Genomic Insight and Optimization of Astaxanthin Production from a New Rhodotorula sp. CP72-2
Abstract
1. Introduction
2. Materials and Methods
2.1. Characterization of Strain CP72-2
2.1.1. Phenotypic Characteristics
2.1.2. Genotypic Characteristics
Genomic DNA Extraction
Sequencing of the 26S rRNA Gene (D1/D2 Domain)
Genome Sequencing, Gene Prediction, and Functional Annotation
2.2. Cultivation for Astaxanthin Production
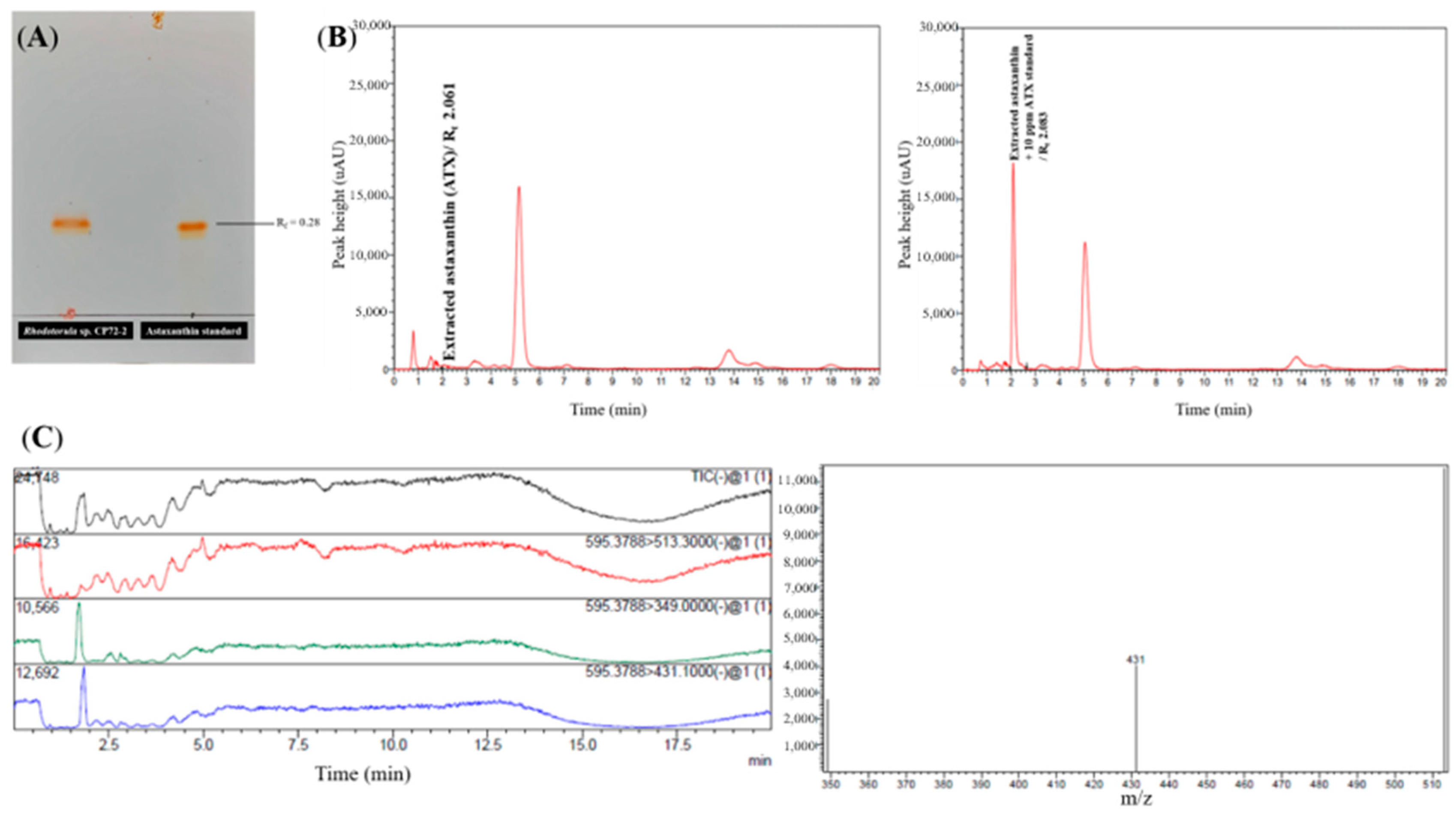
2.2.1. Qualitative Analysis for Astaxanthin Production by Thin-Layer Chromatography (TLC), High-Performance Liquid Chromatography (HPLC), and Liquid Chromatography-Mass Spectrometry (LC-MS)
TLC Identification
HPLC Identification
LC-MS Identification
2.2.2. Quantification Analysis of Astaxanthin Production
2.3. Optimization of Astaxanthin Production
2.4. Statistical Analysis
3. Results
3.1. Characteristics of Strain CP72-2
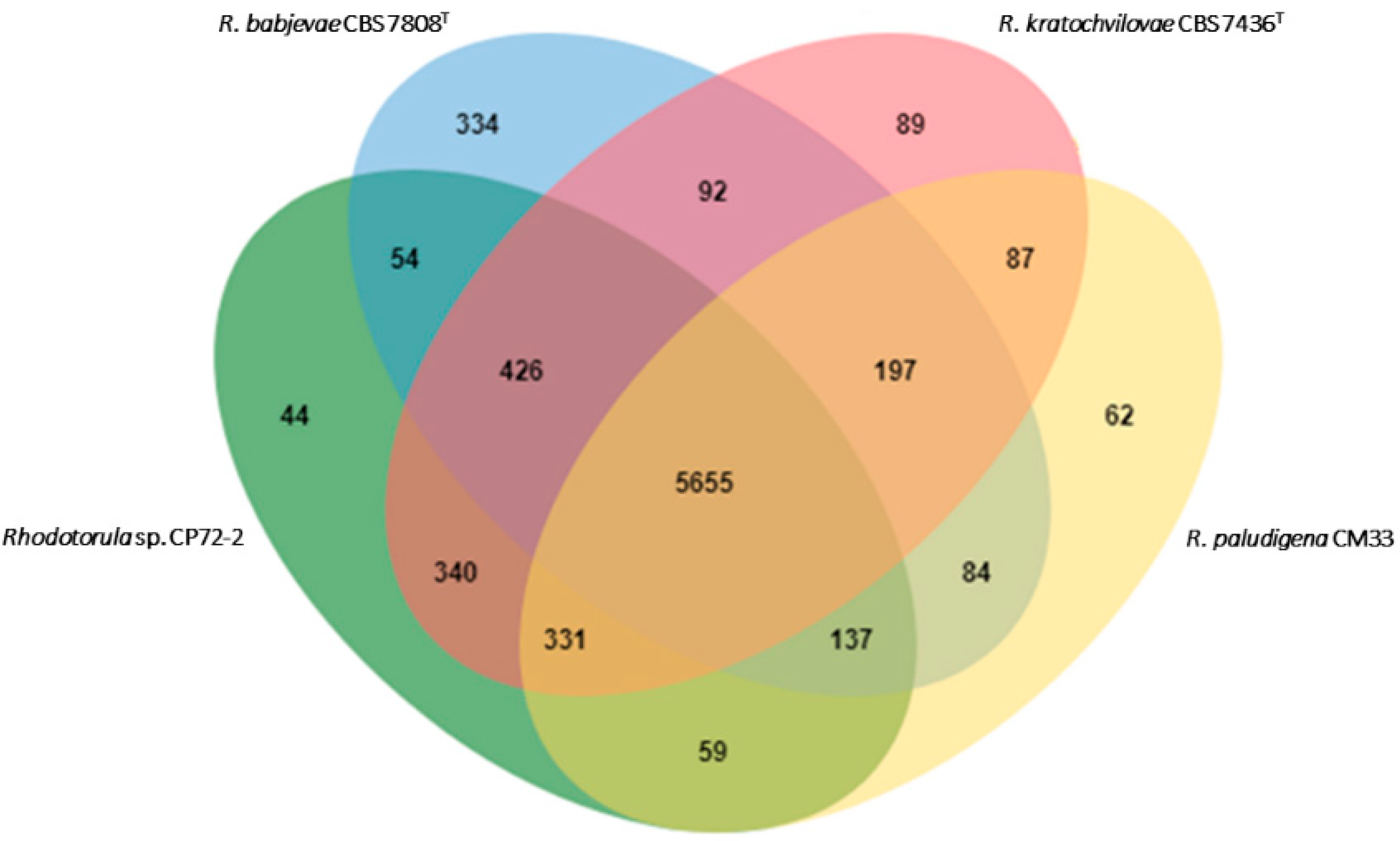
3.2. Genomic Features of Strain CP72-2
3.3. Screening of Astaxanthin Production
3.4. Astaxanthin Biosynthesis-Associated Genes from Strain CP72-2 Genome
3.5. Optimization of Astaxanthin Production
3.5.1. Effects of Different Carbon Sources on Astaxanthin Production
3.5.2. Effect of Glucose Concentration on the Astaxanthin Production
3.5.3. Effect of Initial pH of the AP Medium on the Astaxanthin Production
3.5.4. Effect of Temperature on the Astaxanthin Production
3.5.5. Effect of Incubation Period on the Astaxanthin Production
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Allahkarami, S.; Sepahi, A.A.; Hosseini, H.; Razavi, M.R. Isolation and identification of carotenoid-producing Rhodotorula sp. from Pinaceae forest ecosystems and optimization of in vitro carotenoid production. Biotechnol. Rep. 2021, 32, e00687. [Google Scholar] [CrossRef] [PubMed]
- Dursun, D.; Dalgıç, A.C. Optimization of astaxanthin pigment bioprocessing by four different yeast species using wheat wastes. Biocatal. Agric. Biotechnol. 2016, 7, 1–6. [Google Scholar] [CrossRef]
- Visser, H.; van Ooyen, A.J.J.; Verdoes, J.C. Metabolic engineering of the astaxanthin-biosynthetic pathway of Xanthophyllomyces dendrorhous. FEMS Yeast Res. 2003, 4, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Guerin, M.; Huntley, M.E.; Olaizola, M. Haematococcus astaxanthin: Applications for human health and nutrition. Trends Biotechnol. 2003, 21, 210–216. [Google Scholar] [CrossRef]
- Rodríguez-Sáiz, M.; de la Fuente, J.L.; Barredo, J.L. Xanthophyllomyces dendrorhous for the industrial production of astaxanthin. Appl. Microbiol. Biotechnol. 2010, 88, 645–658. [Google Scholar] [CrossRef]
- Ambati, R.R.; Moi, P.S.; Ravi, S.; Aswathanarayana, R.G. Astaxanthin: Sources, extraction, stability, biological activities and its commercial applications—A review. Mar. Drugs 2014, 12, 128–152. [Google Scholar] [CrossRef]
- Casella, P.; Iovine, A.; Mehariya, S.; Marino, T.; Musmarra, D.; Molino, A. Smart method for carotenoids characterization in Haematococcus pluvialis red phase and evaluation of astaxanthin thermal stability. Antioxidants 2020, 9, 422. [Google Scholar] [CrossRef]
- Kikukawa, H.; Shimizu, C.; Hirono-Hara, Y.; Hara, K.Y. Screening of plant oils promoting growth of the red yeast Xanthophyllomyces dendrorhous with astaxanthin and fatty acid production. Biocatal. Agric. Biotechnol. 2021, 35, 102101. [Google Scholar] [CrossRef]
- Kim, J.-H.; Kang, S.-W.; Kim, S.-W.; Chang, H.-I. High-level production of astaxanthin by Xanthophyllomyces dendrorhous mutant JH1 using statistical experimental designs. Biosci. Biotechnol. Biochem. 2005, 69, 1743–1748. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T. The Yeasts: A Taxonomic Study; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- O’Donnell, K.; Gray, L.E. Phylogenetic relationships of the soybean sudden death syndrome pathogen Fusarium solani f. sp. phaseoli inferred from rDNA sequence data and PCR primers for its identification. Mol. Plant-Microbe Interact. MPMI 1995, 8, 709–716. [Google Scholar] [CrossRef]
- Zerbino, D.R.; McEwen, G.K.; Margulies, E.H.; Birney, E. Pebble and rock band: Heuristic resolution of repeats and scaffolding in the velvet short-read de novo assembler. PLoS ONE 2009, 4, e8407. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef] [PubMed]
- Hunt, M.; Newbold, C.; Berriman, M.; Otto, T.D. A comprehensive evaluation of assembly scaffolding tools. Genome Biol. 2014, 15, R42. [Google Scholar] [CrossRef] [PubMed]
- Boetzer, M.; Pirovano, W. Toward almost closed genomes with GapFiller. Genome Biol. 2012, 13, R56. [Google Scholar] [CrossRef]
- Stanke, M.; Schöffmann, O.; Morgenstern, B.; Waack, S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinform. 2006, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef]
- Ushakumari, U.N.; Ramanujan, R. Isolation of astaxanthin from marine yeast and study of its pharmacological activity. Int. Curr. Pharm. J. 2013, 2, 67–69. [Google Scholar] [CrossRef]
- Sharma, R.; Ghoshal, G. Optimization of carotenoids production by Rhodotorula mucilaginosa (MTCC-1403) using agro-industrial waste in bioreactor: A statistical approach. Biotechnol. Rep. 2020, 25, e00407. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Miao, F.; Geng, Y.; Lu, D.; Zhang, C.; Zeng, M. Accurate quantification of astaxanthin from Haematococcus crude extract spectrophotometrically. Chin. J. Oceanol. Limnol. 2012, 30, 627–637. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef] [PubMed]
- Libkind, D.; Čadež, N.; Opulente, D.A.; Langdon, Q.K.; Rosa, C.A.; Sampaio, J.P.; Gonçalves, P.; Hittinger, C.T.; Lachance, M.A. Towards yeast taxogenomics: Lessons from novel species descriptions based on complete genome sequences. FEMS Yeast Res. 2020, 20, foaa042. [Google Scholar] [CrossRef]
- Fakankun, I.; Fristensky, B.; Levin, D.B. Genome sequence analysis of the oleaginous yeast, Rhodotorula diobovata, and comparison of the carotenogenic and oleaginous pathway genes and gene products with other oleaginous yeasts. J. Fungi 2021, 7, 320. [Google Scholar] [CrossRef]
- Lombard, V.; Ramulu, H.G.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Boraston, A.B.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 2004, 382, 769–781. [Google Scholar] [CrossRef]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 2013, 6, 1–14. [Google Scholar] [CrossRef]
- Li, C.-J.; Zhao, D.; Cheng, P.; Zheng, L.; Yu, G.-H. Genomics and lipidomics analysis of the biotechnologically important oleaginous red yeast Rhodotorula glutinis ZHK provides new insights into its lipid and carotenoid metabolism. BMC Genom 2020, 21, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Barredo, J.L.; García-Estrada, C.; Kosalkova, K.; Barreiro, C. Biosynthesis of astaxanthin as a main carotenoid in the heterobasidiomycetous yeast Xanthophyllomyces dendrorhous. J. Fungi 2017, 3, 44. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Yu, X. Effect of different carbon source on expression of carotenogenic genes and astaxanthin production in Xanthophyllomyces dendrorhous. Adv. J. Food Sci. Technol. 2013, 5, 1375–1379. [Google Scholar] [CrossRef]
- Miao, L.; Chi, S.; Wu, M.; Liu, Z.; Li, Y. Deregulation of phytoene-β-carotene synthase results in derepression of astaxanthin synthesis at high glucose concentration in Phaffia rhodozyma astaxanthin-overproducing strain MK19. BMC Microbiol. 2019, 19, 133. [Google Scholar] [CrossRef]
- Reynders, M.B.; Rawlings, D.E.; Harrison, S.T.L. Demonstration of the Crabtree effect in Phaffia rhodozyma during continuous and fed-batch cultivation. Biotechnol. Lett. 1997, 19, 549–552. [Google Scholar] [CrossRef]
- Sandmann, G. Carotenoid biosynthesis in microorganisms and plants. Eur. J. Biochem. 1994, 223, 7–24. [Google Scholar] [CrossRef]
- Johnson, E.A.; Lewis, M.J. Astaxanthin formation by the yeast Phaffia rhodozyma. J. Gen. Microbiol. 1979, 115, 173–183. [Google Scholar] [CrossRef]
- Meyer, P.S.; Du Preez, J.C. Effect of culture conditions on astaxanthin production by a mutant of Phaffia rhodozyma in batch and chemostat culture. Appl. Microbiol. Biotechnol. 1994, 40, 780–785. [Google Scholar] [CrossRef]
- Fang, T.J.; Cheng, Y.-S. Improvement of astaxanthin production by Phaffia rhodozyma through mutation and optimization of culture conditions. J. Ferment Bioeng. 1993, 75, 466–469. [Google Scholar] [CrossRef]
- Frengova, G.I.; Beshkova, D.M. Carotenoids from Rhodotorula and Phaffia: Yeasts of biotechnological importance. J. Ind. Microbiol. Biotechnol. 2009, 36, 163. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.M.; Acheampong, E.; Patel, T.R.; Chornet, E. Study of growth parameters for Phaffia rhodozyma cultivated in peat hydrolysates. Appl. Biochem. Biotechnol. 1992, 37, 235–241. [Google Scholar] [CrossRef]
- Martin, A.M.; Lu, C.; Patel, T.R. Growth parameters for the yeast Rhodotorula rubra grown in peat extracts. J. Ferment. Bioeng. 1993, 76, 321–325. [Google Scholar] [CrossRef]
- Kusdiyantini, E.; Gaudin, P.; Goma, G.; Blanc, P.J. Growth kinetics and astaxanthin production of Phaffia rhodozyma on glycerol as a carbon source during batch fermentation. Biotechnol. Lett. 1998, 20, 929–934. [Google Scholar] [CrossRef]
- Polulyakh, O.V.; Podoprigora, O.I.; Eliseev, S.A.; Ershov, Y.U.V.; Bykhovskii, V.Y.; Dmitrovskii, A.A. Biosynthesis of torulene and torularhodin by the yeast Phaffia rhodozyma. Appl. Biochem. Microbiol. USA 1992, 27, 411–414. [Google Scholar]
- Ren, Y.; Deng, J.; Huang, J.; Wu, Z.; Yi, L.; Bi, Y.; Chen, F. Using green alga Haematococcus pluvialis for astaxanthin and lipid co-production: Advances and outlook. Bioresour. Technol. 2021, 340, 125736. [Google Scholar] [CrossRef]
- Xiao, R.; Li, X.; Leonard, E.; Tharayil, N.; Zheng, Y. Investigation on the effects of cultivation conditions, fed-batch operation, and enzymatic hydrolysate of corn stover on the astaxanthin production by Thraustochytrium striatum. Algal Res. 2019, 39, 101475. [Google Scholar] [CrossRef]
- Hayman, E.P.; Yokoyama, H.; Chichester, C.O.; Simpson, K.L. Carotenoid biosynthesis in Rhodotorula glutinis. J. Bacteriol. 1974, 120, 1339–1343. [Google Scholar] [CrossRef]
- Naghavi, F.S.; Hanachi, P.; Soudi, M.R.; Saboora, A.; Ghorbani, A. Evaluation of the relationship between the incubation time and carotenoid production in Rhodotorula slooffiae and R. mucilaginosa isolated from leather tanning wastewater. Iran J. Basic Med. Sci. 2013, 16, 1114. [Google Scholar] [CrossRef]


| Strain | Nearest Relatives | Similarity (%) | Length (bp) | Accession No. |
|---|---|---|---|---|
| CP72-2 | R. kratochvilovae CBS 7436T | 98.35 | 604 | NG_042345 |
| R. evergladensis CBS 10880T | 98.18 | 604 | NG_057805 | |
| R. araucariae CBS 6031T | 98.12 | 639 | NG_058387 | |
| R. glutinis CBS 20T | 97.57 | 1350 | NG_055728 | |
| R. babjevae CBS 7808T | 97.52 | 604 | NG_042339 |
| Biosynthetic Pathways | Putative Genes | Gene Product | Scaffold ID |
|---|---|---|---|
| Mevalonate | ACAT | Acetyl-CoA acetyltransferase (EC:2.3.1.9) | scaffold3.g328 |
| HMGCS | Hydroxymethylglutaryl-CoA synthase (EC:2.3.3.10) | scaffold118.g7197 | |
| HMGCR | Hydroxymethylglutaryl-CoA reductase (EC:1.1.1.34) | scaffold69.g5807 | |
| PMVK | Phosphomevalonate kinase (EC:2.7.4.2) | scaffold3.g296 | |
| MVD | Diphosphomevalonate decarboxylase (EC:4.1.1.33) | scaffold42.g4393 | |
| Isoprene biosynthesis | IDI | Isopentenyl-diphosphate delta-isomerase (EC:5.3.3.2) | scaffold3.g272 |
| GGPS | Geranylgeranyl diphosphate synthase (EC:2.5.1.1 2.5.1.10 2.5.1.29) | scaffold33.g3627 | |
| FDPS | Farnesyl diphosphate synthase (EC:2.5.1.1 2.5.1.10) | scaffold10.g1242 | |
| CrtE | Geranylgeranyl pyrophosphate synthase | ||
| Astaxanthin biosynthesis | CrtYB | Phytoene synthase/Lycopene beta-cyclase (EC:2.5.1.32 5.5.1.19) | scaffold16.g1927 |
| CrtI | Phytoene desaturase (EC:1.3.99.30) | scaffold16.g1930 | |
| Astaxanthin biosynthesis | CrtS | Beta-carotene 4-ketolase/3-hydroxylase (also known as astaxanthin synthase) (EC:1.14.99.63 1.14.15.24 1.14.99.-) | scaffold40.g4227 |
| CrtR | Cytochrome P450 reductase (EC:1.14.14.1 1.6.2.4) | scaffold80.g6179 | |
| CrtW | Beta-carotene/zeaxanthin 4-ketolase (EC: EC:1.14.99.63 1.14.99.64) | scaffold38.g4135 | |
| CrtO | Beta-carotene ketolase (EC:1.14.99.63) | scaffold16.g1930 | |
| CrtZ | Beta-carotene hydroxylase (EC:1.14.13.-) | scaffold69.g5865 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kingkaew, E.; Tedsree, N.; Phuengjayaem, S.; Rojsitthisak, P.; Sritularak, B.; Thitikornpong, W.; Thompho, S.; Mhuantong, W.; Tanasupawat, S. Genomic Insight and Optimization of Astaxanthin Production from a New Rhodotorula sp. CP72-2. Fermentation 2023, 9, 501. https://doi.org/10.3390/fermentation9060501
Kingkaew E, Tedsree N, Phuengjayaem S, Rojsitthisak P, Sritularak B, Thitikornpong W, Thompho S, Mhuantong W, Tanasupawat S. Genomic Insight and Optimization of Astaxanthin Production from a New Rhodotorula sp. CP72-2. Fermentation. 2023; 9(6):501. https://doi.org/10.3390/fermentation9060501
Chicago/Turabian StyleKingkaew, Engkarat, Nisachon Tedsree, Sukanya Phuengjayaem, Pornchai Rojsitthisak, Boonchoo Sritularak, Worathat Thitikornpong, Somphob Thompho, Wuttichai Mhuantong, and Somboon Tanasupawat. 2023. "Genomic Insight and Optimization of Astaxanthin Production from a New Rhodotorula sp. CP72-2" Fermentation 9, no. 6: 501. https://doi.org/10.3390/fermentation9060501
APA StyleKingkaew, E., Tedsree, N., Phuengjayaem, S., Rojsitthisak, P., Sritularak, B., Thitikornpong, W., Thompho, S., Mhuantong, W., & Tanasupawat, S. (2023). Genomic Insight and Optimization of Astaxanthin Production from a New Rhodotorula sp. CP72-2. Fermentation, 9(6), 501. https://doi.org/10.3390/fermentation9060501






