Effects of Different Fermentation Methods on the Quality and Microbial Diversity of Passion Fruit Wine
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Determination of Physicochemical Indices during the Fermentation of Passion Fruit Wine
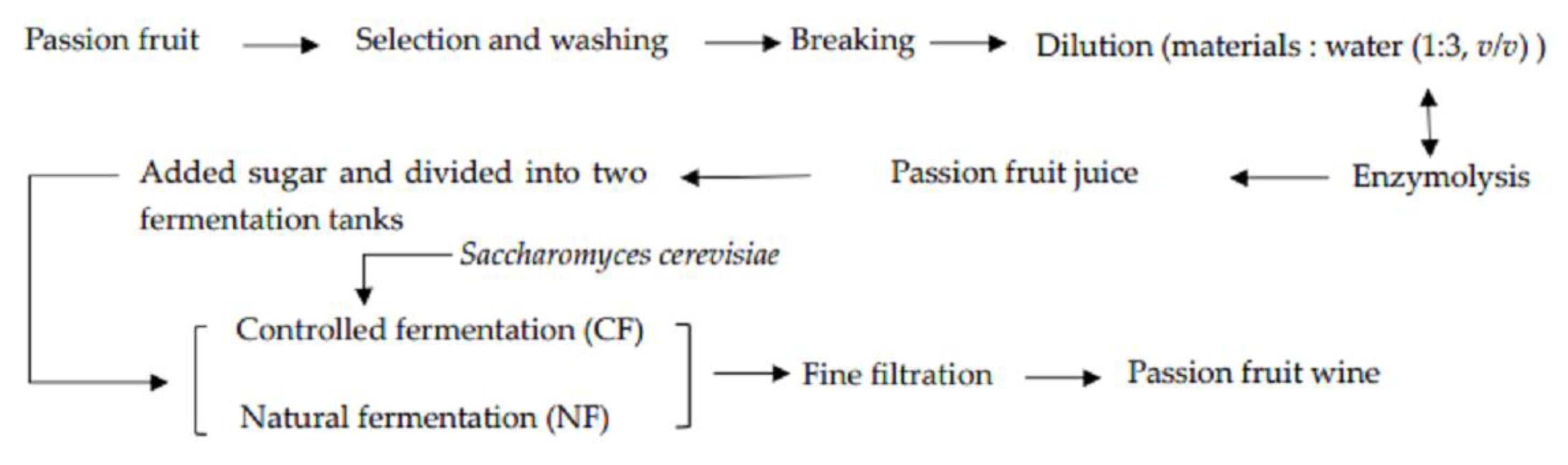
2.2.1. Fermentation Process of Passion Fruit Wine
2.2.2. Determination of Physicochemical Indices
2.3. Determination of Microbial Diversity during the Fermentation of Passion Fruit Wine
2.3.1. Sample Collection
2.3.2. Determination of Microbial Diversity
2.4. Determination of Nonvolatile Metabolites during the Fermentation of Passion Fruit Wine
2.5. Determination of Volatile Flavor Substances during the Fermentation of Passion Fruit Wine
2.6. Data Analysis
3. Results and Discussion
3.1. Changes in Physicochemical Indices during the Fermentation of Passion Fruit Wine
3.2. Analysis of Microbial Diversity during the Fermentation of Passion Fruit Wine
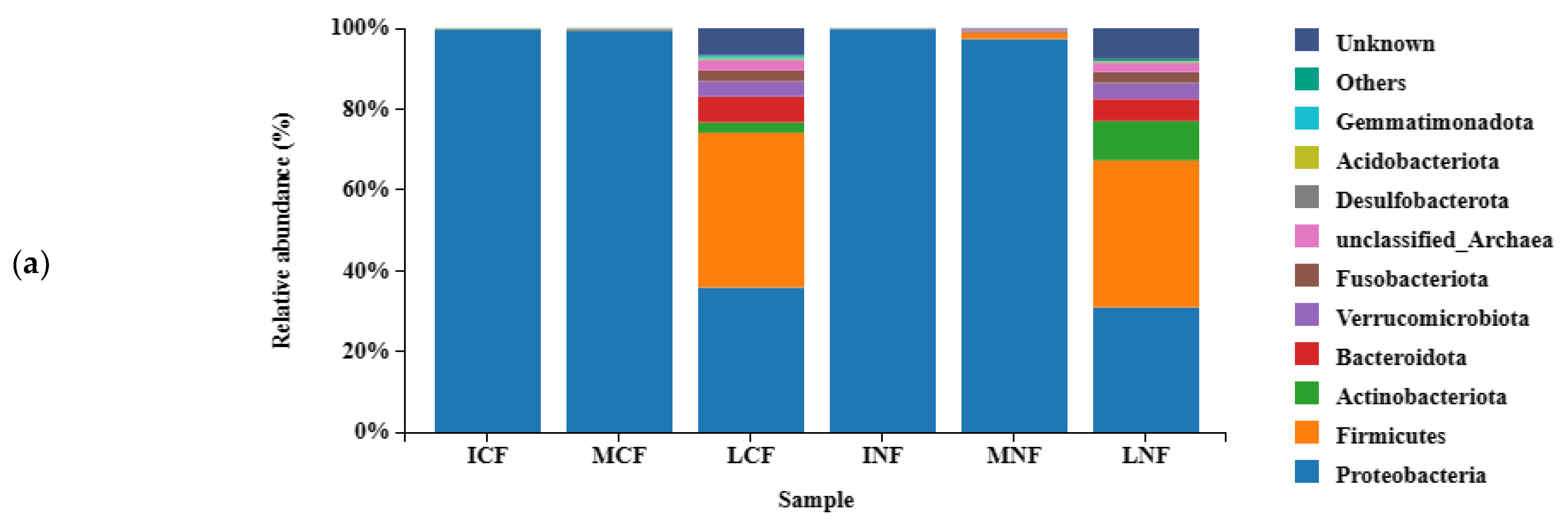
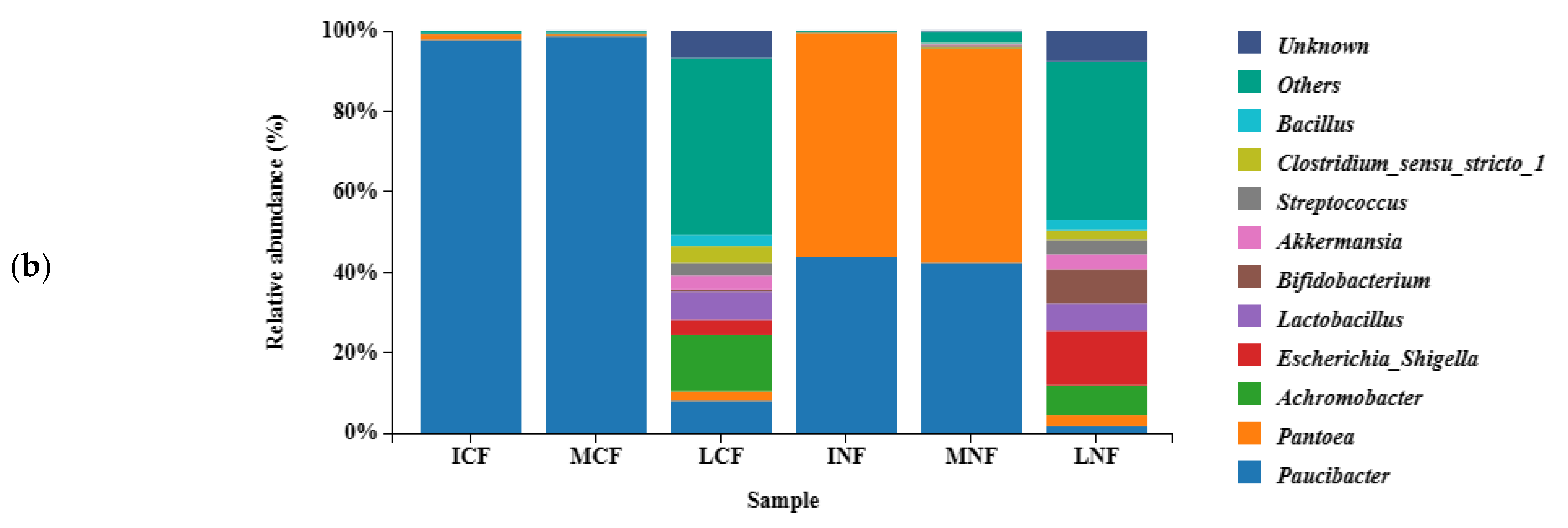
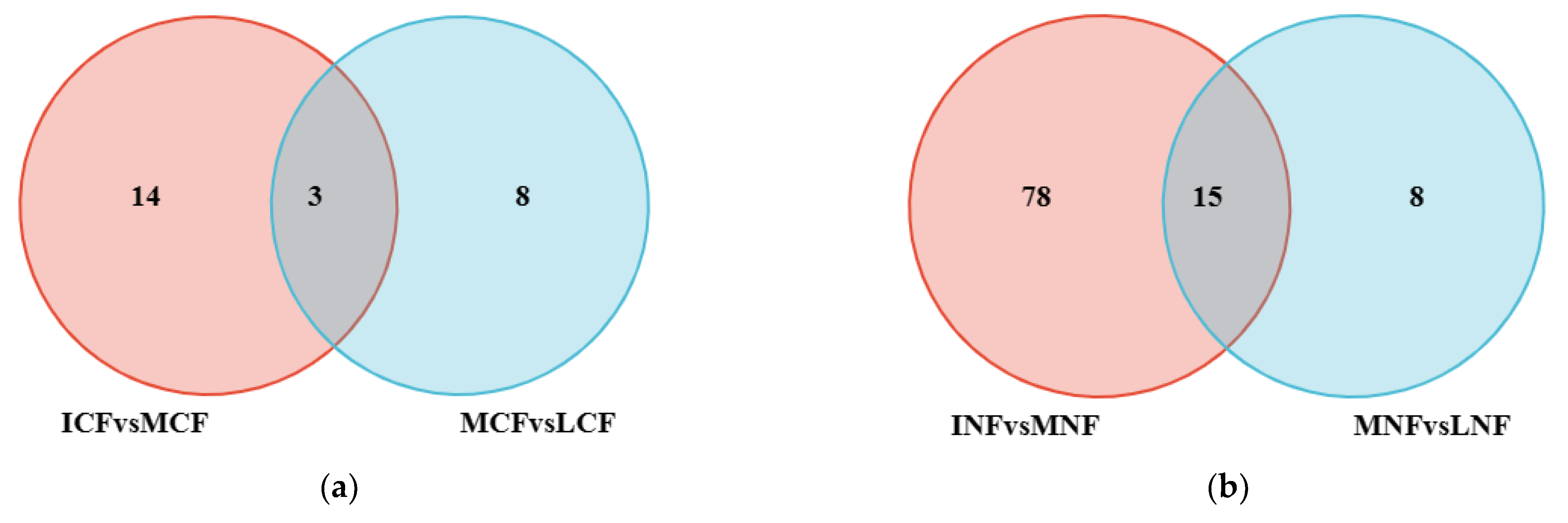
3.2.1. Fungal Population Structure
3.2.2. Bacterial Population Structure
3.3. Analysis of Nonvolatile Metabolites and KEGG (Kyoto Encyclopedia of Genes and Genomes) Enrichment during the Fermentation of Passion Fruit Wine
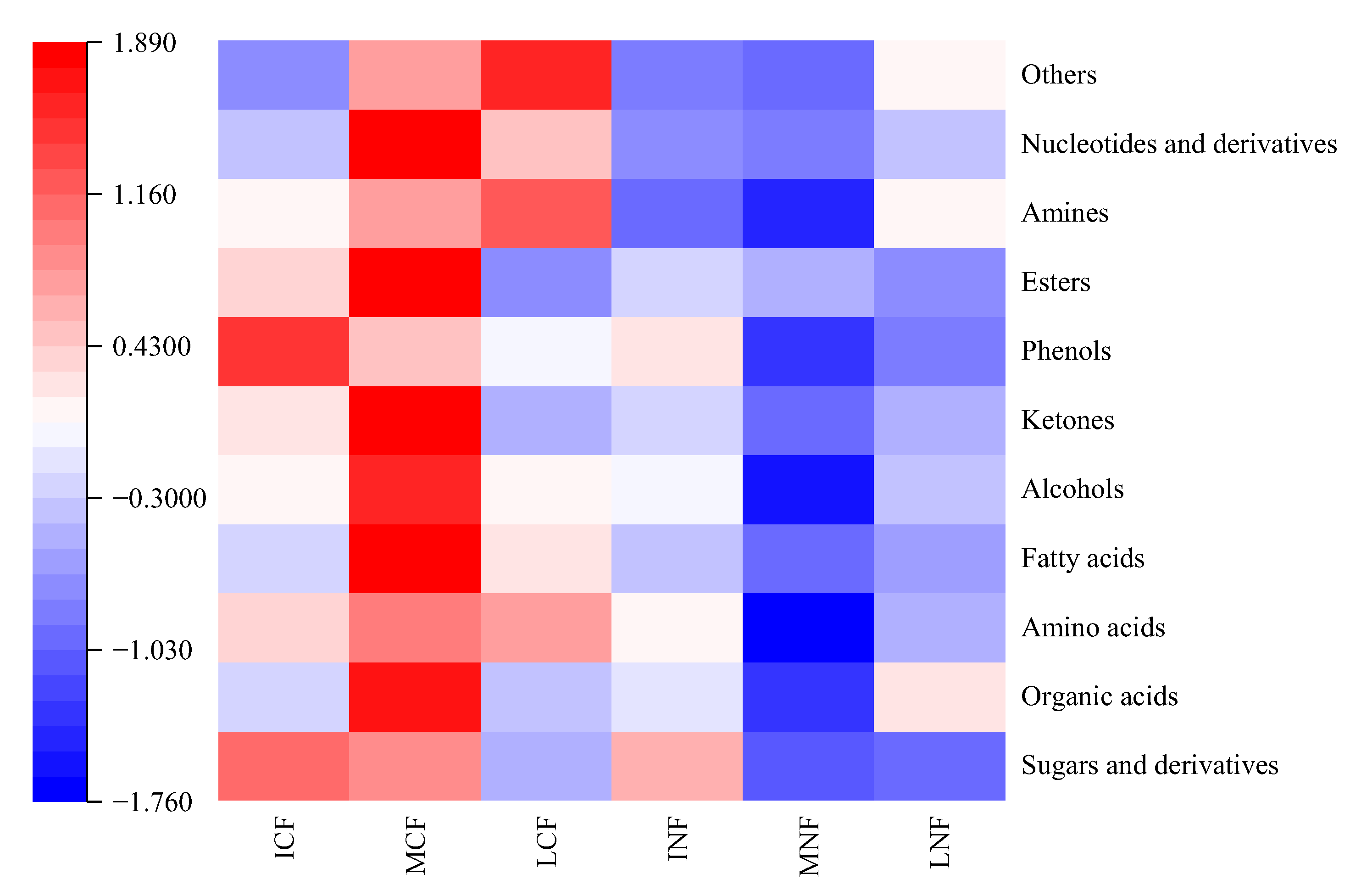
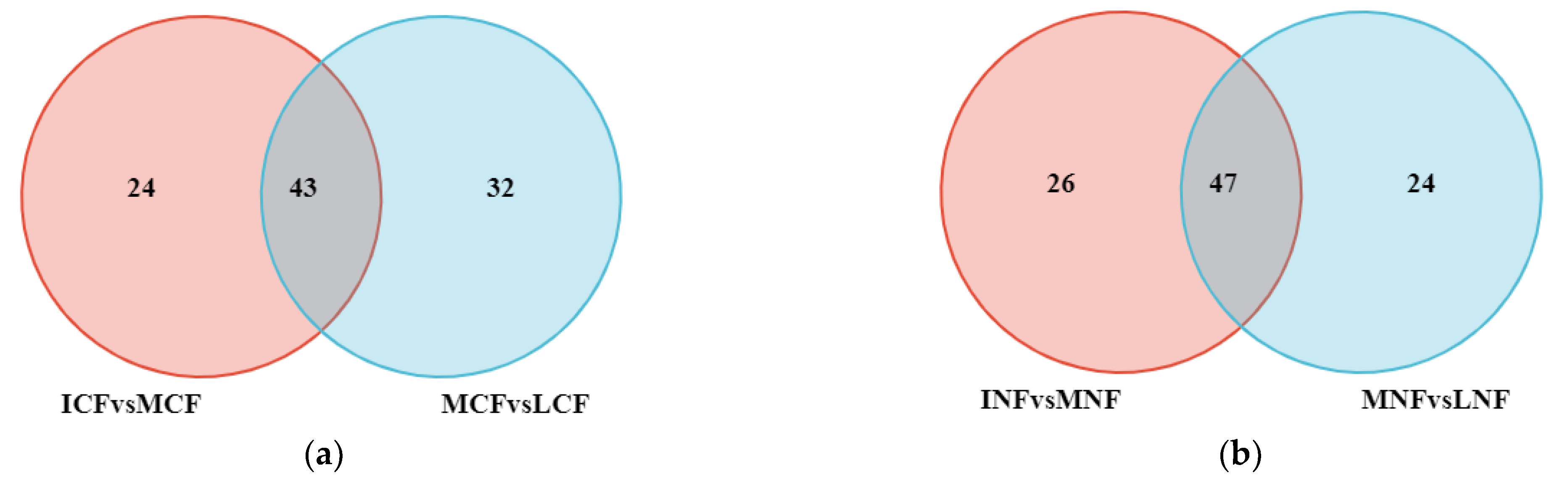
3.3.1. Analysis of Different Types of Nonvolatile Metabolites during Fermentation
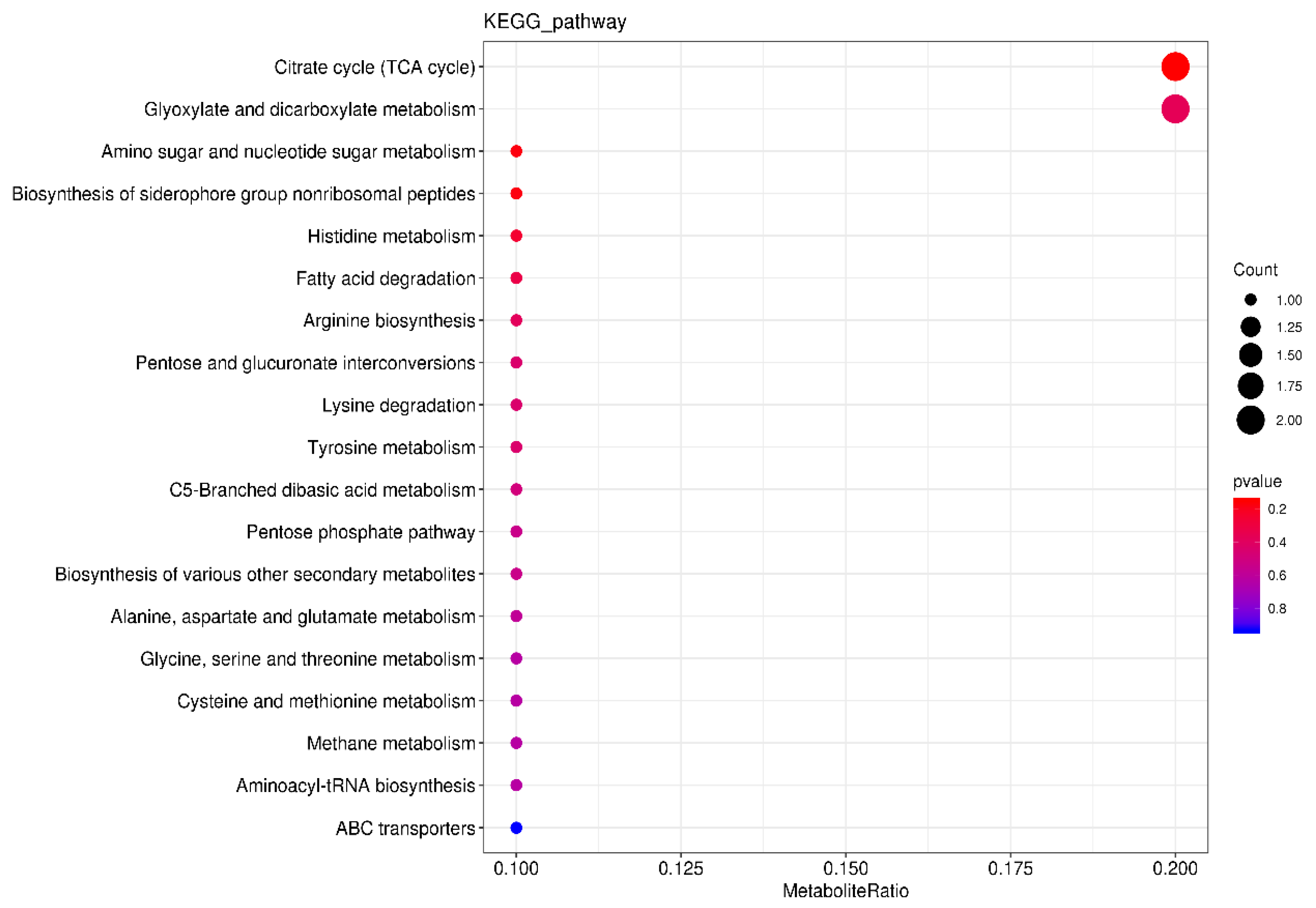
3.3.2. Screening of Key Nonvolatile Metabolites and Analysis of Metabolic Pathways during Fermentation
3.4. Analysis of Volatile Flavor Substances during the Fermentation of Passion Fruit Wine
3.4.1. Volatile Flavor Substances during Fermentation
3.4.2. Screening of Key Volatile Flavor Compounds during Fermentation
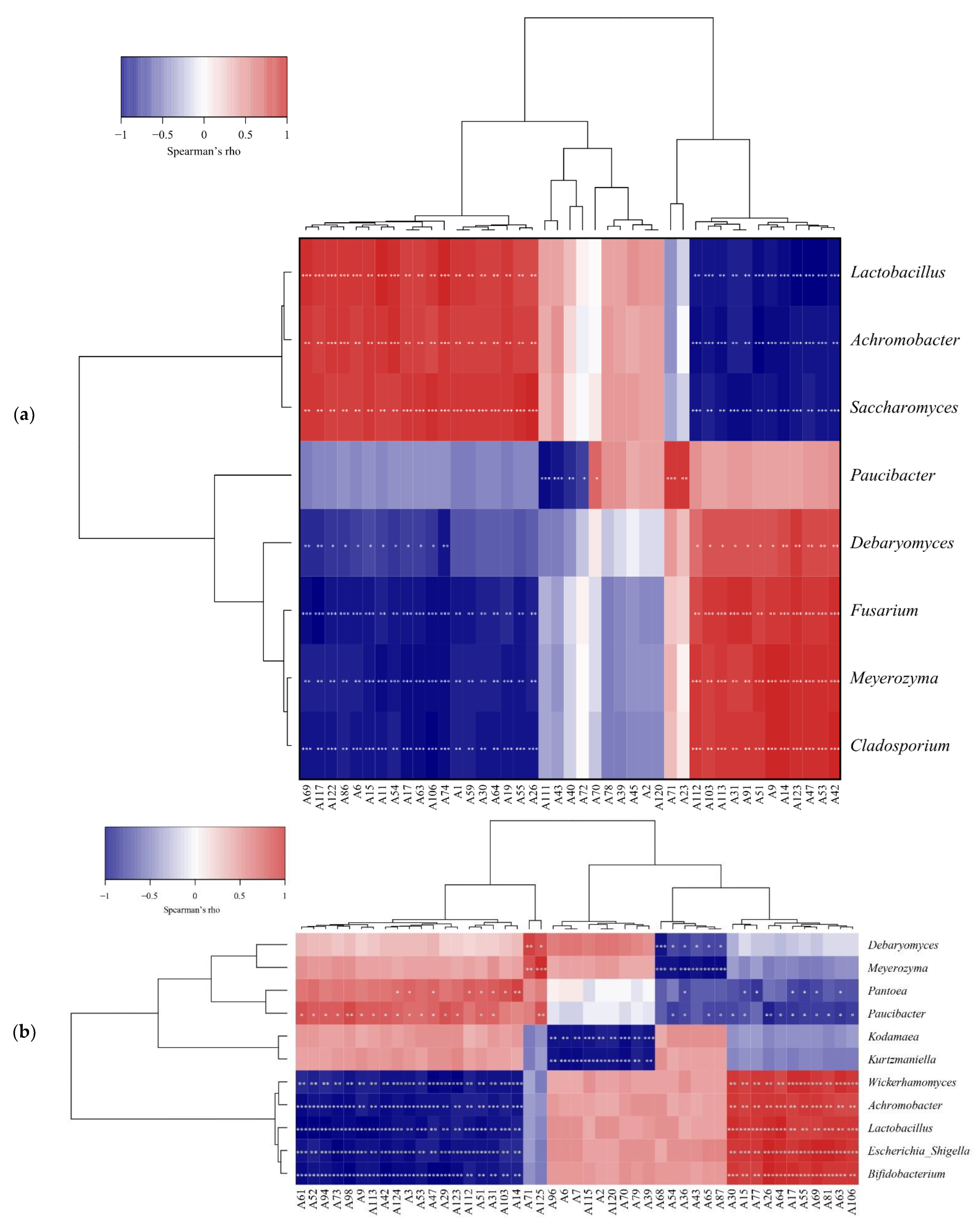
3.5. Correlation between the Microbial Community and Flavor Substances during Fermentation
3.5.1. Correlation between Physicochemical Indices and Microbial Community
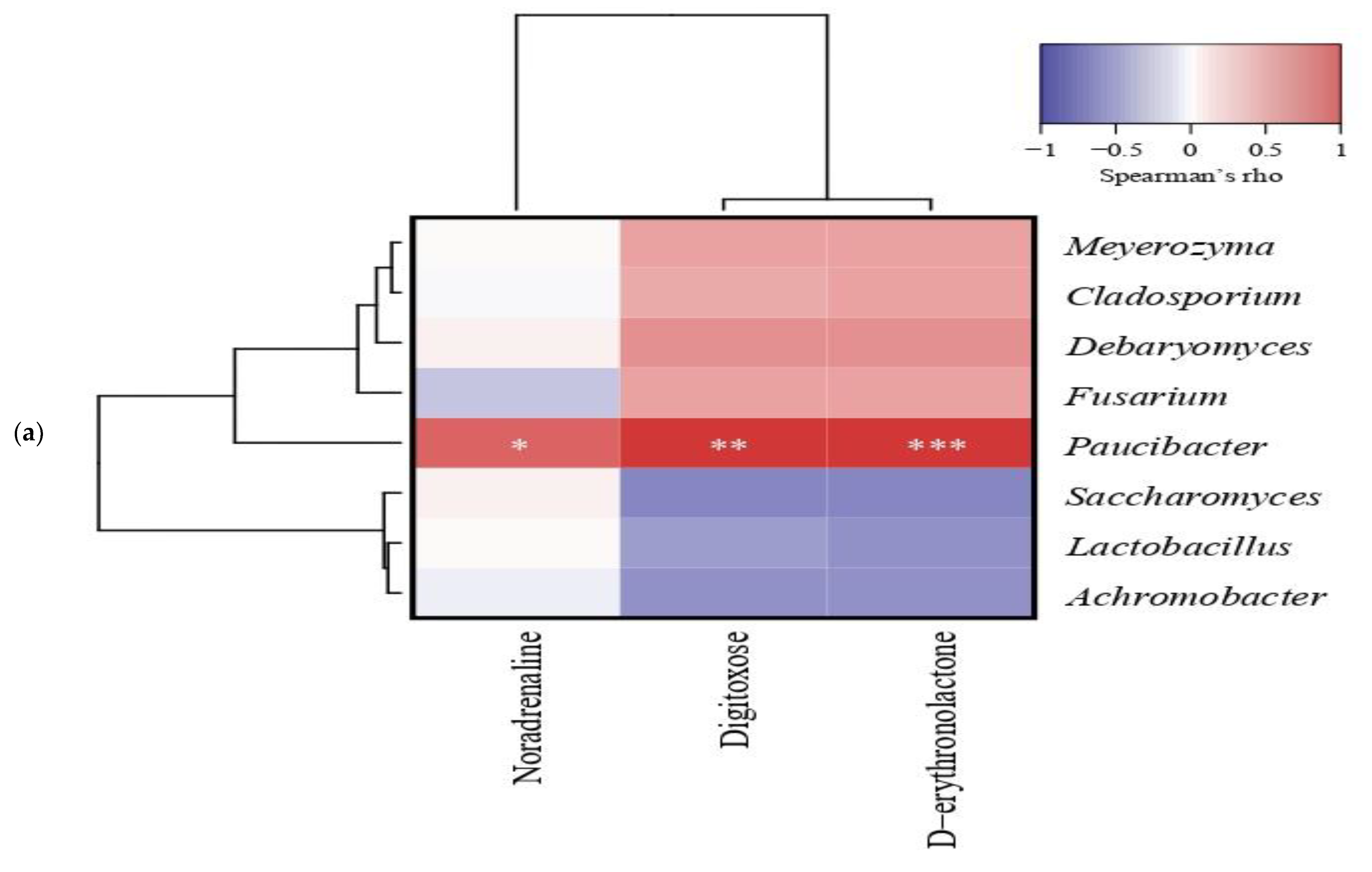
3.5.2. Correlation between the Microbial Community and Nonvolatile Differential Metabolites
3.5.3. Correlation between the Microbial Community and Volatile Flavor Substances
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xinhui, S.; Jun, T.; Xiu, L.; Yiyuan, Y.; Liping, P.; Qiong, Z.; Yongxian, L. Development of Selenium-Rich Passion Fruit Industry in Guangxi. Agric. Biotechnol. 2019, 8, 38–41. [Google Scholar]
- Asadujjaman, M.; Mishuk, A.U.; Hossain, M.A.; Karmakar, U.K. Medicinal potential of Passiflora foetida L. plant extracts: Biological and pharmacological activities. J. Intefgr. Med. 2014, 12, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Niwayama, S.; Higuchi, H. Passion fruit quality under acidic soil conditions. Hortic. J. 2019, 88, 50–56. [Google Scholar] [CrossRef]
- Wijaya, C.J.; Saputra, S.N.; Soetaredjo, F.E.; Putro, J.N.; Lin, C.X.; Kurniawan, A.; Ju, Y.-H.; Ismadji, S. Cellulose nanocrystals from passion fruit peels waste as antibiotic drug carrier. Carbohydr. Polym. 2017, 175, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Dos Reis, L.C.R.; Facco, E.M.P.; Salvador, M.; Flôres, S.H.; de Oliveira Rios, A. Antioxidant potential and physicochemical characterization of yellow, purple and orange passion fruit. J. Food Sci. Technol. 2018, 55, 2679–2691. [Google Scholar] [CrossRef]
- Sanchez, B.; Celestino, S.; de Abreu Gloria, M.; Celestino, I.; Lozada, M.; Júnior, S.; de Alencar, E.; de Oliveira, L.D.L. Pasteurization of passion fruit Passiflora setacea pulp to optimize bioactive compounds retention. Food Chem. X 2020, 6, 100084. [Google Scholar] [CrossRef]
- Nguyen, T.T.T.; Le, T.Q.; Nguyen, T.T.A.; Nguyen, L.T.M.; Nguyen, D.T.C.; Van Tran, T. Characterizations and antibacterial activities of passion fruit peel pectin/chitosan composite films incorporated Piper betle L. leaf extract for preservation of purple eggplants. Heliyon 2022, 8, e10096. [Google Scholar] [CrossRef]
- Urrego, N.; Sepúlveda, P.; Aragón, M.; Ramos, F.A.; Costa, G.M.; Ospina, L.F.; Castellanos, L. Flavonoids and saponins from Passiflora edulis f. edulis leaves (purple passion fruit) and its potential anti-inflammatory activity. J. Pharm. Pharmacol. 2021, 73, 1530–1538. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Katano, Y. Cardiovascular protective effects of polyphenols contained in passion fruit seeds namely piceatannol and scirpusin B: A review. Tokai J. Exp. Clin. Med. 2021, 46, 151–161. [Google Scholar] [PubMed]
- Cabral, B.; Goncalves, T.A.F.; Abreu, L.S.; Andrade, A.W.L.; de Castro, F.D.; Tavares, J.F.; Guerra, G.C.B.; de Rezende, A.A.; de Medeiros, I.A.; Zucolotto, S.M. Cardiovascular effects induced by fruit peels from Passiflora edulis in hypertensive rats and fingerprint analysis by HPLC-ESI-MSn spectrometry. Planta Med. 2022, 88, 356–366. [Google Scholar] [CrossRef]
- Li, W.; Li, C.; Zhang, F.; Huang, B. Research progress on the nutritional quality and functional substances of passionflower and its application. J. China Agric. Univ. 2022, 27, 79–92. [Google Scholar] [CrossRef]
- Chen, D.; Yap, Z.Y.; Liu, S.-Q. Evaluation of the performance of Torulaspora delbrueckii, Williopsis saturnus, and Kluyveromyces lactis in lychee wine fermentation. Int. J. Food Microbiol. 2015, 206, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Sadoudi, M.; Tourdot-Maréchal, R.; Rousseaux, S.; Steyer, D.; Gallardo-Chacón, J.-J.; Ballester, J.; Vichi, S.; Guérin-Schneider, R.; Caixach, J.; Alexandre, H. Yeast–yeast interactions revealed by aromatic profile analysis of Sauvignon Blanc wine fermented by single or co-culture of non-Saccharomyces and Saccharomyces yeasts. Food Microbiol. 2012, 32, 243–253. [Google Scholar] [CrossRef]
- Tofalo, R.; Patrignani, F.; Lanciotti, R.; Perpetuini, G.; Schirone, M.; Di Gianvito, P.; Pizzoni, D.; Arfelli, G.; Suzzi, G. Aroma profile of Montepulciano d’Abruzzo wine fermented by single and co-culture starters of autochthonous Saccharomyces and non-Saccharomyces yeasts. Front. Microbiol. 2016, 7, 610. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, M.; Ye, P.; Lin, F.; Huang, J.; Wang, H.; Zhou, R.; Zhang, S.; Zhou, J.; Cai, L. Characterization of major properties and aroma profile of kiwi wine co-cultured by Saccharomyces yeast (S. cerevisiae, S. bayanus, S. uvarum) and T. delbrueckii. Eur. Food Res. Technol. 2020, 246, 807–820. [Google Scholar] [CrossRef]
- Bagheri, B.; Bauer, F.F.; Setati, M.E. The impact of Saccharomyces cerevisiae on a wine yeast consortium in natural and inoculated fermentations. Front. Microbiol. 2017, 8, 1988. [Google Scholar] [CrossRef] [PubMed]
- Benito, S. The impact of Torulaspora delbrueckii yeast in winemaking. Appl. Microbiol. Biotechnol. 2018, 102, 3081–3094. [Google Scholar] [CrossRef]
- Gu, X.; Li, S.; Yang, L.; Liu, Y.; Ma, Y.; Chen, Z.; Wang, Y.; Liu, X. Analysis of Microbial Diversity, Physicochemical Indexes and Correlation with Volatile Flavor during the Fermentation of Red Millet Huangjiu. Sci. Technol. Food Ind. 2022, 43, 133–143. [Google Scholar]
- Deng, X.; Huang, Z.; Yao, Y.; Jiang, K.; Deng, J.; Ren, Z. Effect of initial acidity on fermentation of Saccharomyces cerevisiae. China Brew. 2023, 42, 115–119. [Google Scholar]
- Santos, R.d.S.; Biasoto, A.; Rybka, A.; Castro, C.d.C.; Aidar, S.d.T.; Borges, G.; Silva, F. Physicochemical characterization, bioactive compounds, in vitro antioxidant activity, sensory profile and consumer acceptability of fermented alcoholic beverage obtained from caatinga passion fruit (Passiflora cincinnata Mast.). LWT 2021, 148, 111714. [Google Scholar] [CrossRef]
- Guo, C.; Gao, T.; Zhao, E. Determination of total sugar from wine by phenol-sulfuric acid coloration method. China Brew. 2008, 22, 84–85. [Google Scholar]
- Lee, S.G.; Vance, T.M.; Nam, T.-G.; Kim, D.-O.; Koo, S.I.; Chun, O.K. Evaluation of pH differential and HPLC methods expressed as cyanidin-3-glucoside equivalent for measuring the total anthocyanin contents of berries. J. Food Meas. Charact. 2016, 10, 562–568. [Google Scholar] [CrossRef]
- Zhu, X.; Zhang, F.; Yu, J.; Hu, J.; Wu, S.; Wang, D. Analysis of microbial community structure and metabolic characteristics of dominant microbes in rice milk for yellow wine. Food Sci. 2017, 38, 82–86. [Google Scholar]
- Jia, M.; Yang, X.; Zhou, G.; Guo, Q.; Zhu, L.; Zheng, Y. Microbial Community Diversity Analysis during Spontaneous Fermentation of Aromatic Lees Made from Huang Jiu (Yellow Rice Wine) Lees by High-Throughput Sequencing. Food Sci. 2022. Available online: http://kns.cnki.net/kcms/detail/11.2206.TS.20220819.0914.018.html (accessed on 16 November 2022).
- Mu, Y.; Jiang, L.; Su, W. Analysis of Microbial Diversity in Three Rice Wine Kojis by Illumina High-Throughput Sequencing. Food Sci. 2019, 40, 115–122. [Google Scholar]
- Liu, Y.; Huang, J.; Liu, J.; Wu, C.; Jin, Y.; Zhou, R. Effect of different inoculation on the quality of greengage wines and microbial diversity. Food Ferment. Ind. 2021, 47, 59–66. [Google Scholar]
- Zhang, S.; Liao, Y.; Ji, N.; Zhou, X. Analysis on microbial diversity of Beijing light-flavor Daqu by high-throughput sequencing. China Brew. 2016, 35, 49–53. [Google Scholar]
- Jeong, D.M.; Yoo, S.J.; Jeon, M.-S.; Chun, B.H.; Han, D.M.; Jeon, C.O.; Eyun, S.-I.; Seo, Y.-J.; Kang, H.A. Genomic features, aroma profiles, and probiotic potential of the Debaryomyces hansenii species complex strains isolated from Korean soybean fermented food. Food Microbiol. 2022, 105, 104011. [Google Scholar] [CrossRef]
- Yan, W.; Gao, H.; Qian, X.; Jiang, Y.; Zhou, J.; Dong, W.; Xin, F.; Zhang, W.; Jiang, M. Biotechnological applications of the non-conventional yeast Meyerozyma guilliermondii. Biotechnol. Adv. 2021, 46, 107674. [Google Scholar] [CrossRef]
- Benito, S.; Morata, A.; Palomero, F.; González, M.; Suárez-Lepe, J. Formation of vinylphenolic pyranoanthocyanins by Saccharomyces cerevisiae and Pichia guillermondii in red wines produced following different fermentation strategies. Food Chem. 2011, 124, 15–23. [Google Scholar] [CrossRef]
- Aplin, J.J.; Paup, V.D.; Ross, C.F.; Edwards, C.G. Chemical and sensory profiles of merlot wines produced by sequential inoculation of Metschnikowia pulcherrima or Meyerzyma guilliermondii. Fermentation 2021, 7, 126. [Google Scholar] [CrossRef]
- Padilla, B.; Gil, J.V.; Manzanares, P. Challenges of the non-conventional yeast Wickerhamomyces anomalus in winemaking. Fermentation 2018, 4, 68. [Google Scholar] [CrossRef]
- Sabel, A.; Martens, S.; Petri, A.; König, H.; Claus, H. Wickerhamomyces anomalus AS1: A new strain with potential to improve wine aroma. Ann. Microbiol. 2014, 64, 483–491. [Google Scholar] [CrossRef]
- Sun, N.; Gao, Z.; Li, S.; Chen, X.; Guo, J. Assessment of chemical constitution and aroma properties of kiwi wines obtained from pure and mixed fermentation with Wickerhamomyces anomalus and Saccharomyces cerevisiae. J. Sci. Food Agric. 2022, 102, 175–184. [Google Scholar] [CrossRef]
- Zhao, J.; Qiu, S.; Wang, X.; Yuan, M.; Zhou, H. Interaction Mechanism of Wickerhamomyces anomalus and Saccharomyces cerevisiae in Mixed Culture Fermentation. Food Sci. 2022, 43, 201–208. [Google Scholar]
- Sieuwerts, S.; Bron, P.A.; Smid, E.J. Mutually stimulating interactions between lactic acid bacteria and Saccharomyces cerevisiae in sourdough fermentation. LWT 2018, 90, 201–206. [Google Scholar] [CrossRef]
- Pina, C.; Santos, C.; Couto, J.A.; Hogg, T. Ethanol tolerance of five non-Saccharomyces wine yeasts in comparison with a strain of Saccharomyces cerevisiae—Influence of different culture conditions. Food Microbiol. 2004, 21, 439–447. [Google Scholar] [CrossRef]
- Rojas, V.; Gil, J.V.; Piñaga, F.; Manzanares, P. Acetate ester formation in wine by mixed cultures in laboratory fermentations. Int. J. Food Microbiol. 2003, 86, 181–188. [Google Scholar] [CrossRef]
- Liu, H.; Sun, B. Effect of fermentation processing on the flavor of Baijiu. J. Agric. Food Chem. 2018, 66, 5425–5432. [Google Scholar] [CrossRef]
- Jiang, W.; Wei, J.; Li, B.; Dong, J.; Wang, X.; Su, Z.; Han, X.; Huangfu, J.; Luan, C.; Hao, J.; et al. Analysis of Characteristic Flavor Compounds in Single-Grain Chinese Baijiu Brewed from Different Raw Materials. Food Sci. 2020, 41, 234–238. [Google Scholar]
- Luo, A.; Xi, X.; Zheng, T.; Yang, Z.; Tian, Y.; Ma, X.; Zhao, H. Analysis on diversity of the microbial community of light-flavor Baijiu distiller’s grains based on Illumina MiSeq high-throughput sequencing. China Brew. 2022, 41, 98–102. [Google Scholar]
- Nam, Y.H.; Choi, A.; Hwang, J.M.; Yim, K.J.; Kim, J.-H.; Choi, G.-G.; Chung, E.J. Paucibacter aquatile sp. nov. isolated from freshwater of the Nakdong River, Republic of Korea. Arch. Microbiol. 2018, 200, 877–882. [Google Scholar] [CrossRef]
- Barata, A.; Malfeito-Ferreira, M.; Loureiro, V. The microbial ecology of wine grape berries. Int. J. Food Microbiol. 2012, 153, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Aflakpui, F.W.K.; Yu, H.; Luo, L.; Lin, W.-T. Characterization of activity and microbial diversity of typical types of Daqu for traditional Chinese vinegar. Ann. Microbiol. 2015, 65, 2019–2027. [Google Scholar] [CrossRef]
- Tian, F.; Chen, J.; Nian, J.; Huo, G. Study on probiotic properties of Lactobacillus acidophilus and Bifidobacterium. Sci. Technol. Food Ind. 2012, 33, 139–142. [Google Scholar]
- Zhang, Y.; Ping, Y.; Zhou, R.; Wang, J.; Zhang, G. High throughput sequencing-based analysis of microbial diversity in dental unit waterlines supports the importance of providing safe water for clinical use. J. Infect. Public Health 2018, 11, 357–363. [Google Scholar] [CrossRef] [PubMed]
- del Carmen Portillo, M.; Mas, A. Analysis of microbial diversity and dynamics during wine fermentation of Grenache grape variety by high-throughput barcoding sequencing. LWT Food Sci. Technol. 2016, 72, 317–321. [Google Scholar] [CrossRef]
- Youssef, N.; Sheik, C.S.; Krumholz, L.R.; Najar, F.Z.; Roe, B.A.; Elshahed, M.S. Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys. Appl. Environ. Microbiol. 2009, 75, 5227–5236. [Google Scholar] [CrossRef]
- Wang, H.; Hua, J.; Yu, Q.; Li, J.; Wang, J.; Deng, Y.; Yuan, H.; Jiang, Y. Widely targeted metabolomic analysis reveals dynamic changes in non-volatile and volatile metabolites during green tea processing. Food Chem. 2021, 363, 130131. [Google Scholar] [CrossRef]
- Sun, Z.; Xiao, D. Review in Metabolic Modulation of Higher Alcohols in Top-Fermenting Yeast. In Advances in Applied Biotechnology, Proceedings of the 3rd International Conference on Applied Biotechnology (ICAB2016), Tianjin, China, 25–27 November 2016; Springer: Singapore, 2018; pp. 767–773. [Google Scholar]
- Moreira, N.; Araújo, A.M.; Rogerson, F.; Vasconcelos, I.; De Freitas, V.; de Pinho, P.G. Development and optimization of a HS-SPME-GC-MS methodology to quantify volatile carbonyl compounds in Port wines. Food Chem. 2019, 270, 518–526. [Google Scholar] [CrossRef]
- Bell, S.J.; Henschke, P.A. Implications of nitrogen nutrition for grapes, fermentation and wine. Aust. J. Grape Wine Res. 2005, 11, 242–295. [Google Scholar] [CrossRef]
- Jeleń, H.H.; Gaca, A.; Marcinkowska, M. Use of sorbent-based vacuum extraction for determination of volatile phenols in beer. Food Anal. Methods 2018, 11, 3089–3094. [Google Scholar] [CrossRef]
- Radonjić, S.; Prosen, H.; Maraš, V.; Demšar, L.; Košmerl, T. Incidence of volatile phenols in Montenegrin red wines: Vranac, Kratošija and Cabernet sauvignon. Chem. Ind. Chem. Eng. Q. 2020, 26, 337–347. [Google Scholar] [CrossRef]
- Silva, I.; Campos, F.M.; Hogg, T.; Couto, J.A. Factors influencing the production of volatile phenols by wine lactic acid bacteria. Int. J. Food Microbiol. 2011, 145, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wu, Q.; Xu, Y. Core microbiota in Chinese liquor fermentation and associations with environmental factors. Acta Microbiol. Sin. 2018, 58, 142–153. [Google Scholar]
- Fan, G.; Teng, C.; Xu, D.; Fu, Z.; Liu, P.; Wu, Q.; Yang, R.; Li, X. Improving ethyl acetate production in Baijiu manufacture by Wickerhamomyces anomalus and Saccharomyces cerevisiae mixed culture fermentations. BioMed Res. Int. 2019, 2019, 1470543. [Google Scholar] [CrossRef] [PubMed]


| Time (d) | Group | Physicochemical Indices | ||||||
|---|---|---|---|---|---|---|---|---|
| Soluble Solids (%) | Total Sugars (g/L) | Alcohol Content (%) | pH | Total Acids (g/L) | Total Phenols (mg/L) | Anthocyanins (mg/L) | ||
| 0 | 20.50 ± 0.00 | 155.86 ± 0.26 | 0.00 ± 0.00 | 3.55 ± 0.01 | 9.54 ± 0.30 | 306.97 ± 1.01 | 25.83 ± 2.01 | |
| 1 | CF | 18.15 ± 0.08 a | 124.22 ± 2.50 a | 1.50 ± 0.50 b | 3.33 ± 0.01 a | 12.59 ± 0.22 b | 453.10 ± 2.10 b | 8.63 ± 0.92 a |
| NF | 20.16 ± 0.13 b | 151.44 ± 6.09 b | 0.00 ± 0.00 a | 3.56 ± 0.01 b | 9.52 ± 0.28 a | 349.73 ± 4.55 a | 23.04 ± 1.17 b | |
| 2 | CF | 14.57 ± 0.14 a | 75.75 ± 2.27 a | 3.00 ± 0.00 b | 3.31 ± 0.01 a | 15.46 ± 0.29 b | 491.14 ± 6.17 b | 4.06 ± 0.34 a |
| NF | 20.14 ± 0.12 b | 149.54 ± 0.44 b | 0.00 ± 0.00 a | 3.55 ± 0.00 b | 9.52 ± 0.28 a | 318.08 ± 3.64 a | 17.36 ± 0.29 b | |
| 3 | CF | 11.36 ± 0.09 a | 56.87 ± 2.18 a | 4.67 ± 0.58 b | 3.29 ± 0.01 a | 16.10 ± 0.50 b | 498.89 ± 6.62 b | 5.29 ± 0.67 a |
| NF | 19.01 ± 0.01 b | 134.82 ± 1.87 b | 0.83 ± 0.29 a | 3.34 ± 0.02 b | 10.69 ± 0.19 a | 450.07 ± 5.56 a | 7.24 ± 0.10 b | |
| 4 | CF | 9.34 ± 0.10 a | 40.60 ± 0.83 a | 6.67 ± 0.29 b | 3.31 ± 0.02 b | 16.08 ± 0.00 b | 508.32 ± 7.30 b | 4.95 ± 0.35 a |
| NF | 17.35 ± 0.09 b | 113.97 ± 0.23 b | 1.83 ± 0.29 a | 3.23 ± 0.01 a | 12.16 ± 0.37 a | 476.67 ± 13.66 a | 3.45 ± 1.75 a | |
| 5 | CF | 7.96 ± 0.14 a | 16.81 ± 0.15 a | 7.17 ± 0.29 b | 3.31 ± 0.00 b | 16.40 ± 0.55 b | 450.74 ± 9.81 a | 5.51 ± 0.50 b |
| NF | 15.29 ± 0.15 b | 81.44 ± 1.34 b | 2.91 ± 0.08 a | 3.20 ± 0.01 a | 14.40 ± 0.24 a | 501.92 ± 5.34 b | 1.72 ± 0.48 a | |
| 6 | CF | 7.09 ± 0.16 a | 13.79 ± 0.09 a | 8.33 ± 0.29 b | 3.30 ± 0.01 b | 16.96 ± 0.37 b | 430.54 ± 6.49 a | 6.62 ± 0.39 b |
| NF | 13.20 ± 0.20 b | 69.83 ± 1.43 b | 4.50 ± 0.14 a | 3.21 ± 0.01 a | 15.34 ± 0.24 a | 454.44 ± 2.67 b | 1.89 ± 0.26 a | |
| 8 | CF | 6.36 ± 0.06 a | 3.91 ± 0.13 a | 10.17 ± 0.29 b | 3.31 ± 0.01 b | 17.52 ± 0.48 a | 428.52 ± 2.10 a | 6.18 ± 0.33 b |
| NF | 8.55 ± 0.08 b | 40.17 ± 1.50 b | 6.16 ± 0.29 a | 3.21 ± 0.01 a | 16.90 ± 0.25 a | 453.77 ± 2.10 b | 1.78 ± 0.39 a | |
| 10 | CF | 6.36 ± 0.07 a | 3.74 ± 0.09 a | 10.33 ± 0.29 a | 3.31 ± 0.01 b | 17.28 ± 0.42 a | 400.91 ± 5.34 a | 6.23 ± 0.34 b |
| NF | 6.56 ± 0.10 a | 7.90 ± 0.10 b | 9.22 ± 0.70 a | 3.23 ± 0.01 a | 16.91 ± 0.31 a | 395.19 ± 5.56 a | 2.39 ± 0.82 a | |
| 12 | CF | 6.33 ± 0.11 a | 3.68 ± 0.05 a | 10.33 ± 0.29 b | 3.31 ± 0.01 b | 17.36 ± 0.73 a | 358.15 ± 4.21 a | 6.79 ± 0.42 b |
| NF | 6.35 ± 0.13 a | 3.74 ± 0.05 a | 9.16 ± 0.29 a | 3.25 ± 0.01 a | 16.93 ± 0.33 a | 373.30 ± 5.56 b | 3.56 ± 0.42 a | |
| 14 | CF | 6.26 ± 0.11 a | 3.68 ± 1.45 a | 10.33 ± 1.15 a | 3.30 ± 0.01 a | 17.54 ± 0.46 a | 370.94 ± 8.16 b | 6.85 ± 0.17 b |
| NF | 6.28 ± 0.15 a | 3.79 ± 0.09 a | 9.16 ± 0.29 a | 3.30 ± 0.01 a | 16.88 ± 1.00 a | 350.40 ± 7.89 a | 2.34 ± 0.87 a | |
| 16 | CF | 6.20 ± 0.00 a | 3.74 ± 0.59 a | 10.33 ± 0.29 a | 3.31 ± 0.01 a | 17.44 ± 0.14 a | 346.70 ± 5.18 a | 6.46 ± 0.42 b |
| NF | 6.20 ± 0.19 a | 3.71 ± 1.64 a | 9.16 ± 0.29 a | 3.30 ± 0.01 a | 16.82 ± 0.44 a | 352.76 ± 5.83 a | 2.39 ± 0.19 a | |
| 18 | CF | 6.07 ± 0.12 a | 3.79 ± 1.65 a | 9.33 ± 0.58 a | 3.31 ± 0.01 a | 17.22 ± 0.53 a | 317.74 ± 8.23 a | 3.56 ± 0.48 a |
| NF | 6.07 ± 0.12 a | 3.76 ± 0.69 a | 9.16 ± 0.76 a | 3.30 ± 0.01 a | 16.78 ± 0.39 a | 316.73 ± 4.08 a | 5.73 ± 0.19 b | |
| 20 | CF | 6.07 ± 0.12 a | 2.96 ± 0.35 a | 9.50 ± 0.50 a | 3.31 ± 0.00 a | 17.04 ± 0.48 a | 327.17 ± 8.02 b | 4.51 ± 0.44 b |
| NF | 6.07 ± 0.12 a | 3.65 ± 1.69 a | 8.67 ± 0.58 a | 3.31 ± 0.01 a | 16.48 ± 0.37 a | 280.37 ± 7.71 a | 2.73 ± 0.67 a | |
| Number | Compounds | Relative Content (%) | |||||
|---|---|---|---|---|---|---|---|
| ICF | MCF | LCF | INF | MNF | LNF | ||
| Esters | |||||||
| A1 | Decanoic acid, ethyl ester | 0.02 ± 0.01 a | 0.39 ± 0.08 a | 1.13 ± 0.14 b | nd | 0.45 ± 0.08 a | 0.62 ± 0.03 a |
| A2 | Octanoic acid, 3-methylbutyl ester | nd | 0.03 ± 0.00 a | 0.02 ± 0.00 a | nd | 0.03 ± 0.00 a | 0.02 ± 0.00 a |
| A3 | Terpinyl formate | 0.07 ± 0.01 a | 0.15 ± 0.01 a | 0.06 ± 0.01 a | 0.07 ± 0.00 a | 0.14 ± 0.00 a | 0.08 ± 0.01 b |
| A4 | Octanoic acid, ethyl ester | 0.02 ± 0.01 a | 0.34 ± 0.04 a | 0.24 ± 0.04 a | nd | 0.29 ± 0.03 a | 0.26 ± 0.04 a |
| A5 | Butylphosphonic acid, dodecyl isohexyl ester | 0.02 ± 0.00 a | 0.03 ± 0.00 a | 0.01 ± 0.00 a | 0.03 ± 0.00 a | 0.03 ± 0.00 a | 0.01 ± 0.00 a |
| A6 | Dodecanoic acid, ethyl ester | 0.01 ± 0.00 a | 0.24 ± 0.03 a | 0.39 ± 0.06 b | nd | 0.27 ± 0.01 a | 0.13 ± 0.01 a |
| A7 | O-Methyl-DL-serine, N-dimethylaminomethylene-, butyl ester | nd | 0.04 ± 0.01 a | 0.06 ± 0.02 b | nd | 0.05 ± 0.00 a | 0.02 ± 0.00 a |
| A8 | Ethyl 6-methylpyridine-2-carboxylate | 0.23 ± 0.01 a | 0.23 ± 0.01 a | 0.09 ± 0.00 a | 0.24 ± 0.01 b | 0.22 ± 0.00 a | 0.12 ± 0.01 b |
| A9 | 2-(4-Methoxyphenyl)ethyl acetate | 0.09 ± 0.00 a | 0.05 ± 0.01 a | 0.03 ± 0.00 b | 0.09 ± 0.01 a | 0.04 ± 0.00 a | 0.02 ± 0.00 a |
| A10 | Vinyl caprylate | 0.01 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.02 ± 0.01 a |
| A11 | Butylphosphonic acid, di(2-phenylethyl) ester | 0.01 ± 0.00 a | 0.08 ± 0.01 a | 0.15 ± 0.01 a | 0.01 ± 0.00 a | 0.11 ± 0.02 b | 0.13 ± 0.00 a |
| A12 | Succinic acid, ethyl 3-pentyl ester | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.03 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.13 ± 0.01 b |
| A13 | 2-Methylpropionic acid, 4-cyanophenyl ester | nd | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| A14 | Hexanoic acid, 3-hydroxy-, ethyl ester | 0.09 ± 0.00 a | 0.04 ± 0.00 b | nd | 0.09 ± 0.01 a | 0.03 ± 0.00 a | 0.02 ± 0.00 b |
| A15 | Ethylene glycol acetate formate | nd | nd | 0.07 ± 0.01 b | nd | nd | 0.04 ± 0.00 a |
| A16 | Propanoic acid, 2-hydroxy-, ethyl ester | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.02 ± 0.00 b |
| A17 | 2-Pentanol, acetate | nd | 0.02 ± 0.01 a | 0.09 ± 0.02 a | nd | 0.04 ± 0.01 a | 0.12 ± 0.01 b |
| A18 | Benzeneacetic acid, 4-ethenyl-, ethyl ester | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.03 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.03 ± 0.00 a |
| A19 | Hexanoic acid, ethyl ester | 0.01 ± 0.00 b | 0.02 ± 0.00 a | 0.04 ± 0.01 b | nd | 0.02 ± 0.01 a | 0.03 ± 0.00 a |
| A20 | Acetic acid benzo[1,2,5]thiadiazol-5-yl ester | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.01 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a |
| A21 | Methyl 5(3)-methyl-4-hydroxy-3(5)-pyrazolecarboxylate | 11.47 ± 0.63 a | 12.67 ± 0.66 b | 11.29 ± 0.50 b | 11.63. ± 0.65 a | 10.83 ± 0.44 a | 10.04 ± 1.00 a |
| A22 | Tetrahydrofurfuryl propionate | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.02 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.02 ± 0.01 a |
| A23 | 2,4-Pentadienoic acid, 1-cyclopenten-3-on-1-yl ester | 0.04 ± 0.00 a | 0.05 ± 0.00 b | 0.04 ± 0.00 a | 0.04 ± 0.00 a | 0.04 ± 0.00 a | 0.03 ± 0.01 a |
| A24 | Cyclohexyl methyl methylphosphonate | 0.01 ± 0.00 a | 0.01 ± 0.00 a | nd | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| Total | 12.28 ± 0.63 a | 14.51 ± 0.68 b | 13.85 ± 0.79 b | 12.41 ± 0.65 a | 12.71 ± 0.48 a | 11.96 ± 1.08 a | |
| Alchols | |||||||
| A25 | Morphinan-14-ol, 6-azido-4,5-epoxy-3-methoxy-17-methyl-, (5.alpha.,6.beta.)- | 0.12 ± 0.04 a | 0.09 ± 0.03 a | 0.20 ± 0.02 a | 0.08 ± 0.01 a | 0.11 ± 0.04 a | 0.18 ± 0.05 a |
| A26 | Phenylethyl Alcohol | 0.62 ± 0.09 a | 5.85 ± 0.16 a | 7.88 ± 0.61 a | 0.69 ± 0.05 a | 6.74 ± 0.14 b | 9.52 ± 0.20 b |
| A27 | Benzenemethanol, .alpha.-methyl-.alpha.-(1-methyl-2-propenyl)- | 0.07 ± 0.00 a | 0.07 ± 0.00 a | 0.05 ± 0.00 a | 0.07 ± 0.00 a | 0.06 ± 0.00 a | 0.05 ± 0.00 a |
| A28 | 2-Furanmethanol, 5-ethenyltetrahydro-.alpha.,.alpha.,5-trimethyl-, cis- | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.01 ± 0.00 a | 0.02 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| A29 | Cyclohexanol, 3-(acetyloxymethyl)-2,2,4-trimethyl- | 6.76 ± 0.12 a | 6.73 ± 0.16 b | 4.82 ± 0.09 b | 6.66 ± 0.24 a | 5.89 ± 0.16 a | 4.36 ± 0.09 a |
| A30 | 1-Butanol, 3-methyl- | 0.32 ± 0.10 a | 6.55 ± 0.10 a | 10.21 ± 0.63 a | 0.19 ± 0.01 a | 6.44 ± 0.20 a | 10.52 ± 0.58 a |
| A31 | 1-Pentanol, 4-methyl- | 0.09 ± 0.00 a | 0.07 ± 0.00 a | 0.05 ± 0.01 a | 0.11 ± 0.00 b | 0.10 ± 0.00 b | 0.09 ± 0.00 b |
| A32 | 1-Hexanol | 0.09 ± 0.01 a | 0.07 ± 0.00 a | 0.05 ± 0.01 a | 0.11 ± 0.00 b | 0.10 ± 0.01 b | 0.08 ± 0.01 b |
| A33 | Ethanol, 2-(2-hydroxyethoxy)-, 1-nitrate | 0.01 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.01 ± 0.00 a | 0.02 ± 0.00 a | 0.01 ± 0.00 a |
| A34 | Ethanol, 2-(di-2-propenylamino)- | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | nd |
| A35 | Benzyl alcohol | 1.01 ± 0.03 a | 1.00 ± 0.04 a | 1.47 ± 0.11 a | 3.25 ± 0.15 b | 3.01 ± 0.21 b | 6.02 ± 0.24 b |
| Total | 9.13 ± 0.38 a | 20.47 ± 0.16 a | 24.76 ± 1.04 a | 11.20 ± 0.36 b | 22.50 ± 0.16 b | 30.85 ± 0.91 b | |
| Ketones | |||||||
| A36 | 3-Hexanone-2,2,4,4-d4 | 0.30 ± 0.01 b | 0.27 ± 0.01 b | 0.33 ± 0.01 a | 0.28 ± 0.02 a | 0.24 ± 0.01 a | 0.32 ± 0.00 a |
| A37 | 3-Buten-2-one, 4-(2,6,6-trimethyl-2-cyclohexen-1-yl)- | 0.02 ± 0.00 a | 0.03 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a |
| A38 | Ketone, methyl 2,4,5-trimethylpyrrol-3-yl | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.02 ± 0.00 b | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| A39 | Ethanone, 1-(2,4-dihydroxyphenyl)- | nd | 0.01 ± 0.00 a | nd | 0.04 ± 0.00 b | 0.46 ± 0.01 b | 0.34 ± 0.01 b |
| A40 | 2,2-Dimethyl-4(1H,3H)-quinazolinone, 2Me derivative | 0.04 ± 0.00 a | 0.03 ± 0.00 a | 0.04 ± 0.00 a | 0.04 ± 0.00 a | 0.03 ± 0.01 a | 0.04 ± 0.01 a |
| A41 | 4H-Naphtho[1,2-b]pyran-4-one, 2-methyl- | 0.03 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 b | 0.03 ± 0.01 a | 0.02 ± 0.00 a | nd |
| A42 | 2-Buten-1-one, 1-(2,6,6-trimethyl-1,3-cyclohexadien-1-yl)- | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.01 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.01 ± 0.00 a |
| A43 | 2-Butanone, 3-ethoxy-3-methyl- | 0.13 ± 0.01 b | 0.11 ± 0.00 b | 0.14 ± 0.00 a | 0.12 ± 0.00 a | 0.10 ± 0.00 a | 0.14 ± 0.00 a |
| A44 | 2-Pyrrolidinone, 1-methyl- | 0.86 ± 0.01 b | 0.35 ± 0.01 a | 0.32 ± 0.02 a | 0.78 ± 0.05 a | 0.32 ± 0.04 a | 0.30 ± 0.02 a |
| A45 | 1,4-Dioxane-2,5-dione | nd | 0.02 ± 0.00 b | 0.01 ± 0.00 a | nd | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| A46 | Pyrrolizidine-1-one, 7-acetylmethyl- | 0.01 ± 0.00 a | 0.01 ± 0.00 a | nd | nd | nd | 0.01 ± 0.00 a |
| A47 | 4(3H)-Pyrimidinone, 3-methyl- | 11.56 ± 0.41 b | 8.72 ± 0.13 b | 6.56 ± 0.18 a | 11.11 ± 0.22 a | 7.60 ± 0.42 a | 6.11 ± 0.18 a |
| A48 | Methanesulfonylacetone | 0.02 ± 0.01 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.02 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| Total | 13.09 ± 0.40 b | 9.62 ± 0.13 b | 7.50 ± 0.20 a | 12.47 ± 0.27 a | 8.86 ± 0.47 a | 7.31 ± 0.17 a | |
| Acids | |||||||
| A49 | 3,4-Methylenedioxycinnamic acid | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| A50 | 4-tert-Butyl-2-nitrophenol, acetate | 22.91 ± 0.12 a | 22.76 ± 0.51 b | 17.52 ± 0.61 b | 22.69 ± 0.85 a | 21.60 ± 0.66 a | 15.76 ± 0.33 a |
| A51 | 2-Chloro-6H-thieno[2,3-b]pyrrole-5-carboxylic acid, N,O-bis-methyl- | 0.18 ± 0.00 a | 0.09 ± 0.01 a | 0.05 ± 0.01 b | 0.17 ± 0.03 a | 0.08 ± 0.00 a | 0.03 ± 0.00 a |
| A52 | 3-Propoxy-benzoic acid | 0.04 ± 0.01 a | 0.03 ± 0.00 a | 0.02 ± 0.00 b | 0.04 ± 0.01 a | 0.02 ± 0.00 a | nd |
| A53 | 2-[(2-Aminoethyl)sulfanyl]acetic acid | 0.09 ± 0.00 a | 0.05 ± 0.01 b | nd | 0.09 ± 0.01 a | 0.03 ± 0.00 a | 0.02 ± 0.00 b |
| A54 | p-Tolylacetic acid | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 b | 0.01 ± 0.00 a | 0.03 ± 0.00 b |
| A55 | Acetic acid, (aminooxy)- | 0.01 ± 0.00 b | 0.06 ± 0.01 b | 0.08 ± 0.01 b | nd | 0.04 ± 0.00 a | 0.07 ± 0.00 a |
| A56 | Butanoic acid, 3-amino- | nd | nd | 0.06 ± 0.00 a | nd | nd | 0.06 ± 0.01 a |
| A57 | 1H-Pyrazole-4-carboxylic acid, 1-methyl- | 0.06 ± 0.00 b | 0.04 ± 0.00 a | 0.04 ± 0.01 a | 0.05 ± 0.01 a | 0.04 ± 0.00 a | 0.04 ± 0.00 a |
| A58 | (S)-2-Chloro-3-methylbutyric acid | 0.04 ± 0.00 a | 0.05 ± 0.01 a | 0.08 ± 0.01 a | 0.11 ± 0.03 b | 0.10 ± 0.02 b | 0.18 ± 0.00 b |
| Total | 23.36 ± 0.14 a | 23.09 ± 0.54 b | 17.87 ± 0.59 b | 23.17 ± 0.90 a | 21.93 ± 0.67 a | 16.20 ± 0.33 a | |
| Alkanes and alkenes | |||||||
| A59 | Tricarbomethoxyethylene | nd | 0.02 ± 0.00 a | 0.06 ± 0.01 b | nd | 0.02 ± 0.00 a | 0.03 ± 0.00 a |
| A60 | Octadecane | 0.01 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| A61 | 3-Cyano-5,5-dimethoxycarbonyl-N-methylisoxazolidine | 0.03 ± 0.00 a | 0.02 ± 0.00 a | 0.01 ± 0.00 b | 0.02 ± 0.00 a | 0.02 ± 0.00 a | nd |
| A62 | Pentadecane, 8-hexyl- | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.02 ± 0.00 b | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| A63 | Ethane, 1,1-diethoxy- | nd | 0.10 ± 0.01 a | 0.19 ± 0.02 a | nd | 0.13 ± 0.01 b | 0.24 ± 0.01 b |
| A64 | Azetidine | 0.05 ± 0.02 a | 0.83 ± 0.04 a | 1.25 ± 0.08 a | 0.03 ± 0.00 a | 0.82 ± 0.02 a | 1.25 ± 0.10 a |
| A65 | 2-Methoxy-1,3-dioxolane | 0.29 ± 0.01 b | 0.27 ± 0.00 b | 0.31 ± 0.00 a | 0.27 ± 0.01 a | 0.23 ± 0.01 a | 0.32 ± 0.00 a |
| A66 | Propane, 1,1′-sulfonylbis- | 0.06 ± 0.01 a | 0.04 ± 0.01 a | 0.03 ± 0.01 a | 0.05 ± 0.01 a | 0.04 ± 0.00 a | 0.02 ± 0.01 a |
| A67 | Propane, 1-(diisopropylphosphino)-3-(diisopropylphosphinyl)- | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | nd | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| A68 | Propane, 1,1′-[ethylidenebis(oxy)]bis- | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.02 ± 0.00 b | 0.01 ± 0.00 a | 0.02 ± 0.00 b |
| A69 | Methane, triiodo- | nd | nd | 0.01 ± 0.00 a | nd | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| Total | 0.46 ± 0.01 a | 1.33 ± 0.05 a | 1.92 ± 0.10 a | 0.41 ± 0.03 a | 1.30 ± 0.01 a | 1.92 ± 0.09 a | |
| Benzene rings | |||||||
| A70 | Phenol, 4-ethyl- | 0.02 ± 0.00 a | 0.09 ± 0.00 a | 0.02 ± 0.00 a | 0.47 ± 0.02 b | 1.53 ± 0.04 b | 1.38 ± 0.02 b |
| A71 | 3-Acetyl-2,5,6-trimethylhydroquinone | 4.98 ± 0.06 b | 5.28 ± 0.11 b | 4.83 ± 0.17 b | 4.73 ± 0.05 a | 4.97 ± 0.03 a | 4.66 ± 0.05 a |
| A72 | 2,4-Di-tert-butylphenol | 14.16 ± 1.12 a | 8.41 ± 0.46 a | 13.65 ± 1.21 b | 14.96 ± 1.02 a | 10.31 ± 1.27 a | 10.50 ± 1.55 a |
| A73 | p-(Benzylideneamino)phenol | 0.05 ± 0.00 a | 0.03 ± 0.00 a | 0.03 ± 0.01 b | 0.05 ± 0.01 a | 0.03 ± 0.00 a | 0.01 ± 0.00 a |
| A74 | 1,4-Di(methyl-d3)benzene-d4 | 2.55 ± 0.11 a | 2.80 ± 0.07 b | 3.62 ± 0.02 a | 2.54 ± 0.12 a | 2.45 ± 0.09 a | 3.53 ± 0.30 a |
| A75 | Phenol | 2.96 ± 0.08 b | 1.72 ± 0.16 b | 1.50 ± 0.11 a | 2.65 ± 0.02 a | 1.49 ± 0.08 a | 1.38 ± 0.07 a |
| A76 | Benzene, 3-butenyl- | 0.04 ± 0.00 a | 0.05 ± 0.01 a | 0.02 ± 0.00 a | 0.03 ± 0.01 a | 0.04 ± 0.00 a | 0.03 ± 0.00 b |
| A77 | Benzene, 1,4-dimethyl-2,5-bis(1-methylethyl)- | 0.04 ± 0.00 b | 0.04 ± 0.00 a | 0.05 ± 0.00 a | 0.03 ± 0.00 a | 0.04 ± 0.00 a | 0.05 ± 0.00 a |
| A78 | 4-Methoxy-2-allylphenol | nd | 0.01 ± 0.00 a | nd | 0.02 ± 0.00 b | 0.06 ± 0.01 b | 0.07 ± 0.01 b |
| Total | 24.79 ± 1.13 a | 18.44 ± 0.46 a | 23.72 ± 1.33 b | 25.48 ± 0.93 a | 20.91 ± 1.12 b | 21.60 ± 1.31 a | |
| Aldehydes | |||||||
| A79 | Benzaldehyde, 3,4-dimethyl- | 0.42 ± 0.03 a | 0.81 ± 0.14 a | 0.59 ± 0.11 a | 0.35 ± 0.02 a | 0.91 ± 0.03 a | 0.60 ± 0.08 a |
| A80 | 1-Methylimidazole-5-carboxaldehyde | 1.10 ± 0.02 a | 0.58 ± 0.03 a | 0.52 ± 0.02 a | 1.11 ± 0.07 a | 0.53 ± 0.04 a | 0.53 ± 0.04 a |
| A81 | Benzaldehyde | 0.35 ± 0.05 a | 0.36 ± 0.01 a | 0.41 ± 0.04 a | 0.30 ± 0.03 a | 0.51 ± 0.03 b | 1.00 ± 0.04 b |
| Total | 1.87 ± 0.06 a | 1.76 ± 0.12 a | 1.52 ± 0.14 a | 1.76 ± 0.09 a | 1.95 ± 0.07 b | 2.13 ± 0.06 b | |
| Amines | |||||||
| A82 | 3,4-Dichloro-N-methylaniline | nd | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| A83 | (Methylsulfamoyl)amine | 0.66 ± 0.06 b | 0.61 ± 0.01 a | 0.36 ± 0.09 b | 0.42 ± 0.10 a | 0.52 ± 0.02 a | 0.16 ± 0.02 a |
| A84 | N,N-Diethyl-1-cyclopropyl-pentanamine | 0.06 ± 0.00 b | 0.04 ± 0.00 a | 0.04 ± 0.00 a | 0.05 ± 0.00 a | 0.04 ± 0.00 a | 0.04 ± 0.01 a |
| A85 | 6-Isopropyl-benzothiazol-2-ylamine | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| A86 | 1,2,4,5-tetrazin-3-amine, N-(2-methoxyethyl)-6-(methylthio)- | nd | 0.01 ± 0.00 a | 0.03 ± 0.00 b | nd | 0.01 ± 0.00 a | 0.02 ± 0.00 a |
| A87 | Methanamine, 1-methoxy-N-methyl-N-nitroso- | 0.29 ± 0.01 b | 0.26 ± 0.01 b | 0.32 ± 0.01 a | 0.27 ± 0.01 a | 0.23 ± 0.01 a | 0.32 ± 0.00 a |
| A88 | N-Vinylformamide | 0.06 ± 0.01 a | 0.02 ± 0.01 a | 0.02 ± 0.00 a | 0.05 ± 0.01 a | 0.02 ± 0.00 a | 0.01 ± 0.00 a |
| A89 | 4H-1,2,4-Triazol-3-amine, 4-methyl- | 0.70 ± 0.03 b | 0.28 ± 0.02 a | 0.27 ± 0.02 a | 0.65 ± 0.03 a | 0.27 ± 0.03 a | 0.26 ± 0.02 a |
| A90 | 5-Methyl-1,2,4-oxadiazol-3-amine | 0.19 ± 0.01 a | 0.21 ± 0.01 b | 0.27 ± 0.01 a | 0.19 ± 0.01 a | 0.18 ± 0.00 a | 0.27 ± 0.03 a |
| A91 | Ethanediamide | 0.05 ± 0.00 a | 0.02 ± 0.00 a | nd | 0.05 ± 0.00 a | 0.02 ± 0.00 a | 0.01 ± 0.00 b |
| Total | 2.02 ± 0.06 b | 1.48 ± 0.01 b | 1.33 ± 0.11 b | 1.69 ± 0.06 a | 1.31 ± 0.05 a | 1.11 ± 0.03 a | |
| Furan, pyrazine and pyridine compounds | |||||||
| A92 | 5-Methyl-2-(2-methyl-2-tetrahydrofuryl)tetrahydrofuran | 0.01 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.01 a |
| A93 | 3-Methyl-5-hydrazinopyrazine | 0.07 ± 0.02 a | 0.04 ± 0.01 a | 0.04 ± 0.01 a | 0.06 ± 0.03 a | 0.03 ± 0.01 a | 0.03 ± 0.01 a |
| A94 | 2,4-Diamino-6-cyanamino-1,3,5-triazine | 0.11 ± 0.00 b | 0.08 ± 0.00 b | 0.05 ± 0.01 b | 0.09 ± 0.01 a | 0.06 ± 0.00 a | 0.03 ± 0.00 a |
| A95 | 1,3,5-Triazine, 2,4,6-trimethyl- | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| A96 | 2-Isoamyl-6-methylpyrazine | 0.04 ± 0.00 a | 0.06 ± 0.01 b | 0.05 ± 0.00 a | 0.11 ± 0.01 b | 0.26 ± 0.00 a | 0.22 ± 0.02 b |
| A97 | 2(1H)-Pyridinone, 3-hydroxy- | 0.75 ± 0.02 b | 0.53 ± 0.01 b | 0.49 ± 0.02 b | 0.66 ± 0.03 a | 0.47 ± 0.02 a | 0.43 ± 0.01 a |
| A98 | 2(1H)-Pyridinone, 1-methyl- | 0.30 ± 0.02 b | 0.19 ± 0.03 a | 0.17 ± 0.01 a | 0.25 ± 0.02 a | 0.19 ± 0.02 a | 0.15 ± 0.01 a |
| A99 | 1,4-Dihydro-4-oxopyridazine | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.01 ± 0.00 a |
| A100 | 4-(2-Aminoethyl)pyridine | 0.02 ± 0.00 a | 0.02 ± 0.01 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a | 0.01 ± 0.00 a |
| A101 | Pyridinium, 1-methyl-, iodide | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| A102 | Furan, 2-[(ethylthio)methyl]- | 0.07 ± 0.00 a | 0.06 ± 0.00 a | 0.03 ± 0.00 a | 0.07 ± 0.01 a | 0.05 ± 0.00 a | 0.03 ± 0.00 a |
| A103 | Furan, 2-methyl-5-(methylthio)- | 4.76 ± 0.09 b | 2.86 ± 0.12 b | 2.18 ± 0.12 a | 4.16 ± 0.21 a | 2.45 ± 0.13 a | 2.01 ± 0.06 a |
| A104 | 2-Methyl-1,2,4-triazolo(2,3-a)pyrazine | 0.03 ± 0.00 a | 0.03 ± 0.01 a | 0.03 ± 0.00 a | 0.03 ± 0.00 a | 0.03 ± 0.00 a | 0.03 ± 0.00 a |
| Total | 6.20 ± 0.12 b | 3.93 ± 0.15 b | 3.11 ± 0.12 a | 5.50 ± 0.29 a | 3.62 ± 0.16 a | 3.01 ± 0.03 a | |
| Others | |||||||
| A105 | Hexanenitrile, 3-(1-pyrrolidinylmethylene)- | nd | nd | nd | nd | nd | nd |
| A106 | (S)-9-[(R)-2-(Hydroxymethyl)pyrrolidin-1-yl]-3-methyl-3,4-dihydro-2H-benzo[b][1,4,5]oxathiazepine 1,1-dioxide | nd | 0.01 ± 0.00 a | 0.03 ± 0.01 a | nd | 0.02 ± 0.00 a | 0.03 ± 0.00 a |
| A107 | Urea, N,N-dimethy′-N′-buty′-N′-(2-ethylhexyl)- | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| A108 | 1H-Tetrazol-5-amine | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.01 a |
| A109 | 4-(Diethylamino)benzonitrile | nd | nd | nd | nd | 0.01 ± 0.00 b | 0.01 ± 0.00 b |
| A110 | 1-(1-(Methylthio)propyl)-2-propyldisulfane | 0.03 ± 0.02 a | 0.01 ± 0.01 a | 0.03 ± 0.01 a | 0.02 ± 0.01 a | 0.02 ± 0.01 a | 0.02 ± 0.01 a |
| A111 | 6-Phenyl-5,6-dihydro-5,6-azaboruracil | 0.04 ± 0.00 a | 0.03 ± 0.00 a | 0.05 ± 0.00 b | 0.04 ± 0.00 a | 0.03 ± 0.01 a | 0.03 ± 0.00 a |
| A112 | 5-Amino-1-(2-cyano-ethyl)-1H-pyrazole-4-carbonitrile | 0.08 ± 0.00 a | 0.04 ± 0.00 a | 0.03 ± 0.00 b | 0.08 ± 0.01 a | 0.04 ± 0.00 a | 0.01 ± 0.00 a |
| A113 | Benzofurazan, 4,6-dinitro-, 1-oxide | 0.07 ± 0.00 a | 0.04 ± 0.00 a | 0.03 ± 0.01 b | 0.07 ± 0.01 a | 0.03 ± 0.00 a | 0.01 ± 0.00 a |
| A114 | 1,2,4-Thiadiazole-5(2H)-thione, 3-methyl- | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| A115 | Benzyl methyl sulfide | nd | nd | nd | nd | nd | 0.03 ± 0.00 b |
| A116 | Acetic anhydride | nd | 0.02 ± 0.00 b | 0.02 ± 0.00 a | nd | 0.01 ± 0.00 a | 0.02 ± 0.00 a |
| A117 | Carbonyl sulfide | 0.01 ± 0.00 b | 0.02 ± 0.00 a | 0.03 ± 0.00 a | nd | 0.02 ± 0.00 a | 0.03 ± 0.01 a |
| A118 | 1H-Imidazole-2-methanol, 1-methyl- | 4.61 ± 0.16 b | 3.22 ± 0.11 b | 3.00 ± 0.09 b | 4.14 ± 0.08 a | 2.94 ± 0.11 a | 2.71 ± 0.04 a |
| A119 | Naphthalene, 6-(1,1-dimethylethyl)-1,2,3,4-tetrahydro- | 0.01 ± 0.00 b | nd | 0.01 ± 0.00 a | nd | nd | nd |
| A120 | 3(2H)-Thiophenone, dihydro-2-methyl- | nd | 0.02 ± 0.00 b | 0.01 ± 0.00 a | nd | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| A121 | 1-Methyl-3-nitropyrazole | 0.40 ± 0.04 b | 0.29 ± 0.04 a | 0.25 ± 0.00 a | 0.34 ± 0.01 a | 0.25 ± 0.03 a | 0.25 ± 0.01 a |
| A122 | 2,2′-Biphenylylenephosphoric acid chloride | 0.01 ± 0.00 a | 0.02 ± 0.00 a | 0.03 ± 0.00 b | 0.01 ± 0.00 a | 0.02 ± 0.00 a | 0.02 ± 0.00 a |
| A123 | 5-Cyano-1,2,3-thiadiazole | 0.17 ± 0.00 a | 0.16 ± 0.00 b | 0.09 ± 0.01 a | 0.18 ± 0.01 a | 0.14 ± 0.00 a | 0.08 ± 0.01 a |
| A124 | 3-Imidazol-1-ylpropanenitrile | 0.08 ± 0.01 a | 0.08 ± 0.01 a | 0.03 ± 0.01 a | 0.08 ± 0.00 a | 0.07 ± 0.00 a | 0.04 ± 0.00 b |
| A125 | Ethylene glycol diglycidyl ether | 1.26 ± 0.07 b | 1.36 ± 0.04 b | 0.74 ± 0.05 b | 0.91 ± 0.04 a | 1.20 ± 0.03 a | 0.54 ± 0.04 a |
| A126 | 1H-Indene, 2,3-dihydro-1,1,4,6-tetramethyl- | nd | 0.01 ± 0.00 a | nd | nd | 0.01 ± 0.00 a | 0.01 ± 0.00 b |
| A127 | Phosphine oxide, cyclohexyl methyl menthyloxy- | nd | nd | nd | nd | nd | nd |
| A128 | 3,5-Dimethylisoxazole-4-sulfonyl chloride | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | nd | 0.01 ± 0.00 a | 0.01 ± 0.00 a |
| Total | 6.81 ± 0.13 b | 5.39 ± 0.10 b | 4.42 ± 0.06 b | 5.90 ± 0.03 a | 4.91 ± 0.13 a | 3.90 ± 0.08 a | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, X.; Zhang, X.; Hao, L.; Lin, Q.; Bao, Y. Effects of Different Fermentation Methods on the Quality and Microbial Diversity of Passion Fruit Wine. Fermentation 2023, 9, 439. https://doi.org/10.3390/fermentation9050439
Ye X, Zhang X, Hao L, Lin Q, Bao Y. Effects of Different Fermentation Methods on the Quality and Microbial Diversity of Passion Fruit Wine. Fermentation. 2023; 9(5):439. https://doi.org/10.3390/fermentation9050439
Chicago/Turabian StyleYe, Xiaofang, Xinyong Zhang, Lifen Hao, Qi Lin, and Yuanyuan Bao. 2023. "Effects of Different Fermentation Methods on the Quality and Microbial Diversity of Passion Fruit Wine" Fermentation 9, no. 5: 439. https://doi.org/10.3390/fermentation9050439
APA StyleYe, X., Zhang, X., Hao, L., Lin, Q., & Bao, Y. (2023). Effects of Different Fermentation Methods on the Quality and Microbial Diversity of Passion Fruit Wine. Fermentation, 9(5), 439. https://doi.org/10.3390/fermentation9050439





