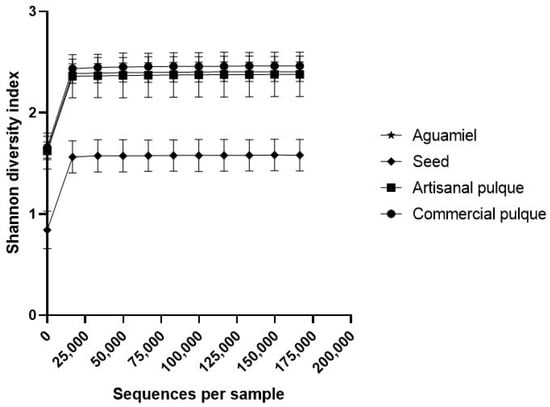
Figure 3.
Shannon diversity index in aguamiel, the seed, artisanal and adulterated pulque. Rarefaction plot of alpha diversity using the Shannon index in aguamiel (cross), seed (diamond), artisanal (square) and adulterated (circle) pulque. Each datum point represents the average of three independent replicates.
Finally, when comparing the seed samples to the rest of the research matrices, the former presented a significantly lower diversity score (less than 2) with a similar richness index. This phenomenon occurs because, of the total number of microorganisms identified in the seed, 83.37% belonged to the Lactobacillus genus, which has an effect on diversity, but not on abundance (Figure 1). These results are similar to those reported by Gamba et al. [36] after evaluating the bacterial richness and diversity in sugary kefir and its grains, since the Chao1 index was similar in all samples after seven fermentation cycles, while diversity was higher during the early stages as a result of changes in growth dynamics after metabolite accumulation and microbial interactions [11].
3.3. Interpretation of Statistics and Graphs for Principal Component Analysis
The principal component analysis indicates biome-specific differences among the bacterial communities in the studied samples. For the determination of the principal component (PC), data of the eigenvalue, proportion, accumulation, and sedimentation graph were considered. The accumulative PC values of the first two principal components explained 99.20% of the variance (as PCs must explain at least 90% of the variance), while the sedimentation graph showed that the first two PCs have eigenvalues greater than, equal to or close to 1. Based on the above, only PC1 (2.9851) explained 74.60% of the variation in the data, while PC2 explained only 24.60% of the total variation, and PC3 0.08%. In addition, the magnitude and direction of the coefficients of the original variables were examined, considering that the higher the absolute value of the coefficient is, the more important the corresponding variable is in the calculation of the component. The eigenvector analysis is shown in Figure 4, where each symbol represents one sample, and the percentage of the explained variance for each principal coordinate (PC) is indicated on the corresponding axis. In this sense, the seed, artisanal, and commercial pulque have large positive influences on PC1, which brings to view that the seed plays an important role as an inoculum in the diversity and microbial load during the fermentation process of aguamiel to pulque. Meanwhile, aguamiel has a large negative influence on PC2 and a very low positive influence on PC1, which means that the highest percentage of microbial load that is found in artisanal and commercial pulque must come from the seed.
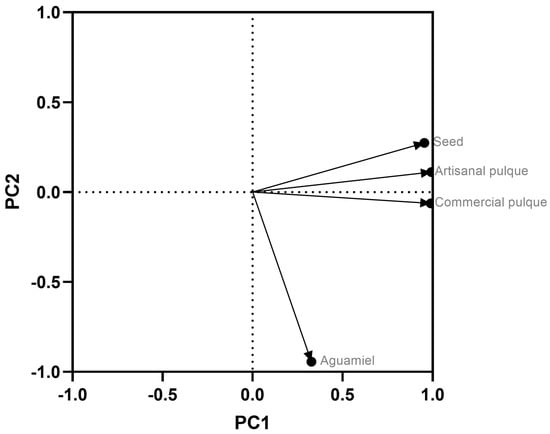
Figure 4.
Principal coordinate analysis (PCoA) plot using the weighted UniFrac metric between all the samples. Aguamiel, seed, artisanal and commercial pulque samples are represented. Each symbol represents one sample. The percentage of explained variance for each principal coordinate (PC) is indicated on the corresponding axis.
Even though aseptic conditions during pulque production have been questioned in the past [35,36], the results of this research confirm the prevalence of beneficial lactic acid bacteria over pathogenic microorganisms, since their presence does not exceed 24, 4, 3, and 2% of the population screened in the seed, aguamiel, artisanal, and commercial pulque, respectively (Figure 5). This is mainly due to the accumulation of various fermentation by-products, such as organic acids, hydrogen peroxide, and bacteriocins, which act as biopreservative agents leading to the eradication of putrefactive and pathogenic bacteria, thus improving food safety [37,38]. For example, the antimicrobial potential of the heterofermentative bacteria Lactobacillus kefiri isolated from a fresh sample of kefir has been reported, since its surface proteins exert a protective action against Salmonella enterica invasion [31]. However, not all fermentative conditions favor the prevalence of LAB over pathogenic microorganisms. This was demonstrated by Choi et al. [25], as they discovered that kimchi could be a vector for pathogens because it is often consumed before sufficient fermentation has taken place, so bacteria such as E. coli O157:H7, Salmonella enteritidis, Staphylococcus aureus, and Listeria monocytogenes can grow and survive on this substrate.
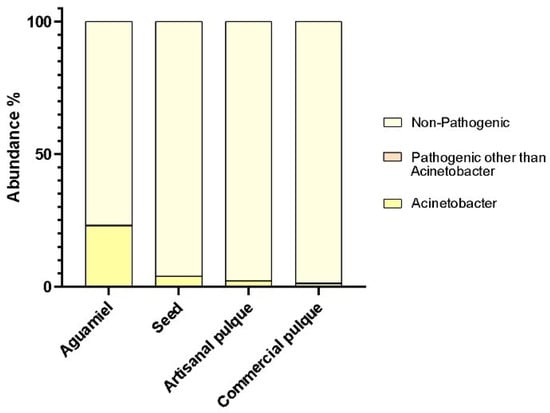
Figure 5.
Abundance of pathogenic and non-pathogenic bacteria in Agave products. The relative abundance of non-pathogenic bacteria, pathogenic bacteria other than Acinetobacter, and Acinetobacter bacteria in aguamiel, the seed, artisanal and commercial pulque is shown.
4. Conclusions
Pulque is a traditional Mexican fermented beverage characterized by its content of probiotic bacteria and prebiotic substrates. However, since it is produced under non-aseptic conditions, in recent years, there have been questions about its safety. Therefore, it is important to identify the microbial composition of fermented foods and beverages to understand their health benefits and eradicate prejudices about their consumption. In this sense, the present study demonstrated that the native microbiota of pulque is constituted mostly of lactic acid bacteria, with the species Lactobacillus, Leuconostoc, and Lactococcus representing almost 95% of the total LAB, while less than 1% corresponded to genera with pathogenic potential, demonstrating that its consumption does not represent a risk to human health. Similarly, the microbiota of aguamiel was mainly constituted by Lactobacillus (5%), Leuconostoc (49%), Lactococcus (22.33%), and Acinetobacter bacteria (23%). However, the microbial composition of pulque was shown to be very similar to that of the seed (previously fermented batch of pulque used as inoculum) when comparing it to the microbial composition of aguamiel, which emphasizes the importance of the preservation of the seed and its use as a quality standard during the production of artisanal and commercial pulque.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation9060564/s1, Table S1: Bacterial genera reported over time in aguamiel and pulque; Table S2: Relative abundance of 16S amplicons assigned to bacterial groups found in aguamiel, seed samples, artisanal and commercial pulque from the Zacatlán region. Data points are presented as the percentage of total generated 16S sequences, and each data point represents the average of three replicates.
Author Contributions
B.P.-A., research leader, provided general supervision and wrote the manuscript. E.G.E.-K. designed and performed the experimental work and wrote the initial version of the manuscript. A.R.H.-S. PhD student, carried out the data analysis, wrote the manuscript and designed the graphic material. E.M.O.-R. MSc student, wrote the manuscript and designed the graphic material. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Original datasets are available in Zenodo (16S Illumina Sequencing of Aguamiel Microbiota, https://doi.org/10.5281/zenodo.7379079, RRID:SCR_004129), accessed on 15 November 2022.
Acknowledgments
The authors would like to acknowledge the valuable contribution of the pulque producers of the Zacatlán region in Puebla.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial relationships that could be construed as potential conflict of interest.
References
- Pérez, B.; Cardoso, G. Traditional fermented beverages in Mexico: Biotechnological, nutritional, and functional approaches. Food Res. Int. 2020, 136, 109307. [Google Scholar] [CrossRef] [PubMed]
- Marco, M.L.; Sanders, M.E.; Gänzle, M.; Arrieta, M.C.; Cotter, P.D.; De Vuyst, L.; Hill, C.; Holzapfel, W.; Lebeer, S.; Merenstein, D.; et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on fermented foods. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Blandino, A.; Al-Aseeri, M.; Pandiella, S.; Cantero, D.; Webb, C. Cereal-based fermented foods and beverages. Food Res. Int. 2003, 36, 527–543. [Google Scholar] [CrossRef]
- Ojeda, C.; Álvarez, G.; Figueredo, C.; Islas, L.; Lappe, P.; Nabjan, G.; Torres, I.; Vallejo, M.; Casas, A. Traditional Fermented Beverages of Mexico: A Biocultural Unseen Foodscape. Foods 2021, 10, 2390. [Google Scholar] [CrossRef]
- Velasco, A.; Pedroza, A. Proceso de elaboración del pulque, su importancia económica y concepción social en Apan, Hidalgo. In Ejercicios Etnográficos. Aprendiendo a Investigar; Ramos, J., Ed.; Instituto Nacional de Antropología e Historia: México Distrito Federal, Mexico, 2013; pp. 59–103. [Google Scholar]
- Romero, M.; Osorio, P.; Flores, A.; Robledo, N.; Mora, R. Composición química, capacidad antioxidante y el efecto prebiótico del aguamiel (Agave atrovirens) durante su fermentación in vitro. Rev. Mex. Ing. Quim. 2015, 14, 281–292. [Google Scholar]
- Escalante, A.; Rodríguez, M.; Martínez, M.; López, A.; Bolivar, F.; Gosset, G. Characterization of bacterial diversity in pulque, a traditional Mexican alcoholic fermented beverage, as determined by 16S rDNA analysis. FEMS Microbiol. Lett. 2004, 19, 273–279. [Google Scholar] [CrossRef]
- Álvarez, M.; García, E.; Suárez, J.; Luna, M.; Rodríquez, M. Conocimiento tradicional, cultivo y aprovechamiento del maguey pulquero en municipios de Puebla y Tlaxcala. Polibotánica 2018, 45, 205–222. [Google Scholar]
- Mora-López, J.L.; Reyes-Agüero, J.A.; Flores-Flores, J.L.; Peña-Valdivia, C.B.; Aguirre-Rivera, J.R. Variación morfológica y humanización de la sección Salmianae del género Agave. Agrociencia 2011, 45, 465–477. [Google Scholar]
- Cervantes, M. El pulque: Características microbiológicas y contenido alcohólico mediante espectroscopia Raman. Nova 2007, 5, 135–146. [Google Scholar] [CrossRef]
- Escalante, A.; López, D.; Velázquez, J.; Giles, M.; Bolivar, F.; López, A. Pulque, a traditional Mexican alcoholic fermented beverage: Historical, microbiological, and technical aspects. Front. Microbiol. 2016, 7, 1026. [Google Scholar] [CrossRef]
- Loyola-Montemayor, E. La Industria del Pulque; Banco de México: Ciudad de México, Mexico, 2013. [Google Scholar]
- Nazhand, A.; Souto, E.B.; Lucarini, M.; Souto, S.B.; Durazzo, A.; Santini, A. Ready to Use Therapeutical Beverages: Focus on Functional Beverages Containing Probiotics, Prebiotics and Synbiotics. Beverages 2020, 6, 26. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, X.; Jiang, Y.; Zhao, W.; Guo, T.; Cao, Y.; Teng, J.; Hao, X.; Zhao, J.; Yang, Z. Antioxidant status and gut microbiota change in an aging mouse model as influenced by exopolysaccharide produced by Lactobacillus plantarum YW11 isolated from Tibetan kefir. Dairy Sci. 2017, 100, 6025–6041. [Google Scholar] [CrossRef]
- Torres, E.; Lenoir, M.; Mayorga, L.; Allain, T.; Sokol, H.; Langella, P.; Sánchez, M.; Bermúdez, L. Identification of novel anti-inflammatory probiotic strains isolated from pulque. Appl. Microbiol. Biotechnol. 2016, 100, 385–396. [Google Scholar] [CrossRef]
- Giles, M.; Sandoval, J.; Matus, V.; Campos, I.; Bolivar, F.; Escalante, A. In vitro and in vivo probiotic assessment of Leuconostoc mesenteroides P45 isolated from pulque, a Mexican traditional alcoholic beverage. SpringerPlus 2016, 5, 708. [Google Scholar] [CrossRef]
- Jeong, K.; Kim, M.; Jeon, S.A.; Kim, Y.H.; Lee, S. A randomized trial of Lactobacillus rhamnosus IDCC 3201 tyndallizate (RHT3201) for treating atopic dermatitis. Pediatr. Allergy Immunol. 2020, 31, 783–792. [Google Scholar] [CrossRef]
- Oliviero, F.; Spinella, P. Benefits of Probiotics in Rheumatic Disease. Front. Nutr. 2020, 7, 157. [Google Scholar] [CrossRef] [PubMed]
- Escalante, A.; Giles, M.; Hernández, G.; Córdova, M.; López, A.; Gosset, G.; Bolicar, F. Analysis of bacterial community during the fermentation of pulque, a traditional Mexican alcoholic beverage, using a polyphasic approach. Int. J. Food Microbiol. 2008, 12, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Villareal, S.; Enríquez, M.; Michel, M.; Flores, C.; Montañez, J.; Aguilar, C.; Rodriguez, R. Metagenomic microbial diversity in aguamiel from two Agave species during 4-year seasons. Food Biotechnol. 2019, 33, 1–16. [Google Scholar] [CrossRef]
- Rocha, C.; Espinal, A.; Martinez, S.; Caballero, J.; Alcaraz, L.; Cruz, A. Deep microbial community profiling along the fermentation process of pulque, a biocultural resource of Mexico. Microbiol. Res. 2020, 241, 126593. [Google Scholar] [CrossRef]
- Rojas, E.; Viesca, F.; Espeitx, E.; Quintero, B. El maguey, el pulque y las pulquerías de Toluca, Estado de México, ¿patrimonio Gastronómico turístico? PASOS 2016, 14, 1199–1215. [Google Scholar] [CrossRef]
- Enríquez-Salazar, M.I.; Veana, F.; Aguilar, C.N.; De la Garza, I.M.; López, M.G.; Rutiaga-Quiñones, O.M.; Morlett-Chávez, J.A.; Rodriguez-Herrera, R. Microbial diversity and biochemical profile of aguamiel collected from Agave salmiana and A. atrovirens during different seasons of year. Food Sci. Biotechnol. 2017, 26, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M. Hafnia alvei. Rev. Chil. Infectol. 2009, 26, 355. [Google Scholar] [CrossRef]
- Choi, Y.; Lee, S.; Kim, H.J.; Lee, H.; Kim, S.; Lee, J.; Ha, J.; Oh, H.; Choi, K.-H.; Yoon, Y. Pathogenic Escherichia coli and Salmonella Can Survive in Kimchi during Fermentation. J. Food Prot. 2018, 81, 17–459. [Google Scholar] [CrossRef]
- Carasi, P.; Díaz, M.; Racedo, S.M.; De Antoni, G.; Urdaci, M.C.; Serradell, M.A. Safety Characterization and Antimicrobial Properties of Kefir-Isolated Lactobacillus kefiri. BioMed Res. Int. 2014, 2014, 208974. [Google Scholar] [CrossRef] [PubMed]
- Aguado-Santacruz, G.A.; Aguado-Rodríguez, D.L.; Moreno-Gómez, B.; Arroyo-González, D.; Centeno-Jamaica, D.; Aguirre-Mancilla, C.; García-Moya, E. Endomicrobiota bacteriana de agave pulquero (Agave salmiana): Aislamiento, frecuencia e identificación molecular. Rev. Fitotec. Mex. 2022, 45, 243. [Google Scholar] [CrossRef]
- Gómez-Aldapa, C.A.; Díaz-Cruz, C.A.; Villarruel-López, A.; Torres-Vitela, R.; Añorve-Morga, J.; Rangel-Vargas, E.; Cerna-Cortes, J.F.; Vigueras-Ramírez, J.G.; Castro-Rosas, J. Behavior of Salmonella Typhimurium, Staphylococcus aureus, Listeria monocytogenes, and Shigella flexneri and Shigella sonnei during production of pulque, a traditional Mexican beverage. J. Food Prot. 2011, 74, 580–587. [Google Scholar] [CrossRef]
- Huezo, A. Efecto Antagónico de Bebida Synbiotic Elaborada a Base de Aguamiel. Master’s Thesis, Universidad Popular Autónoma del Estado de Puebla, Puebla, Mexico, 2020. [Google Scholar]
- Gavrilova, E.; Anisimova, E.; Gabdelkhadieva, A.; Nikitina, E.; Vafina, A.; Tarullina, D.; Bogachev, M.; Katumov, A. Newly isolated lactic acid bacteria from silage targeting biofilms of foodborne pathogens during milk fermentation. BMC Microbiol. 2019, 19, 248. [Google Scholar] [CrossRef]
- Álvarez, G.; Pacheco, F.; Figueredo, C.; Casas, A. Management, morphological and genetic diversity of domesticated agaves in Michoacán, México. J. Ethnobiol. 2020, 16, 3. [Google Scholar] [CrossRef]
- Chacón, K.; Torres, J.; Giles, M.; Escalante, A.; Gibbson, J. Genomic profiling of bacterial and fungal communities and their productive functionally during pulque fermentation by whole-genome shotgun sequencing. Sci. Rep. 2020, 10, 15115. [Google Scholar] [CrossRef]
- Vieco, N.; Belguesmia, Y.; Raspoet, R.; Auclair, E.; Gancel, F.; Kempf, I.; Drider, D. Benefits and inputs from lactic acid bacteria and their bacteriocins as alternatives to antibiotic growth promoters during food-animal production. Front. Microbiol. 2019, 10, 57. [Google Scholar] [CrossRef]
- Margalef, R. Homage to Evelyn Hutchinson, or why there is an upper limit to diversity. Open J. For. 1972, 44, 211–235. [Google Scholar]
- Sepulveda, S. Evaluación del Efecto Estacional Sobre El Microbioma Involucrado en la Fermentación del Aguamiel Y SU Influencia en Las Pro-Piedades Químicas del Pulque. Master’s Thesis, Universidad Autónoma de Chihuahua, Chihuahua, Mexico, 2020. [Google Scholar]
- Gamba, R.R.; Koyanagi, T.; Peláez, A.L.; De Antoni, G.; Enomoto, T. Changes in Microbiota during Multiple Fermentation of Kefir in Different Sugar Solutions Revealed by High-Throughput Sequencing. Curr. Microbiol. 2021, 78, 2406–2413. [Google Scholar] [CrossRef] [PubMed]
- Valadez, M. Pulque limpio/Pulque sucio: Disputas en torno a la legitimidad y la producción social del valor. Rev. Colomb. Antropol. 2014, 50, 14–18. [Google Scholar] [CrossRef]
- Ortiz-Basurto, R.I.; Pourcelly, G.; Doco, T.; Williams, P.; Dornier, M.; Belleville, M.-P. Analysis of the Main Components of the Aguamiel Produced by the Maguey-Pulquero (Agave mapisaga) throughout the Harvest Period. J. Agric. Food Chem. 2008, 56, 3682–3687. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).