Abstract
Trichoderma is one of the fungi commonly used in fermentation engineering. The hydrolytic enzymes secreted by Trichoderma have great economic value. Trichoderma guizhouense NJAU4742 is a branch of Trichoderma harzianum, which also has application potential. Lactose can induce fungi to secrete cellulase. Unfortunately, neither the lactose-inducing effect nor the mechanism of lactose metabolism in the study of Trichoderma guizhouense NJAU4742 is clear. Our study showed that carbon sources such as glucose, galactose, and sucrose could not induce cellulase secretion from Trichoderma guizhouense NJAU4742. Lactose induced the filter paper activity of the cellulase secreted by Trichoderma to reach 4.13 ± 0.11 U·mL−1. The ratio of 0.4% lactose–0.6% straw is the best way to induce cellulase and is better than adding only straw or lactose. TgRas family genes respond differently to different carbon sources at the gene level, and these proteins may be involved in different carbon source metabolisms. The results of transcriptional responses under different growth conditions showed that TgRas1 occupies a dominant position among TgRas family genes. The growth of the ΔTgRas1 mutant on the plate was inhibited, and the hyphae were dense, thick, and swollen. Under the condition of lactose, the biomass of ΔTgRas1 was severely inhibited in liquid fermentation, and its biomass decreased by 91.43% compared with WT. The liquid fermentation of ΔTgRas1 under other carbon source conditions was not affected.
1. Introduction
Trichoderma is a fungus with high utilization value and is often used in fermentation engineering [1]. The secondary metabolites of Trichoderma also have the effect of inhibiting the growth of pathogenic microorganisms and stimulating plant growth [2,3,4]. Trichoderma harzianum and Trichoderma reesei can secrete a variety of lignocellulose-decomposing enzymes, which are widely used in the production of biological products such as cellulase [5]. Trichoderma guizhouense NJAU4742, which evolutionarily belongs to one of the clades of Trichoderma harzianum, was isolated from soil samples in Guizhou Province, China [6]. Trichoderma guizhouense NJAU4742 has been elucidated to promote root growth and development and activate plant innate immune response [7]. In addition, Trichoderma guizhouense NJAU4742 retains the straw-degrading characteristics of Trichoderma, which also has application value. The expression and secretion mechanisms of cellulases have been studied for decades [8]. Studies have shown that the expression of cellulase genes is not only dependent on the induction of cellulose, but can also be induced by other carbon sources [9]. Lactose is an inexpensive inducer that has attracted widespread attention because of its low cost.
Lactose is composed of D-galactose and D-glucose linked by β-1,4 glycosidic linkages. Previous studies have shown that lactose is an inexpensive carbon source. Not only can it be used for fermentation to produce cellulase [10], but it is also the only cheap and soluble substrate currently available for Trichoderma reesei fermentation engineering [11,12]. Lactose as a carbon source has also been shown to enhance the ability of Vibrio cellulolyticus C-1 to produce cellulase [13]. Although the cheap properties of lactose can be used in microbial fermentation technology, not all microorganisms can effectively utilize lactose, which is one of the reasons that limits the development of the fermentation industry.
Ras family genes are ubiquitous in mammalian and eukaryotic cells. Ras genes play a key role in fungal carbon metabolism. A large number of studies have shown that Ras protein is involved in the process of fungal growth and development. There are two types of Ras proteins in Candida albicans; knocking out Ras1 can inhibit the hyphal production and the overexpression of Ras can enhance the hyphal formation [14]. There are two Ras genes in corn smut (U. maydis). The Ras1 mutants have been reported to be lethal, and Δras2 exhibits a reduced ability to form hyphae with a round shape [15]. It has been reported that Ras genes in Aspergillus and Aspergillus fumigatus regulate growth and development more extensively. The RasA gene of both filamentous fungi is involved in the process of mitosis, as well as processes that regulate the asexual developmental cycle [16]. The Ras1 and Ras2 in Cryptococcus neoformans not only participate in the regulation of high temperature growth process, but also participate in mycelium formation and type II transformation, which affects the pathogenicity [17,18]. Ras regulates cellular carbon sensing by regulating its affinity for GTP and GDP. Therefore, Ras may also play an important role in fungal lactose metabolism. This study aimed to investigate the feasibility of lactose-induced cellulase in Trichoderma guizhouense NJAU4742 and the effect of TgRas1 gene on lactose metabolism.
2. Methods and Materials
2.1. Determination of Enzymatic Activity of T. guizhouense NJAU4742 Solid Fermentation
Determination of endoglucanase activity: Take 480 µL 0.5% carboxymethyl cellulose solution (CMC-Na) in a 2 mL centrifuge tube, add 500 µL 50 mM acetic acid buffer, and finally add 20 µL crude enzyme solution. After mixing evenly, react in a 50 °C water bath for 10 min, then add 1 mL of DNS reagent in a boiling water bath for 10 min and cool down. Its absorbance value is measured at a wavelength of OD520 nm. The amount of enzyme required to produce 1 µmol of reducing sugar per 1 min is defined as 1 unit of enzyme activity (U).
Determination of exoglucanase activity: Take 10 µL 5 mM pNPC and add it to the 96-well plate, add 40 µL crude enzyme solution and 50 µL 50 mM acetic acid buffer, and use the inactivated crude enzyme solution as a blank control. The 96-well plate is reacted in a 50 °C water bath for 10 min, and 100 µL 1 M of Na2CO3 solution is added to terminate the reaction. Its absorbance value is measured at OD402 nm. The content of p-nitrophenol is calculated based on the marking. The amount of enzyme required to produce 1 µmol of p-nitrophenol per minute for 1 min is defined as one unit of enzyme activity (U).
Determination of xylanase activity: Take 480 µL of xylanase in a 2 mL centrifuge tube, add 500 µL of 50 mM acetic acid buffer, and finally add 20 µL of crude enzyme solution. After mixing evenly, react in a 50 °C water bath for 20 min, then add 1 mL of DNS reagent in a boiling water bath for 10 min and cool down. Its absorbance value is measured at OD520 nm. The amount of enzyme required to produce 1 µmol of reducing sugar per 1 min is defined as 1 unit of enzyme activity (U).
Determination of enzyme activity of filter paper: Take 50 µL of crude enzyme solution, add 500 µL of 50 mM acetic acid buffer and 450 µL of deionized water, and put two small discs of filter paper (Whatman No. 1 filter paper; use a hole puncher to obtain a small disc of filter paper; approximately 10 mg) and add it to a 2 mL centrifuge tube. After the reaction in a water bath at 50 °C for 20 min, add 1 mL of DNS reagent and cool in a boiling water bath for 10 min. Its absorbance value is measured at OD520 nm. The amount of enzyme required to produce 1 µmol of reducing sugar per 1 min is defined as 1 unit of enzyme activity (U).
The crude enzyme solution inactivated in a 100 °C water bath for 10 min is used as the comparison solution, and the remaining steps are the same. Enzyme activity refers to the results calculated after deducting the reducing sugar in the comparison solution. Drawing of glucose standard curve: Add different volumes of glucose standard solution, deionized water, and DNS reagents to each centrifuge tube and mix well, then place each centrifuge tube in a boiling water bath at the same time and react for 10 min. After cooling, the absorbance value is measured at OD520 nm, with glucose content as the abscissa and OD value as the ordinate, and the standard curve of glucose is drawn. The enzyme inactivated in a 100 °C water bath for 10 min is used as the control solution, and the remaining steps are the same. The production of 1 µmol of reducing sugar per minute is defined as 1 enzyme activity (unit U).
Drawing of the standard curve of p-nitrophenol: Different volumes of standard solution of p-nitrophenol (1 mg·mL−1), deionized water and Na2CO3 solution are drawn in a 96-well plate, and the absorbance is measured at OD402 nm after mixing. The content of p-nitrophenol is used as the abscissa, and the OD value is used as the ordinate to make a standard curve of p-nitrophenol.
2.2. Identification of TgRas Gene in the Genome of T. guizhouense NJAU4742
The TgRas protein sequence was retrieved from the Trichoderma guizhouense NJAU4742 database using the accession number search keywords obtained from the Saccharomyces cerevisiae database, while the Ras gene family hidden Markov model domain (PF00071) was downloaded from the Pfam database. The NCBI BLASTP homepage (http://www.ncbi.nlm.nih.gov (accessed on 30 March 2023)) was used to perform homologous alignment with the T. guizhouense NJAU4742 genome database, and at the same time use HMMER software was used to perform domain alignment and identify all potential Ras genes. All TgRas gene family members of T. guizhouense NJAU4742 were subsequently determined using the online tools CDD (https://www.ncbi.nlm.nih.gov/cdd/ (accessed on 30 March 2023)) and SMART (http://smart.embl.de/ (accessed on 30 March 2023)). The software DNAMAN (https://www.lynnon.com/ (accessed on 30 March 2023)) was used to compare the primary structure of the TgRas family proteins and the ExPASy online tool was used to calculate the chemical characteristics of the TgRas protein in T. guizhouense NJAU4742 (http://web.expasy.org/protparam/ (accessed on 30 March 2023)), such as theoretical isoelectric point (pI), amino acid number, and molecular weight (Da).
2.3. Phylogenetic Analysis of TgRas Gene, Conserved Motif Analysis, Gene Structure Analysis
MUSCLE software (https://www.ebi.ac.uk/Tools/msa/muscle/ (accessed on 30 March 2023)) was used to perform multiple sequence alignment of the amino acid sequence of TgRas protein, MEGA v7 was used to select the maximum likelihood method for phylogenetic analysis and construct the phylogenetic tree, where the bootstrap value was set to 1000, and finally the online interactive tool iTOL was used for data visualization (https://itol.embl.de/ (accessed on 30 March 2023)). The online tool MEME (http://memesuite.org/ (accessed on 30 March 2023)) was used to identify the conserved protein structure of the TgRas family protein, with the maximum motif value set to 10, and the software TBtools was then used to analyze the motif structure of the TgRas family protein to visualize. The exon/intron and CDs structures of the TgRas gene were predicted using the online tool GSDS (http://gsds.cbi.pku.edu.cn/ (accessed on 30 March 2023)).
2.3.1. Cultivation, Determination of Biomass, and Preparation of Intracellular Material
T. guizhouense NJAU4742 was selected for this study, and it was cultured on PDA medium for conidia production and incubated at 28 °C under static cultivation conditions for seven days. The conidia were then harvested by washing the plate with 10 mL of sterile double-distilled water (ddH2O) followed by removal of mycelia by filtration through four layers of gauze. The conidia were resuspended, and the concentration was adjusted to 1 × 107 conidia·mL−1. Mandel’s salt solution without organic components (1.4 g·L−1 (NH4)2SO4, 2.0 g·L−1 KH2PO4, 0.3 g·L−1 CaCl2, 0.3 g·L−1 MgSO4, 5 mg·L−1 FeSO4·7H2O, 20 mg L−1 CoCl2, 1.6 mg L−1 MnSO4, and 1.4 mg L−1 ZnSO4) was supplemented with 1% (w/v) lactose for liquid fermentation. The biomass in submerged cultures was determined by filtering the cultures onto preweighed filter paper (Whatman No. 1), and the harvested biomass was dried at 70 °C for 24 h before being quantified. After being washed once with sterile water, the fresh mycelia were disrupted using glass beads (0.1 mm) and liquid nitrogen and then centrifuged at 10,000× g for 10 min to remove cell debris. The supernatant was frozen in liquid nitrogen and stored at –20 °C for subsequent experiments.
2.3.2. Construction of Functional Fractions for Gene Deletion Complementation
A fragment of approximately 1 kb upstream and downstream of the target gene from the genome of NJAU4742 (NCBI accession number: GQ337429) and a hygromycin fragment from the vector of PcDNA13.1 (Invitrogen, Waltham, MA, USA) were amplified with primers. After purification with the AxyPrep DNA Pure Cycle Kit (Axygen, Hangchow, China), the PCR products of these three genes were ligated together by the HiFi enzyme (KAPA Biosystems, KK2102, Wilmington. MA, USA). After agarose gel electrophoresis analysis, the fusion-completed fragments were recovered by an AxyPrep DNA gel purification and extraction kit (Axygen, Hangchow, China). The gene deletion cassette was obtained with the primer’s gene-up-F and gene-down-R (Table S1).
Seven-day-old spores were harvested from PDA solid medium with 10 mL of freshly sterilized water. The spore suspension was filtered through four layers of sterile medical gauze and then collected in a 1.5 mL sterile centrifuge tube for subsequent experiments. Fifty microliters of the spore solution were spread on the cellophane-covered plates and then incubated at 28 °C for 16 h. After incubation, the cellophane was transferred into a new Petri dish, and then 5 mL of solution A (0.1 M KH2PO4, 1.2 M sorbitol, pH 5.6, filter sterilized 0.15 g lyase (Sigma-Aldrich, St. Louis, MO, USA)) was used to wash the germinated spores on cellophane to make protoplast suspensions. The protoplast was extracted as follows: the harvested suspension was incubated at 28 °C for 100 min and then centrifuged at 100 rpm, after which the cells were gently disrupted with autoclaved tweezer tips; the suspension was filtered with four layers of sterile medical gauze in a 50 mL sterile centrifuge tube, and then the protoplast suspension was obtained by centrifugation at 2000 rpm at 4 °C for 10 min; the supernatant was discarded, and the pelleted protoplasts were resuspended with 5 mL of solution B (1 M sorbitol, 50 mM CaCl2, 10 mM Tris-HCl, pH 7.50) and then centrifuged at 2000 rpm for 10 min at 4 °C. The supernatant was then discarded, and the pelleted protoplasts were resuspended with 200 µL of solution B and kept at 4 °C for the subsequent experiment. The functional fragments were transferred into the protoplasts of Trichoderma guizhouense NJAU4742 by the polyethylene glycol (PEG)-mediated method according to Miao et al. [19], with some modifications. The mixture containing 200 µL protoplasts, 10 µL ligated fragments (concentration should be greater than 250 ng·µL−1), and 50 µL PEG (1 M PEG 6000, 50 mM CaCl2, pH 7.50) was placed on the ice bath for 20 min, and then an additional 2 mL of PEG (room temperature) was added and mixed gently. After 5 min of incubation at room temperature, 3 mL of solution B was added to the system and mixed gently, and then 0.5 mL of the mixture was spread on PDA medium containing 1 M sucrose and incubated at 28 °C for 16 h. The germinated protoplasts were overlaid with PDA medium containing 200 ng·mL−1 hygromycin B and incubated at 28 °C for 72 h. Some single colonies were selected and then transferred into new PDA medium containing 200 ng·mL−1 hygromycin B and incubated at 28 °C for 48 h. The mutants were directly screened for positive deletion mutants by PCR with primer validation (Table S1) and purified into single spores. The construction method of the gene complementary strain is similar to the previous gene-knockout method.
3. Results
3.1. The Combination of Straw and Lactose Can Effectively Promote the Secretion of Cellulase by T. guizhouense NJAU4742
Firstly, the differences in cellulase secretion in Trichoderma guizhouense NJAU4742 under different carbon source conditions were compared. The enzymatic activities of various cellulase enzymes in the liquid fermentation process were measured to show the induction effect of carbon source. The whole process is based on MM medium, to which 1% glucose, galactose, sucrose, lactose, and straw carbon sources are added, respectively. After culturing for 4 days, the extracellular enzyme solution was taken to measure the enzyme activity. It can be seen from Figure 1 that glucose, galactose, and sucrose cannot induce the secretion of cellulase. Both rice straw and lactose could induce Trichoderma guizhouense NJAU4742 to secrete cellulase. Under lactose conditions, the filter paper enzyme activity of the cellulase secreted by Trichoderma guizhouense NJAU4742 reached 4.13 ± 0.11 U·mL−1, which was slightly lower than that induced by straw. In addition, under the condition of lactose, the activity of endoglucanase reached 3.66 ± 0.17 U·mL−1, the activity of exoglucanase reached 3.54 ± 0.22 U·mL−1, and the activity of xylanase activity reached 3.60 ± 0.25 U·mL−1.

Figure 1.
Enzyme activity of cellulase measured under different conditions. (a) Cellulase activity measured under different carbon sources; (b) cellulase activity measured under different concentrations of lactose; (c) cellulase activity measured under different lactose and straw ratios. The “abcde” shows significant differences separately. Different letters represent significant differences from each other.
In addition, we also obtained the best lactose-inducing concentration of Trichoderma guizhouense NJAU4742 to secrete cellulase. We added 0.2–1.4% lactose to MM medium. The enzyme activity data showed that both endoglucanase and exoglucanase had the maximum enzymatic activity under the condition induced by 1% lactose, which were 4.84 ± 0.05 U·mL−1 and 7.22 ± 0.06 U·mL−1, respectively. At this time, xylanase also had a high activity (7.48 ± 0.15 U·mL−1). It is worth noting that cellulase is often a mixture of various enzymes. We further measured the enzyme activity of filter paper to comprehensively measure the effect of cellulase. The results showed that when the lactose concentration was 0.8%, 1%, and 1.2%, the filter paper enzyme activities were 4.80 ± 0.09 U·mL−1, 5.37 ± 0.08 U·mL−1, and 4.72 ± 0.08 U·mL−1, respectively. Therefore, the addition of lactose can induce Trichoderma guizhouense NJAU4742 to secrete cellulase. When the amount of lactose added was approximately 1%, the induction effect was most obvious. However, too much lactose may affect cellulase induction.
Both straw and lactose induction data showed a high activity of cellulase. We then adjusted the ratio of lactose to straw to try to obtain the best solution. The total content of lactose and straw was still 1% in the MM medium, and the content of individual components fluctuated from 0% to 1%. The results showed that the endoglucanase activity of adding 0.4% lactose–0.6% rice straw was not significantly different from that of 0.2% lactose–0.8% rice straw, which were 6.83 ± 0.07 U·mL−1 and 6.79 ± 0.06 U·mL−1, respectively. However, the enzyme activities induced by 1% straw increased by 9.22% and 8.69%, respectively. For exoglucanase, only the combination of 0.4% lactose–0.6% rice straw had the maximum induction effect, and the enzyme activity reached 9.68 ± 0.11 U·mL−1. Compared with adding 1% rice straw, the enzyme activity increased by 10.95%. Adding an appropriate proportion of lactose can also improve the activity of xylanase. When 0.2% lactose–0.8% straw was added, the enzyme activity reached 9.26 ± 0.06 U·mL−1, an increase of 11.56% (compared with 1% straw). Among many combinations, the combination of 0.4% lactose and 0.6% rice straw had the largest filter paper enzyme activity, reaching 7.30 ± 0.05 U·mL−1. Compared with the treatment of 1% rice straw, the activity increased by 18.52%. In summary, the induction effect of lactose and straw mixed was significantly better than that of single straw or lactose. When proportioned, it can most effectively increase the enzymatic activity of hydrolytic enzymes such as cellulase.
3.2. Functional Identification of TgRas Family Genes in T. guizhouense NJAU4742
Previous studies have shown that the Ras gene family is involved in the growth and development of organisms, including the metabolism of various carbon sources. The TgRas family gene sequence was downloaded from the T. guizhouense NJAU4742 database. TgRas family protein domains were further analyzed by searching candidate TgRas family proteins in HMMER and NCBI databases. Twenty-two candidate TgRas genes were identified in the genome of Trichoderma guizhouense NJAU4742. They were sorted according to homology. Table 1 shows information such as chromosome location, amino acid (length), molecular weight, and isoelectric point (pI). The results showed that the length of TgRas family genes ranged from 765 (TgRas9) to 1260 (TgRas14) bases. TgRas family proteins range in length from 194 (TgRas14) to 336 (TgRas22) amino acids. The PI values were between 4.79 (TgRas11) and 9.61 (TgRas22), and the protein molecular weights were between 20.761 kDa (TgRas4) and 36.096 kDa (TgRas22) (Table 1).

Table 1.
General information of TgRas family in Trichoderma guizhouense NJAU4742.
Next, the structures of 22 TgRas genes were analyzed. The blue squares represent the upstream and downstream segments of the gene. The yellow squares represent exons of genes. Introns are indicated by black lines. The results showed that the exon numbers of 22 genes varied widely, ranging from one (TgRas6) to eight (TgRas3). The difference in the number of exons in the TgRas family may occur during the evolution of the TgRas gene, which may also be one of the reasons for the different functions of the TgRas family (Figure 2a). In addition, we identified 10 conserved motifs associated with TgRas. The identified motifs range in length from 11 to 50 amino acids. The color of each square in the figure represents a conserved motif. As can be seen in Figure 2b, the motif combinations of different TgRas proteins are not the same. The conserved motifs and distribution order of TgRas1 and TgRas2 are consistent, which means that the similarity is high. TgRas4, TgRas14, and TgRas18 have specific Motif 6. It is worth noting that some TgRas proteins also have Motif 8, and all of them are located at the amino acid terminal. Motif 8 is a CAAX motif sequence consisting of a cysteine residue followed by two aliphatic residues and a C-terminal X residue. X can be C, S, M, Q, or A. CAAX motifs are premodified targets for protein transport and have important biological functions in organisms. The results showed that the end of TgRas1–4 has a CAAX structure (orange Motif8), which is closely related to the membrane localization function. Figure 2c shows the analysis of the amino acid composition of each functional domain. An amino acid with a larger font size (such as C in Motif 8) means that among the 22 TgRas proteins, more TgRas proteins have amino acid C (such as C in Motif 8) at this position. For Ras family proteins, this motif appears to contribute to membrane localization and signal transduction.
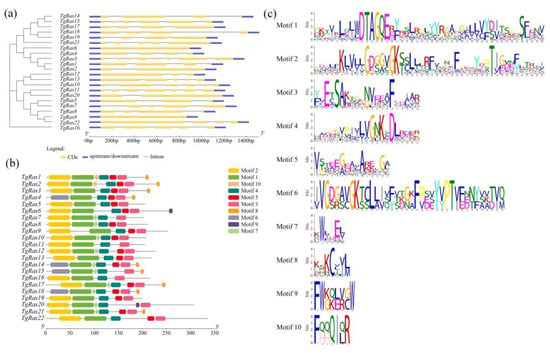
Figure 2.
Identification and analysis of TgRas family genes and TgRas family proteins. (a) Gene structure of TgRas genes; (b) distribution of conserved motif of TgRas proteins; (c) motif analysis of TgRas proteins.
We analyzed the expression profile of TgRas family genes when glucose or rice straw was used as a carbon source. The heatmap shows the quantitative results for 22 TgRas genes. The results showed that the type of carbon source did not affect the high-level expression of TgRas1, TgRas2, TgRas3, TgRas9, TgRas21, and TgRas22. Only under the condition of glucose as a carbon source did TgRas5, TgRas7, TgRas8, TgRas10, TgRas11, TgRas13, TgRas16, TgRas17, TgRas18, and TgRas19 show higher expression levels. TgRas14 and TgRas15 only showed higher expression levels on a rice straw medium. In summary, TgRas family genes respond to different types of carbon sources at the gene level, and these proteins may be involved in different carbon source metabolisms (Figure 3).

Figure 3.
Expression profile of TgRas family genes for different carbon sources and times. Darker red indicates higher gene expression and darker blue indicates lower gene expression.
3.3. The T. guizhouense NJAU4742 Mutant Has a Reduced Ability to Utilize Lactose, Resulting in Abnormal Growth
Based on the TgRas family analysis results of T. guizhouense NJAU4742, we selected TgRas1, TgRas2, TgRas3, and TgRas4 genes for follow-up research. The methods included construction of mutants and validation. These four mutants were named ΔTgRas1, ΔTgRas2, ΔTgRas3, and ΔTgRas4. We compared the transcriptional responses of TgRas family genes in wild-type strains under different temperatures and carbon sources.
The results showed that the expression of TgRas1 had the highest expression level regardless of the carbon source. The results induced by lactose and sucrose were 54.13% and 62.93% higher than those induced by glucose, respectively. The expression of TgRas4 was low, while the expression of TgRas2 and TgRas3 was very low (Figure 4a). Therefore, TgRas1 may be the key gene of carbon source metabolism in T. guizhouense NJAU4742. The wild type was cultured under liquid conditions with ammonium sulfate, ammonium nitrate, and sodium nitrate as nitrogen sources, respectively. The expression of TgRas1 was still the highest, and there was no significant difference between different nitrogen sources. It is worth noting that TgRas4 showed an increased expression level, which was close to that of TgRas1, when nitrate was used as a nitrogen source (Figure 4b). This means that TgRas4 may be involved in the regulation of nitrogen sources. For the results of liquid culture at different temperatures, the expression level of TgRas1 was still high, and the change of temperature seemed to have no effect on TgRas family genes (Figure 4c). In addition, under the growth conditions of pH 9 and pH 3, the expression of TgRas1 gene was significantly lower than that of pH 6, which decreased by 44.71% and 36.33%, respectively (Figure 4d). Interestingly, the expression of TgRas1 tended to increase under the light-only culture condition, but that may also be due to experimental error (Figure 4e). In short, the results suggest that in T. guizhouense NJAU4742, TgRas1 may play a leading role and perform a major function in the organism.

Figure 4.
Transcriptional responses of TgRas family genes in wild-type strains under different carbon sources, nitrogen sources, temperatures, pH levels, and light conditions. (a) Transcriptional responses of TgRas family genes in wild-type strains under different carbon sources; (b) transcriptional responses of TgRas family genes in wild-type strains under different nitrogen sources; (c) transcriptional responses of TgRas family genes in wild-type strains under different temperatures; (d) transcriptional responses of TgRas family genes in wild-type strains under different pH levels; (e) transcriptional responses of TgRas family genes in wild-type strains under different light conditions. The “*” shows significant differences separately.
Furthermore, we evaluated the effect of TgRas family genes on the growth of T. guizhouense NJAU4742. Wild types (WT) ΔTgRas1, ΔTgRas2, ΔTgRas3, and ΔTgRas4 were inoculated on PDA plates for one week, and the daily growth was recorded, as shown in Figure 5a. There was no significant difference in growth between ΔTgRas3 and WT during 7 days of continuous culture. The phenotype of ΔTgRas1 had the greatest influence. The hyphae of ΔTgRas1 could not fill the entire plate, and the colony edge grew irregularly and the colony size was small. The growth of ΔTgRas2 was slow and the colony edge grew irregularly, indicating that the growth polarity of this strain may be limited. ΔTgRas4 was less affected but had slower growth and greater spore production. Irregular colony edges were simultaneously observed in ΔTgRas1, ΔTgRas2, and ΔTgRas4, suggesting that the main function of TgRas family genes may be to affect polar growth (Figure 5b).
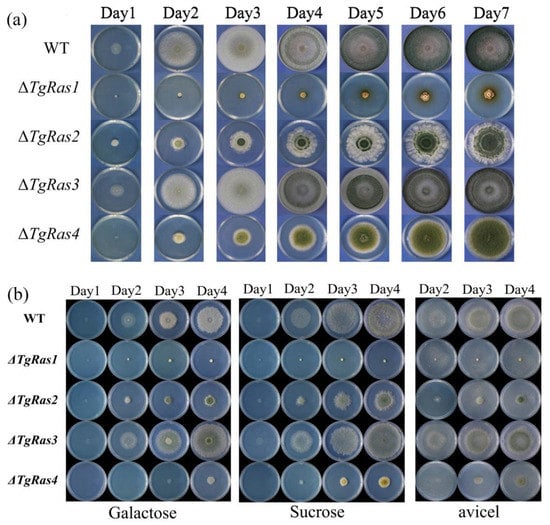
Figure 5.
Growth of single-knockout strains of the TgRas family gene on the plate. (a) Growth of TgRas family single-knockout strains on PDA; (b) growth of TgRas family single-knockout strains on galactose, sucrose, and microcrystalline cellulose.
Further, we observed the hyphal growth and spore germination of each mutant strain on the plate. We observed the fungal border phenotype at 24 h and 48 h time points. ΔTgRas2, ΔTgRas3, and ΔTgRas4 all showed denser hyphae numbers with more mycelial branches. ΔTgRas1 exhibited a fully dysplastic fungal hyphal morphology from the very beginning, with swollen and smooth edges (Figure 6a). The spore germination observed under the microscope showed that as the germination time passed (12–18 h), the hyphae of ΔTgRas2, ΔTgRas3, and ΔTgRas4 were similar to those of WT, and all of them could obviously produce hyphae. However, the germination process of ΔTgRas1 spores was affected, and the spores were large, and no obvious mycelial growth could be seen (Figure 6b). We measured the length of the hyphae to represent the growth rate of the fungus. The results showed that the radial growth rate of ΔTgRas1 was the slowest, and there were almost no elongated hyphae within 72 h. ΔTgRas2 and ΔTgRas4 had slower growth rates, but ΔTgRas3 had little difference in growth rate from WT (Figure 6c,d). Colony edge growth and spore germination also confirmed that the deletion of the TgRas gene had an effect on the polar growth of hyphae, and TgRas1 played a major role in the TgRas gene.

Figure 6.
Mycelial growth and spore germination of TgRas family single-knockout strains growing on PDA. (a) Mycelial growth at the colony edge of the TgRas family single-knockout strain; (b) spore germination of the TgRas family single-knockout strain; (c) mycelium length of single-knockout strains of TgRas family genes. The “ab” shows significant differences separately. Different letters represent significant differences from each other.; (d) radial growth of mycelium of single-knockout strains of the TgRas family genes.
3.4. Liquid Fermentation of TgRas1 Mutants from other Carbon Sources Was Not Affected
The above experimental results have shown that TgRas1 is the main functional gene. Therefore, we observed the growth of wild-type and TgRas1 mutants (ΔTgRas1) in a Biolog-FF medium and investigated the response of ΔTgRas1 to different carbon sources. Each legend and number in Figure 7 represent an individual carbon source. The unmarked numbers indicate that neither wild type nor ΔTgRas1 can grow on this carbon source. The carbon source corresponding to each number is listed in detail in the appendix. The thickened light green line in Figure 7 represents the growth of glucose, the thickened black line represents the growth of lactose, and the thickened dark blue line represents the growth of arabinose. The wild-type strain grew rapidly under the condition of glucose, but after 96 h, the growth of the wild-type strain under the lactose condition gradually increased (Figure 7a). However, during the growth process of ΔTgRas1, the growth of ΔTgRas1 under lactose conditions was much weaker than that under glucose conditions (Figure 7b), which formed a significant contrast with WT. The results showed that ΔTgRas1 could not utilize lactose well, but glucose utilization was not affected (Figure 7).

Figure 7.
Growth of WT and ΔTgRas1 on Biolog-FF plates. (a) WT; (b) ΔTgRas1.
According to the growth and development of ΔTgRas1 on Biolog-FF plates, glucose and lactose were selected as carbon sources for liquid fermentation. The dry weight of the mycelia was measured after 4 days of culture. The results showed that the growth of WT and ΔTgRas1 was basically the same under glucose liquid culture conditions. After standing still for 10 min, the liquid was still cloudy, which was due to the floating of hyphae scattered throughout the medium. Under the condition of lactose liquid culture, the growth status of WT was not inhibited, and the medium was always turbid. Interestingly, the ΔTgRas1 liquid medium became clear after standing for 10 min. ΔTgRas1 sank to the bottom of the medium, and a small amount of granular ΔTgRas1 could be clearly observed. This indicated that the hyphae of ΔTgRas1 might be stunted in growth and more likely to sink to the bottom. Under the glucose carbon source condition, the biomass of ΔTgRas1 increased by 16.36% compared with WT, which is an interesting phenomenon. However, when lactose was used as the sole carbon source, the biomass of ΔTgRas1 was severely suppressed, decreasing by 91.43% compared with WT (Figure 8a,b).

Figure 8.
Growth of ΔTgRas1 strain under different carbon source conditions. (a,b) Growth of ΔTgRas1 strain under different carbon source conditions selected according to the results of Biolog-FF plate; (c,d) growth of ΔTgRas1 strain under different common carbon source conditions. The “*” shows significant differences separately. Different letters represent significant differences from each other. The “**” represents a very significant difference.
In the process of utilizing carbon sources, fungi may secrete relevant hydrolytic enzymes to decompose carbohydrates. In order to avoid the influence of the type of polysaccharides on the bacteria, we additionally selected common carbohydrates (glucose, galactose, lactose, sucrose, soluble starch) for verification. This cultivation was carried out under liquid conditions. WT was turbid in all media, and mycelia were evenly distributed in the media. For carbon source media that do not affect the growth of ΔTgRas1, ΔTgRas1 will naturally settle to the bottom of the bottle. The growth of ΔTgRas1 in lactose medium was still inhibited. We further measured the dry weight of mycelia. ΔTgRas1 exhibited increased biomass under glucose, galactose, sucrose, and starch conditions, which increased by 39.52%, 24.17%, 60.71%, and 36.33%, respectively, compared with WT. Similarly to the previous experimental results, under lactose conditions, the growth of ΔTgRas1 was significantly inhibited, and its biomass was reduced by 94.12% compared with WT. The reason for the biomass growth of ΔTgRas1 under other carbon source conditions is unclear. However, it can be clearly shown that only lactose can severely inhibit the growth of ΔTgRas1, and TgRas1 may be involved in the lactose metabolic pathway (Figure 8c,d).
3.5. Growth in Liquid Fermentation and Solid Fermentation with Straw as the Sole Carbon Source
For fungi, liquid culture conditions allow easy access to nutrients for growth. In the above liquid fermentation results, the growth of ΔTgRas1 under lactose-free conditions was better than that of the wild type, so we further compared the performance of ΔTgRas1 in liquid fermentation and solid fermentation. The only carbon source in the medium was straw. The results showed that WT could grow normally on straw under solid fermentation, and mycelium extended to the whole surface and crevices of straw, while ΔTgRas1 only grew on the surface of straw without mycelium growth. ΔTgRas1 clustered tightly on the straw surface, similarly to the growth form on agar plates (Figure 9a). In liquid culture, the biomass of ΔTgRas1 was slightly higher than that of WT. However, the growth of ΔTgRas1 in solid culture was inhibited, and the biomass was 17.91% lower than that of WT (Figure 9b).
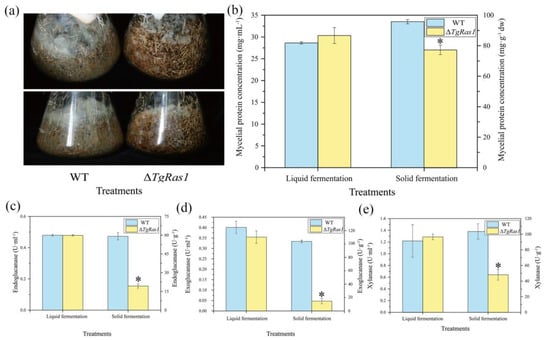
Figure 9.
ΔTgRas1 cultured under liquid and solid conditions with rice straw as the sole carbon source, respectively. (a) Growth morphology of bacteria during solid fermentation; (b) biomass of ΔTgRas1 cultured in liquid and solid conditions with rice straw as the sole carbon source; (c) en-doglucanase of ΔTgRas1 cultured under liquid and solid conditions with rice straw as the sole carbon source; (d) exoglucanase of ΔTgRas1 cultured under liquid and solid conditions with rice straw as the sole carbon source; (e) xylanase of ΔTgRas1 cultured under liquid and solid conditions with rice straw as the sole carbon source. The “*” shows significant differences separately. Different letters represent significant differences from each other.
Next, the activities of cellulose-related enzymes were measured. Under liquid fermentation, there was no significant difference in the endoglucanase activities of WT and ΔTgRas1. However, under solid fermentation, the enzymatic activity of ΔTgRas1 was 70.83% lower than that of WT (Figure 9c). Likewise, there was no significant difference in exoglucanase activity between WT and ΔTgRas1 in liquid fermentation. In solid fermentation, the enzymatic activity of ΔTgRas1 was 85.45% lower than that of WT (Figure 9d). In the liquid fermentation condition, the xylanase activity of WT and ΔTgRas1 did not show a significant difference. Under solid fermentation conditions, the enzyme activity of ΔTgRas1 decreased by 54.29% compared with WT (Figure 9e).
4. Discussion
Most microbial cellulases are inducible hydrolases that are secreted by microorganisms growing on cellulosic substrates [20,21]. Different microorganisms can utilize agricultural residues to produce cellulases and xylanases [22,23]. Straw and bran from cereals, corncobs, and bagasse are the most popular substrates in different regions [24,25]. Enzyme yields can be increased by using a mixture of different substrates instead of a single substrate [26,27]. Lactose is a low-cost additive used to induce fungal secretion of cellulase and xylanase [28]. Lactose has been reported to regulate the secretion of hydrolases from Trichoderma reesei, including xyn1, xyn2, cbh1, cbh2, and egl1-encoded xylanase I, xylanase II, cellulosic biohydrolase II, and endoglucan carbohydrase I [29]. Despite their positive impact on cellulase production, the addition of such additives in inappropriate amounts can lead to inhibitory effects. In summary, the secretion of cellulase depends on the medium and culture conditions, and the types of inducers will have different effects.
In almost all eukaryotes, Ras proteins play a key role in sensing and responding to environmental signals [30,31], and the Ras gene is highly conserved in evolution [32]. In particular, the genome of Saccharomyces cerevisiae contains highly homologous genes to the mammalian Ras gene, which contains two Ras genes (Ras1 and Ras2), one Rap (Rsr1/Bud1), and one Rheb (Rhb1) direct homologue. It is significantly homologous to the mammalian Ras gene, with Ras1 and Ras2 containing 309 and 322 amino acids, respectively, and the first 180 amino acids of these two proteins are nearly 90% homologous [33,34,35].
There have been few reports of obtaining mutants of deleted genes in filamentous fungi, suggesting that Ras activity in some species is essential [36]. In the reported studies, Ras1 deletion mutants were obtained in Aspergillus fumigatus, but there were no successful cases in other filamentous fungi. Ras mutants from other families are easier to obtain [37,38]. In this study, we successfully obtained TgRas1-deleted mutants. It was found that the deletion of the TgRas1 gene affected the polar growth of hyphae and the production and germination of conidia. The marked growth inhibition caused by the ΔTgRas1 mutant may be due to the critical role of TgRas1 in maintaining the survival of filamentous fungi. Two proteins in Saccharomyces cerevisiae, Ras1 and Ras2, are thought to be involved in a range of fungal physiological processes in an antagonistic or cooperative manner, such as asexual development, polarization morphogenesis, and pathogenesis [39]. Fungal hyphae are usually able to maintain their growth direction for a considerable period of time, and from time to time they establish new growth axes, such as lateral branching. Fungal hyphae typically grow in polarity, and once polarity is established, they spread at an alarming rate.
In fact, conidial germination and mycelial growth of ΔTgRas1 were affected to varying degrees on agar solid plates, especially the polar growth of mycelium. Similarly, RasA1 mutants in yeast severely delay polarity establishment, which is characterized by the hyperswelling of conidia [40]. Previously, the ΔAoRas2 mutant of Arthrobotrys oligospora exhibited hyphal swelling, a phenotype similar to that of ΔTgRas1 [41]. The abnormal spore characteristic of ΔTgRas1 is manifested by the swelling of nascent hyphae during germination. These results may imply that the function of TgRas1 is more focused on the growth and survival of Trichoderma guizhouense NJAU4742. Mature hyphae were also found in the ΔTgRas1 mutant, but the hyphae growth rate was significantly inhibited. The growth speed of the other three TgRas mutants also showed obvious differences.
We observed that ΔTgRas1 hyphae showed edge swelling during the growth process, and the growth direction of ΔTgRas1 hyphae was affected. Ras mutants in filamentous fungi such as Neurospora crassa and Schizophyllum exhibit defects in twisted mycelial growth. In Neurospora crassa, there are ropy-like mutants. By destabilizing the apical vesicle supply center, it leads to a deviating growth trajectory [42].
Growth inhibition of the ΔTgRas1 strain further revealed inappropriate development of conidia structure. TgRas in this study also appears to be involved in a pathway that regulates conidia production. The knockout of the TgRas1 gene affected the conidial germination and hyphal development of Trichoderma and changed the colony morphology. This is similar to previous reports of the effects of Ras on other fungi. For example, disruption of Ras2 had no effect on the germination pattern of germ tubes in Fusarium graminearum but slowed conidia germination [43]. RasB knockout mutants in A. fumigatus exhibit delayed initiation of germination, but subsequent processes are not observed, in contrast to the slowed germination in the dominant-negative form of RasA. Germination was not affected in dominant-active forms of RasA or RasB [44]. However, in N. crassa, the germination rate of ΔRas2 did not change significantly [45]. In this study, spore germination was significantly delayed in all four TgRas single-knockout mutants. In ΔTgRas1 with the most obvious phenotype, not only the germination of spores was delayed but the production of conidia was also greatly delayed. WT will produce green conidia in about 4 days, but the conidia of ΔTgRas1 will be delayed to about 10 to 14 days. Obviously, the germination of spores and the generation of conidia in Trichoderma guizhouense NJAU4742 are positively regulated by TgRas family genes. In addition to being involved in the positive regulation of sporulation, TgRas1 was also found to alter mycelial branching in Trichoderma guizhouense NJAU4742. Two or more germ tubes were observed in ΔTgRas1. Fungal germination usually causes germ tubes to sprout from the apical rather than lateral sides of the conidia [46]. However, in our study, multiple germ tubes aberrantly sprouted laterally from the conidia and were dysplastic in ΔTgRas1, which shows disordered branching and slow growth. This phenomenon may be due to the instability of the polar axis of cell development. This phenomenon is very similar to the hyperbranched or serrated hyphae observed in mutants of RasB of A. fumigatus, Ras2 of Neurospora crassa, and Ras of A. gossypii [47].
In this study, ΔTgRas1 was cultured on solid plates with different carbon sources, and a reduction in radial growth, serrated colony edges, and aerial hyphae were observed. This occurs regardless of the type of carbon source. Although the ΔTgRas1 mutant had reduced radial growth on plates, its growth rate in liquid culture was indistinguishable from its wild type. This situation is similar to that seen in A. fumigatus ΔRasB mutants. Interestingly, liquid-fermented ΔTgRas1 showed severe inhibition of mycelial cell growth under a lactose carbon source, but the growth of ΔTgRas1 under other carbon source liquid fermentation conditions returned to normal levels, and polar growth inhibition was also observed. The above results indicate that TgRas1 may be related to the lactose metabolism pathway in Trichoderma guizhouense NJAU4742. In short, the TgRas family genes are critical to the growth of fungi, and most of the proteins in this family have important effects on cell wall integrity, growth site selection, polarity establishment, and maintenance. At the same time, the results of this study complement the evidence that TgRas family genes play an indispensable role in fungal survival and lactose metabolism.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation9050440/s1, Table S1: Primers used to obtain mutant strains.
Author Contributions
Conceptualization, J.M. and D.L.; methodology, J.M. and D.L.; software, C.C. and H.Z.; validation, C.C., Y.G. and J.M.; resources, Q.S.; data curation, Y.G.; writing—original draft preparation, H.G.; writing—review and editing, D.L.; supervision, Q.S.; project administration, D.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
This research was financially supported by the National Natural Science Foundation of China (31972513 and 32172680) and the Fundamental Research Funds for the Central Universities (KYZ201716). All authors agreed to publish. There is no conflict of interest between the authors.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Brotman, Y.; Kapuganti, J.G.; Viterbo, A. Trichoderma. Curr. Biol. 2010, 20, R390–R391. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Cornejo, H.A.; Lopez-Bucio, J.S.; Mendez-Bravo, A.; Macias-Rodriguez, L.; Ramos-Vega, M.; Guevara-Garcia, A.A.; Lopez-Bucio, J. Mitogen-Activated Protein Kinase 6 and Ethylene and Auxin Signaling Pathways Are Involved in Arabidopsis Root-System Architecture Alterations by Trichoderma atroviride. Mol. Plant-Microbe Interact. 2015, 28, 701–710. [Google Scholar] [CrossRef]
- Contreras-Cornejo, H.A.; Macias-Rodriguez, L.; Vergara, A.G.; Lopez-Bucio, J. Trichoderma Modulates Stomatal Aperture and Leaf Transpiration Through an Abscisic Acid-Dependent Mechanism in Arabidopsis. J. Plant Growth Regul. 2015, 34, 425–432. [Google Scholar] [CrossRef]
- Zin, N.A.; Badaluddin, N.A. Biological functions of Trichoderma spp. for agriculture applications. Ann. Agric. Sci. 2020, 65, 168–178. [Google Scholar] [CrossRef]
- Peterson, R.; Nevalainen, H. Trichoderma reesei RUT-C30–thirty years of strain improvement. Microbiology 2012, 158, 58–68. [Google Scholar] [CrossRef]
- Li, Q.R.; Tan, P.; Jiang, Y.L.; Hyde, K.D.; McKenzie, E.H.C.; Bahkali, A.H.; Kang, J.C.; Wang, Y. A novel Trichoderma species isolated from soil in Guizhou, T. guizhouense. Mycol. Prog. 2013, 12, 167–172. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, J.; Shao, J.H.; Feng, H.C.; Zhang, R.F.; Shen, Q.R. Extracellular proteins of Trichoderma guizhouense elicit an immune response in maize (Zea mays) plants. Plant Soil 2020, 449, 133–149. [Google Scholar] [CrossRef]
- Saloheimo, M.; Wang, H.; Valkonen, M.; Vasara, T.; Huuskonen, A.; Riikonen, M.; Pakula, T.; Ward, M.; Penttila, M. Characterization of secretory genes ypt1/yptA and nsf1/nsfA from two filamentous fungi: Induction of secretory pathway genes of Trichoderma reesei under secretion stress conditions. Appl. Environ. Microbiol. 2004, 70, 459–467. [Google Scholar] [CrossRef]
- Sternberg, D.; Mandels, G. Induction of cellulolytic enzymes in Trichoderma reesei by sophorose. J. Bacteriol. 1979, 139, 761–769. [Google Scholar] [CrossRef]
- Ivanova, C.; Bååth, J.A.; Seiboth, B.; Kubicek, C.P. Systems analysis of lactose metabolism in Trichoderma reesei identifies a lactose permease that is essential for cellulase induction. PLoS ONE 2013, 8, e62631. [Google Scholar] [CrossRef]
- Porciuncula, J.D.; Furukawa, T.; Shida, Y.; Mori, K.; Kuhara, S.; Morikawa, Y.; Ogasawara, W. Identification of Major Facilitator Transporters Involved in Cellulase Production during Lactose Culture of Trichoderma reesei PC-3-7. Biosci. Biotechnol. Biochem. 2013, 77, 1014–1022. [Google Scholar] [CrossRef]
- Seiboth, B.; Hofmann, G.; Kubicek, C. Lactose metabolism and cellulase production in Hypocrea jecorina: The gal7 gene, encoding galactose-1-phosphate uridylyltransferase, is essential for growth on galactose but not for cellulase induction. Mol. Genet. Genom. 2002, 267, 124–132. [Google Scholar] [CrossRef]
- Karaffa, L.; Fekete, E.; Gamauf, C.; Szentirmai, A.; Kubicek, C.P.; Seiboth, B. d-Galactose induces cellulase gene expression in Hypocrea jecorina at low growth rates. Microbiology 2006, 152, 1507–1514. [Google Scholar] [CrossRef]
- Yaar, L.; Mevarech, M.; Koltint, Y. A Candida albicans RAS-related gene (CaRSRl) is involved in budding, cell morphogenesis and hypha development. Microbiology 1997, 143, 3033–3044. [Google Scholar] [CrossRef]
- Weeks, G.; Spiegelman, G.B. Roles played by Ras subfamily proteins in the cell and developmental biology of microorganisms. Cell Signal. 2003, 15, 901–909. [Google Scholar] [CrossRef] [PubMed]
- Martin-Vicente, A.; Souza, A.C.O.; Al Abdallah, Q.; Ge, W.; Fortwendel, J.R. SH3-class Ras guanine nucleotide exchange factors are essential for Aspergillus fumigatus invasive growth. Cell Microbiol. 2019, 21, e13013. [Google Scholar] [CrossRef]
- Fortwendel, J.R.; Seitz, A.E.; Askew, D.S.; Rhodes, J.C. Aspergillus Fumigatus rasA: A Non-Essential Gene that Regulates Germination, Mitosis and Hyphal Morphology. Ph.D. Thesis, University of Cincinnati, Cincinnati, Ohio, 1999. [Google Scholar]
- Zhu, Z.; Ma, G.; Yang, M.; Tan, C.; Yang, G.; Wang, S.; Li, N.; Ge, F.; Wang, S. Ras subfamily GTPases regulate development, aflatoxin biosynthesis and pathogenicity in the fungus Aspergillus flavus. Environ. Microbiol. 2021, 23, 5334–5348. [Google Scholar] [CrossRef]
- Miao, Y.; Xia, Y.; Kong, Y.; Zhu, H.; Mei, H.; Li, P.; Feng, H.; Xun, W.; Xu, Z.; Zhang, N.; et al. Overcoming diverse homologous recombinations and single chimeric guide RNA competitive inhibition enhances Cas9-based cyclical multiple genes coediting in filamentous fungi. Environ. Microbiol. 2021, 23, 2937–2954. [Google Scholar] [CrossRef]
- Kubicek, C.P.; Messner, R.; Gruber, F.; Mach, R.L.; Kubicek-Pranz, E.M. The Trichoderma cellulase regulatory puzzle: From the interior life of a secretory fungus. Enzym. Microb. Technol. 1993, 15, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Mach, R.; Zeilinger, S. Regulation of gene expression in industrial fungi: Trichoderma. Appl. Microbiol. Biotechnol. 2003, 60, 515–522. [Google Scholar] [CrossRef]
- Techapun, C.; Poosaran, N.; Watanabe, M.; Sasaki, K. Thermostable and alkaline-tolerant microbial cellulase-free xylanases produced from agricultural wastes and the properties required for use in pulp bleaching bioprocesses: A review. Process Biochem. 2003, 38, 1327–1340. [Google Scholar] [CrossRef]
- Delabona, P.d.S.; Pirota, R.D.P.B.; Codima, C.A.; Tremacoldi, C.R.; Rodrigues, A.; Farinas, C.S. Using Amazon forest fungi and agricultural residues as a strategy to produce cellulolytic enzymes. Biomass Bioenergy 2012, 37, 243–250. [Google Scholar] [CrossRef]
- Mussatto, S.I.; Ballesteros, L.F.; Martins, S.; Teixeira, J.A. Use of agro-industrial wastes in solid-state fermentation processes. Ind. Waste 2012, 274. [Google Scholar] [CrossRef]
- Santos, F.A.; Carvalho-Gonçalves, L.C.T.d.; Cardoso-Simões, A.L.d.C.; Santos, S.F.d.M. Evaluation of the production of cellulases by Penicillium sp. FSDE15 using corncob and wheat bran as substrates. Technol. Rep. 2021, 14, 100648. [Google Scholar] [CrossRef]
- Adsul, M.G.; Bastawde, K.B.; Varma, A.J.; Gokhale, D.V. Strain improvement of Penicillium janthinellum NCIM 1171 for increased cellulase production. Bioresour. Technol. 2007, 98, 1467–1473. [Google Scholar] [CrossRef]
- Steudler, S.; Werner, A.; Walther, T. It is the mix that matters: Substrate-specific enzyme production from filamentous fungi and bacteria through solid-state fermentation. Solid State Ferment. 2019, 169, 51–81. [Google Scholar]
- Singhania, R.R.; Sukumaran, R.K.; Patel, A.K.; Larroche, C.; Pandey, A. Advancement and comparative profiles in the production technologies using solid-state and submerged fermentation for microbial cellulases. Enzym. Microb. Technol. 2010, 46, 541–549. [Google Scholar] [CrossRef]
- Stricker, A.R.; Steiger, M.G.; Mach, R.L. Xyr1 receives the lactose induction signal and regulates lactose metabolism in Hypocrea jecorina. FEBS Lett. 2007, 581, 3915–3920. [Google Scholar] [CrossRef]
- Fortwendel, J.R.; Juvvadi, P.R.; Rogg, L.E.; Asfaw, Y.G.; Burns, K.A.; Randell, S.H.; Steinbach, W.J. Plasma Membrane Localization Is Required for RasA-Mediated Polarized Morphogenesis and Virulence of Aspergillus fumigatus. Eukaryot. Cell 2012, 11, 966–977. [Google Scholar] [CrossRef]
- Arkowitz, R.A.; Bassilana, M. Regulation of hyphal morphogenesis by Ras and Rho small. GTPases. Fungal Biol. Rev. 2015, 29, 7–19. [Google Scholar] [CrossRef]
- Yang, L.; Li, X.; Xie, M.; Bai, N.; Yang, J.; Jiang, K.; Zhang, K.-Q.; Yang, J. Pleiotropic roles of Ras GTPases in the nematode-trapping fungus Arthrobotrys oligospora identified through multi-omics analyses. iScience 2021, 24, 102820. [Google Scholar] [CrossRef] [PubMed]
- Casas-Flores, S.; Rios-Momberg, M.; Rosales-Saavedra, T.; Martinez-Hernandez, P.; Olmedo-Monfil, V.; Herrera-Estrella, A. Cross talk between a fungal blue-light perception system and the cyclic AMP signaling pathway. Eukaryot. Cell 2006, 5, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Tamanoi, F. Ras Signaling in Yeast. Genes Cancer 2011, 2, 210–215. [Google Scholar] [CrossRef]
- Knapp, G.S.; McDonough, K.A. Cyclic AMP Signaling in Mycobacteria. Microbiol. Spectr. 2014, 2, 2. [Google Scholar] [CrossRef]
- Nogueira, K.M.V.; Costa, M.d.N.; de Paula, R.G.; Mendonça-Natividade, F.C.; Ricci-Azevedo, R.; Silva, R.N. Evidence of cAMP involvement in cellobiohydrolase expression and secretion by Trichoderma reesei in presence of the inducer sophorose. BMC Microbiol. 2015, 15, 195. [Google Scholar] [CrossRef]
- Som, T.; Kolaparthi, V.S. Developmental decisions in Aspergillus nidulans are modulated by Ras activity. Mol. Cell Biol. 1994, 14, 5333–5348. [Google Scholar] [CrossRef]
- Boyce, K.J.; Hynes, M.J.; Andrianopoulos, A. The Ras and Rho GTPases genetically interact to co-ordinately regulate cell polarity during development in Penicillium marneffei. Mol. Microbiol. 2005, 55, 1487–1501. [Google Scholar] [CrossRef] [PubMed]
- Lengeler Klaus, B.; Davidson Robert, C.; D’Souza, C.; Harashima, T.; Shen, W.-C.; Wang, P.; Pan, X.; Waugh, M.; Heitman, J. Signal Transduction Cascades Regulating Fungal Development and Virulence. Microbiol. Mol. Biol. Rev. 2000, 64, 746–785. [Google Scholar] [CrossRef]
- Brown, N.A.; Ries, L.N.A.; Goldman, G.H. How nutritional status signalling coordinates metabolism and lignocellulolytic enzyme secretion. Fungal Genet. Biol. 2014, 72, 48–63. [Google Scholar] [CrossRef]
- Schuster, A.; Tisch, D.; Seidl-Seiboth, V.; Kubicek, C.P.; Schmoll, M. Roles of Protein Kinase A and Adenylate Cyclase in Light-Modulated Cellulase Regulation in Trichoderma reesei. Appl. Environ. Microbiol. 2012, 78, 2168–2178. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, M.; Roberson, R.W.; McDaniel, D.P.; Bartnicki-García, S. The effects of ropy-1 mutation on cytoplasmic organization and intracellular motility in mature hyphae of Neurospora crassa. Fungal Genet. Biol. 2002, 37, 171–179. [Google Scholar] [CrossRef]
- Bluhm, B.H.; Zhao, X.; Flaherty, J.E.; Xu, J.R.; Dunkle, L.D. RAS2 Regulates Growth and Pathogenesis in Fusarium graminearum. Mol. Plant-Microbe Interactions® 2007, 20, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Fortwendel, J.R.; Panepinto, J.C.; Seitz, A.E.; Askew, D.S.; Rhodes, J.C. Aspergillus fumigatus rasA and rasB regulate the timing and morphology of asexual development. Fungal Genet. Biol. 2004, 41, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Kana-uchi, A.; Yamashiro, C.T.; Tanabe, S.; Murayama, T. A ras homologue of Neurospora crassa regulates morphology. Mol. Gen. Genet. MGG 1997, 254, 427–432. [Google Scholar] [CrossRef]
- D’Enfert, C. Fungal Spore Germination: Insights from the Molecular Genetics ofAspergillus nidulansandNeurospora crassa. Fungal Genet. Biol. 1997, 21, 163–172. [Google Scholar] [CrossRef]
- Bauer, Y.; Knechtle, P.; Wendland, J.; Helfer, H.; Philippsen, P. A Ras-like GTPase is involved in hyphal growth guidance in the filamentous fungus Ashbya gossypii. Mol. Biol. Cell 2004, 15, 4622–4632. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).