Abstract
In humans and animals, probiotics are widely accepted as crucial for host health and growth. The investigation of the probiotic colonization and expression of probiotics in the host is beneficial for proper usage of probiotics and isolation of indigenous probiotics. In this study, we analyzed commonly used probiotic strains in the intestines/rumen of humans and animals by analyzing metagenomic and paired meta-transcriptomic data from the gut or rumen microbiome of humans (n = 13), pigs (n = 6), chickens (n = 6), cattle (n = 14), sheep (n = 10), and mice (n = 8). First, we generated an expression profile based on 192 selected representative probiotic strains from a published database. A total of 58 probiotic strains were not detected in any samples, while 3 strains were presented and expressed in all individuals. Overall, the probiotic expression of probiotics as detected by meta-transcriptome was significantly higher than the relative abundance of probiotic as detected by metagenomics in cattle, sheep, mice, and humans; however, this difference was not significant in pigs and chickens. In total, 17 (cattle), 21 (sheep), 22 (pig), 14 (chicken), 13 (mouse), and 3 (human) probiotic strains were identified as probiotic strains with significantly higher expression levels [Fold Change (FC) ≥ 2, False Discovery Rate (FDR) ≤ 0.05]. Among them, Clostridium butyricum TOA was found to be significantly expressed in the rumen or gut of all host species. In addition, network analysis based on the expression of probiotics as detected by meta-transcriptomics revealed that several probiotic strains were significantly negatively linked with Salmonella spp., Mycoplasma spp., and Escherichia coli. The results in this study provide a useful reference for developing indigenous probiotics.
1. Introduction
Probiotics, which are live beneficial bacteria or yeasts, have gained significant attention in recent years due to their numerous health-promoting properties, such as competitive inhibition of pathogenic bacteria, immune response regulation, assistance in nutrient metabolism, and even improvement of human cognitive function [1,2,3]. The market demand for highly efficient probiotic strains is huge. In the past decades, many probiotic strains have been isolated from different potential sources and widely applied for food fermentation, the improvement of livestock production, and as clinical therapeutics for human diseases [4,5]. However, some probiotic strains are ineffective or less effective than claimed [6]. One of the most common reasons is that probiotic strains are not universally effective across all hosts [7]. The probiotic strains’ genetic and microecological adaptation to their host are among the leading reasons for probiotic ineffectiveness.
The genetic and microecological adaptation of probiotic strains to their host is a major determinant of their effectiveness. Several studies have shown that indigenous probiotic strains isolated from the host could colonize more steadily in the gut and better increase host growth and pathogen resistance than those isolated from other sources [8,9]. Therefore, investigating the colonization and expression activity of indigenous probiotic strains in humans and animals is beneficial for developing highly effective probiotic strains. In addition, gene expression is one of the ways through which probiotics can exert beneficial effects on the host. Surprisingly, this aspect has often been overlooked in past studies aimed at identifying effective probiotic strains. Investigating the gene expression patterns of probiotic strains and their interactions with the host’s genetic makeup can provide valuable insights into the mechanisms through which these strains contribute to host health [10]. By incorporating this knowledge, researchers and industry professionals can develop more targeted and effective probiotic products, ultimately promoting better health outcomes for both humans and animals.
In this study, we collected the metagenomic data and the paired meta-transcriptomic data from a diverse range of hosts, including cattle, sheep, pigs, chickens, mice, and humans, to investigate the distribution and expression level of probiotic strains in the gut or rumen, as well as to evaluate the link between probiotic strains and typical pathogens. This study aimed to overcome the challenge of developing effective and stable probiotics by analyzing the difference at strain level. The results of this study would provide a useful reference for screening and isolating indigenous probiotic strains in the future, and suggest candidate probiotic strains as antibacterial probiotic strains. Moreover, the expression profiles generated in this study may prompt researchers to reevaluate the use of animal models when performing functional verification of probiotic strains, ultimately leading to more effective and targeted probiotic development and application.
2. Materials and Methods
A total number of 6 datasets (57 individuals) containing both metagenomic and paired meta-transcriptomic sequencing data from different host species (chickens [11], pigs [11], cattle [12], sheep [13], mice [14], and humans [15]) were collected from published articles. The data accession numbers and corresponding citations are also provided in Table 1 for each dataset.

Table 1.
Summary table of the datasets used in this study.
In addition, we selected 192 probiotic strains based on Zhang’s recently published study [16]. The genomes of these probiotic strains were downloaded from the NCBI database (Collection date: 15 January 2022, Table S1).
In order to ensure the accuracy and reliability of our analysis, we employed a stringent data preprocessing workflow to filter the collected metagenomic and metatranscriptomic raw data. Firstly, Q-score based quality control and filtering of host contamination were performed on these collected metagenomic and metatranscriptomic raw data using Kneaddata pipeline v0.7.2 (https://bitbucket.org/biobakery/kneaddata (accessed on 7 July 2020)). This pipeline is designed to process high-throughput sequencing data, ensuring that only high-quality reads are retained for downstream analyses. In brief, raw reads were firstly trimmed with Trimmomatic v0.39 [17] by scanning the reads using a 4bp sliding window and trims low-quality regions with a Phred score less than 20. After trimming low-quality regions, the reads shorter than 60 bp were removed from the subsequent analysis. To eliminate host-derived contamination from the raw reads, quality filtered reads were aligned to their respective reference genomes [Accession number: GCF_002263795 (cattle [18]), GCF_002742125 (sheep [19]), GCF_016699485 (chicken [20]), GCF_000003025 (pig [21]), GCF_000001635 (mouse [22]), and GCF_000001405 (human [23])] with the bmtagger (v.3.102.4) software [24] to identify and remove host-contamination reads, leaving behind only microbial sequences for further analysis.
Owing to the fact that the datasets were obtained from various studies, the efficiency of rRNA removal prior to the generation of Illumina libraries was not consistent, which could significantly impact the identification of bacterial strains using meta-transcriptomic datasets. To mitigate the interference from the expressed ribosomal RNA, SortMeRNA software (v4.3.2), and SMR v4.3 sensitive database were used to remove potential ribosomal RNA sequences from metagenomic and paired meta-transcriptomic data with default parameters. Clean reads were acquired for further analysis after the abovementioned raw read processing steps.
The genomes of the 192 probiotic strains were re-constructed for building a custom taxonomic database containing only these probiotic strain genomes using the Kraken2 [25] taxonomic classification software. Then, Kraken2 was used to assign clean reads to probiotic genomes in the custom database to generate the profile of probiotic strains in each sample. QIIME2 version 2021.4 [26] was used to rarefy the reads count table and calculate the relative abundance of probiotics in each sample. For quantification of selected pathogens, Kraken2 version 2.1.2 [25] was used to assign clean reads of meta-transcriptomic data to the pre-built Genome Taxonomy Database release 202 (GTDB 202, Access date: 13 October 2021) [27,28]. After rarefying the count table, we extracted the items belonging to Salmonella, Mycoplasma, and E. coli for further network analysis.
Differential analysis of probiotic strains between metagenomic abundance and meta-transcriptomic expression in different host species was performed using the “DESeq2” package [29]. The p value was adjusted using FDR (false discovery rate). Differential expression probiotic strains (overexpression or suppressed expression) were determined by log2(Fold Change) > 1 and FDR-adjusted p value ≤ 0.05. Network analysis between probiotic strains and pathogen species (Salmonella spp., Mycoplasma spp., and E. coli) was performed using the FastSpar software, which is an implementation of the SparCC algorithm [30]. All pairwise SparCC negative correlations (R ≤ −0.4 and p value ≤ 0.05) between probiotic strains and selected pathogen strains were used and visualized using the Cytoscape software [31]. For all analyses, statistical significance was determined at p ≤ 0.05. All figures were generated using the R package, ggplot2 [32].
3. Results
3.1. Overall Profile of Probiotic Strains in Different Host Species
Whole genomes of the 192 probiotic strains were downloaded from the NCBI RefSeq or GenBank databases. The relative abundance based on metagenomic reads and expression based on meta-transcriptomic reads in the gut/rumen of six host species were measured by taxonomically assigning clean reads to probiotic genomes using the Kraken2 software. Chickens have the most abundant probiotics in the gut, with an average relative abundance of 9.82 ± 6.72%, followed by pigs (2.48 ± 1.03%), humans (1.93 ± 1.08%), sheep (0.43 ± 0.40%), mice (0.29 ± 0.029%), and cattle (0.21 ± 0.026%) (Figure 1A). Based on the meta-transcriptomic sequencing data, the average expression levels of all probiotic strains were assessed within individual host species to determine their overall expression abundance in each respective host: 0.67 ± 0.026% (cattle), 1.16 ± 0.97% (sheep), 3.58 ± 1.32% (pig), 19.74 ± 16.44% (chicken), 0.81 ± 0.17% (mouse), and 3.73 ± 1.90% (human) (Figure 1B). The extremely high relative abundance and expression level of total probiotics in the chicken gut compared to other host species is striking.
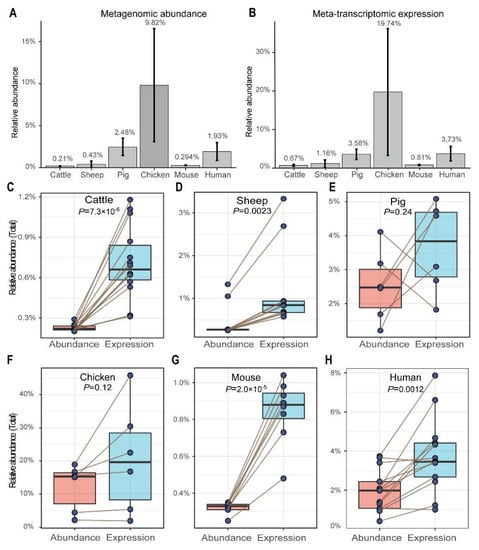
Figure 1.
Relative abundance (A), expression level (B), and probiotic expression activity (C–H) in different host species. Each point represents one sample (metagenomic sample or meta-transcriptomic sample) in (C–H); paired samples from the same animal are linked with a line. Significance between abundance and expression of each host species was determined using a two-tailed paired t-test.
A comparison between the levels of meta-transcriptomic expression and metagenomic abundance was conducted for each individual. It was found that the meta-transcriptomic expression in rumen/gut of cattle (p = 7.3 × 10−6, Figure 1C), sheep (p = 0.0023, Figure 1D), mouse (p = 2.0 × 10−5, Figure 1G), and human (p = 0.0012, Figure 1H) were significantly higher than that of metagenomic abundance. This indicates greater functional activity. The meta-transcriptomic expression in pigs (p = 0.24, Figure 1E) and chickens (p = 0.12, Figure 1F) were higher, although not significantly, than metagenomic abundance.
3.2. An Expression Profile of Probiotic Strains in Humans and Animals
An expression profile of the 192 probiotic strains in different host species was generated (Figure 2). In total, 58 probiotic strains were not detected in any samples, including 13/28 strains of Streptococcus thermophilus, 9/20 strains of Lactococcus lactis subsp. Lactis, 8/18 Bacillus subtilis subsp. subtilis, and 7/12 of Lacticaseibacillus paracasei. In addition, 5/6 Bifidobacterium animalis subsp. lactis strains were not detected in any samples. On the contrary, three probiotic strains, including Bacillus amyloliquefaciens DSM 7, Bdellovibrio bacteriovorus HD100, and Clostridium butyricum TOA, were present and expressed in all samples, indicating they likely play an important role in the basic biological functions in gut or rumen of human and animals. Interesting, we found that four strains (Phaeobacter inhibens DSM 17395, Micrococcus luteus NCTC 2665, Pseudomonas fluorescens A506, and Rhodobacter sphaeroides 2.4.1) that were associated with antimicrobial efficacy in aquaculture were present and expressed in the rumen or gut of partial individuals. For example, Phaeobacter inhibens is a marine microbe and is considered a potential probiotic bacterium in the aquaculture industry for reducing the growth of the pathogen, such as Vibrio anguillarum [33].
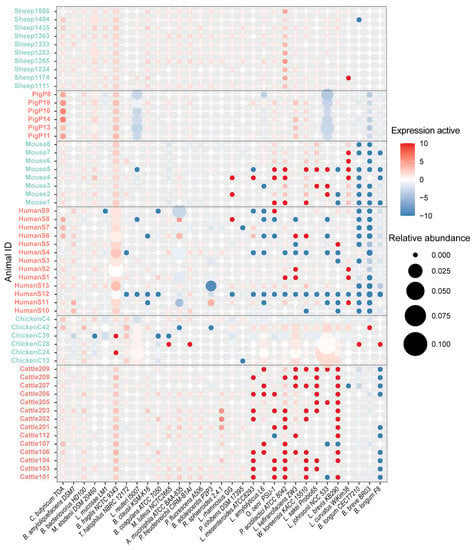
Figure 2.
Expression profile of probiotic strains in the gut/rumen of cattle, sheep, pigs, chickens, mice, and humans. The x-axis and y-axis represent probiotic strain and sample name, respectively. Circle size represents the metagenomic abundance of probiotic strain in the sample. Color scale represents the log2 transformed “expression activity”, which is defined as “meta-transcriptomic expression/metagenomic abundance”.
3.3. The Extremely Highly Expressed Probiotic Strains in the Gut or Rumen
Based on the expression profile, we explored the overexpressed probiotic strains (meta-transcriptomic expression was twice or more than twice than that of metagenomic abundance, FC ≥ 2, FDR ≤ 0.05) and suppressed probiotic strains (meta-transcriptomic expression equal to or less than half of metagenomic abundance, FC ≤ 0.5, FDR ≤ 0.05) using the “DESeq2” R package (Table S2). The fold change of each strain was calculated by metagenomic abundance of probiotic strains divided by meta-transcriptomic expression.
There were 17 (cattle, Figure 3A), 21 (sheep, Figure 3B), 22 (pig, Figure 3C), 14 (chicken, Figure 3D), 13 (mouse, Figure 3E), and 3 (human, Figure 3F) probiotic strains that were identified as overexpressed probiotic strains. Of them, Lactobacillus salivarius CECT 5713 is a probiotic strain that is commonly found in the gut of chickens and other animals. It was found to be the most abundant probiotic strain in this study. In addition, the strain of C. butyricum TOA was the another commonly overexpressed probiotic strains, which was found that significantly high expressed in all host species. It should be noted that there were 9 (cattle), 4 (sheep), 54 (pig), 32 (chicken), 23 (mouse), 31 (human) probiotic strains that demonstrated significantly suppressed expression (Table S2).
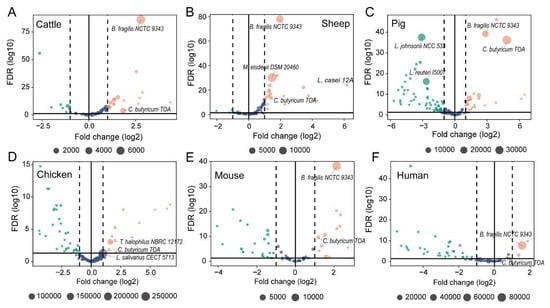
Figure 3.
(A–F) Differential activity of probiotic strains based on metagenomic abundance and meta-transcriptomic expression in host species. Active or suppressed probiotic strains identified by comparing metagenomic abundance and meta-transcriptomic expression of probiotic strains using DESeq2. Fold change between metagenomic abundance (basic) and meta-transcriptomic expression of probiotic strains is shown as log2(FC) (X-axis) and significance (FDR-adjusted) as -log10(FDR) (Y-axis). The circle size in each plot represents the normalized abundance of each strain. Suppressed and active probiotics in different host species were indicated by green and orange colors, respectively.
3.4. Network Analysis at Expression Level Revealed Associations between Probiotics with Pathogens
To explore the relationship between probiotics and pathogens, we generated a negative network plot based on gene expression (meta-transcriptomic data) of probiotics and pathogens to identify potential probiotics that may inhibit pathogenic bacteria (Figure 4). Our results showed that probiotic strains belonging to the genus Lactobacillus (L. curvatus WiKim38, L. reuteri I5007, L. rhamnosus GG ATCC 53103, L. mucosae LM1) were the only species negatively associated with Mycoplasma. Several species of Mycoplasma are pathogenic in humans and animals, such as Mycoplasma californicum. García-Galán’s study reported that the addition of Lactobacillus spp. could negatively affect the viability of Mycoplasma [34]. Here, we identified four candidates of Lactobacillus spp. that may have adverse effects on Mycoplasma. In addition, our results showed that four probiotic strains, including Rhodobacter sphaeroides 2.4.1, Phaeobacter inhibens DSM 17395, Megasphaera elsdenii DSM 20460, and Bacillus coagulans ATCC 7050, were negatively associated with both Salmonella spp. and Escherichia coli. Additionally, nine probiotic strains were negatively linked with Salmonella spp. only. Among them, the four probiotics associated with antimicrobial efficacy in aquaculture as mentioned above (Phaeobacter inhibens DSM 17395, Micrococcus luteus NCTC 2665, and Pseudomonas fluorescens A506) were showed to be negatively associated with either or both Salmonella spp. and E. coli, suggesting that these antimicrobial probiotics may also have an antibacterial effect in humans or livestock.
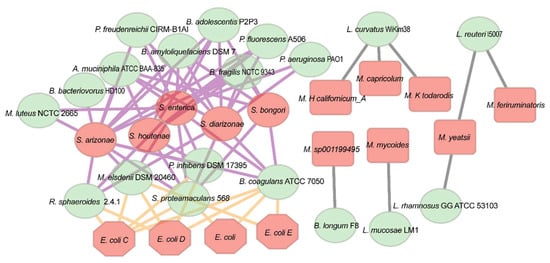
Figure 4.
Network visualizations of the probiotic-pathogen interactions in the gut or rumen of humans and animals. All pairwise SparCC negative correlations (R ≤ −0.4 and p value ≤ 0.05) between probiotic strains (green) and pathogens (red) are shown as edges in the network. The edges between probiotic strains and Salmonella, Mycoplasma, and E. coli are shown with purple, yellow, and grey lines, respectively.
4. Discussion
The global probiotic industry had been greatly expanded in the past decade, and as a result, there is a demand for probiotics with various beneficial functions for human and animal health. Indigenous bacteria, also known as autochthonous or native bacteria, are those that are isolated from a specific host organism. These bacteria often have a symbiotic relationship with their host, contributing to the host’s overall health and well-being [35]. In the context of probiotics, indigenous strains are of particular interest due to their enhanced ability to adapt, colonize, and exert beneficial effects within the host’s gut. Furthermore, indigenous bacteria serve as a vital source for the efficient development of probiotics [36,37]. By studying and harnessing these native strains, researchers can potentially develop more effective probiotic formulations tailored to individual hosts, thereby maximizing the health benefits of probiotic supplementation. In order to develop more efficient probiotic screening strategies in alignment with the concepts of indigenous bacteria, we profiled the expression level of probiotic strains in the gut or rumen of humans and animals by analyzing the metagenomic and meta-transcriptomic sequencing data. In order to investigate the distribution and expression of probiotics in humans, mice, and the main livestock animals, a total of 192 (candidate) probiotic strains reported by published studies were included in this study (Table S1). In the past decades, probiotic research has rapidly increased due to the improvement of multi-omics approaches, such as culturomics, genomics, and transcriptomics [38]. Many microbial strains have been identified to have probiotic properties and are considered probiotics or probiotic candidates. As an example, the NCBI genome database contains and stores 2167 and 1957 publicly available genomes of Bifidobacterium and Lactobacillus, respectively, which are the two most common genera of probiotic bacteria (source: http://www.ncbi.nlm.nih.gov/genome/browse/, accessed on 20 August 2022). In this study, we selected 192 probiotic strains, representing 51 species and 24 genera. Although many potential probiotic strains, especially newly identified strains, were inevitably missed in our study, the selected probiotic strains encompass the main taxonomy of known probiotic strains.
The extremely high relative abundance and expression level of total probiotics in the chicken gut with a relative abundance of 9.82% and 19.74%, compared to other host species, is striking. According to strain level profile of the selected probiotics (Figure 2), Lactobacillus johnsonii NCC 533, Lactobacillus kefiranofaciens ZW3, and Lactobacillus reuteri I5007 occupied the large proportion of the relative abundance of probiotics in chicken gut. A previous publication reported that Lactobacillus, the main source of probiotic strains, was the most abundant genus in chicken feces, with an average relative abundance of 14.9%~17.8% [39]. Several studies have reported that partial Lactobacillus species have positive effects on the growth performance, the immune response, and feed efficiency of chicken [40], such as L. acidophilus [41], L. plantarum [42], L. reuteri [43], and L. paracasei [44]. Here, we found that Lactobacillus johnsonii NCC 533 has a higher relative abundance and expression levels. Studies have shown that the supplementation of L. johnsonii can improve the growth performance of chickens by promoting better feed conversion efficiency and increased weight gain [45,46]. This is particularly beneficial for poultry farmers seeking to maximize their production efficiency. We speculate that the chicken gut is colonized by a large number of probiotic bacteria or genetically similar bacteria that play a critical role in the biological function of the chicken gut. However, some probiotic strains did not make positive function in chicken gut, for example, probiotic products containing Lactobacillus acidophilus used as an alternative to antibiotics in two breeds of roosters demonstrated no significant impact on their growth [47]. Therefore, the expression level should be considered as one factor while we assess the beneficial effectiveness of probiotics in gut.
This study aimed to investigate the expression profiles of 192 probiotic strains across various host species. Interestingly, 58 of these probiotic strains were not detected in any samples, which included several strains from Streptococcus thermophilus, Lactococcus lactis subsp. Lactis, Bacillus subtilis subsp. Subtilis, Lacticaseibacillus paracasei, and Bifidobacterium animalis subsp. Lactis. These findings suggest that not all probiotic strains may be universally beneficial (colonization and expression) across different host species. In contrast, three probiotic strains—Bacillus amyloliquefaciens DSM 7, Bdellovibrio bacteriovorus HD100, and Clostridium butyricum TOA—were found to be present and expressed in all samples, indicating their potential importance in the basic biological functions of the gut or rumen. This observation highlights the potential of these strains as promising candidates for further research on probiotics in various hosts.
Additionally, our study identified four strains (Phaeobacter inhibens DSM 17395, Micrococcus luteus NCTC 2665, Pseudomonas fluorescens A506, and Rhodobacter sphaeroides 2.4.1) associated with antimicrobial efficacy in aquaculture that were present and expressed in the gut or rumen of some individuals. One of these strains, Phaeobacter inhibens, has been considered a potential probiotic bacterium in the aquaculture industry for its ability to reduce the growth of pathogens, such as Vibrio anguillarum [33]. However, its function in the gut/rumen of livestock remains underexplored in previous research. Considering the presence and expression of these bacteria in humans and livestock, as demonstrated by our results, it is crucial to include them when screening antibacterial probiotics for these hosts.
In general, we believe that the highly expressed gut or rumen microorganisms likely play a stronger role in the host, suggesting that more attention to these probiotic strains is warranted in our studies. The most obvious example is Clostridium butyricum, which has been applied as a clinical probiotic for several diseases [48] and as dietary supplements in pigs [49], chickens [50], and ruminants [51,52] for improving economic traits. In this study, Clostridium butyricum TOA was found to be present and expressed in all samples, as well as highly expressed in the rumen or gut of all host species, indicating it likely has high functional activity in the gut or rumen (FC ≥ 2, FDR ≤ 0.05). This finding suggests that this particular probiotic strain may play a crucial role in the gut health of various host species. Previous studies have shown that Clostridium butyricum is a beneficial probiotic due to its ability to produce butyrate, a short-chain fatty acid that provides energy for colonocytes and supports the health of the intestinal epithelium [53,54]. Butyrate has been associated with various health benefits, including anti-inflammatory effects and improved gut barrier function. In pig, Clostridium butyricum TOA was the most abundant probiotic strain of our selected strains and has an extremely high expression level with an average fold change of 4.8. Previous studies have reported that dietary supplementation of Clostridium butyricum significantly increased growth performance and reduced Salmonella typhimurium infection in pigs [49,55], which may be partly explained its high expression level in pig gut according to our results.
Lactobacillus salivarius CECT 5713 was highly expressed in chicken gut, indicating that it plays an important role in the gut microbiome of chickens. Lactobacillus salivarius has been confirmed to have probiotic properties and has been shown to affect performance and immune in animals [56]. For other strains with higher expression levels in gut or rumen, their potential probiotic function in the corresponding host gut or rumen should be further explored.
Here, we also investigate the relationship between probiotics and pathogens by analyzing the gene expression data (meta-transcriptomic) of probiotics and pathogens to identify potential probiotics that may inhibit pathogenic bacteria. A negative network plot was generated (Figure 4), which revealed interesting insights into the associations between probiotics and pathogens. Our results indicated that probiotic strains belonging to the genus Lactobacillus, such as L. curvatus WiKim38, L. reuteri I5007, L. rhamnosus GG ATCC 53103, and L. mucosae LM1, were the only species negatively associated with Mycoplasma [57]. Mycoplasma species, such as Mycoplasma californicum, which causes bovine mastitis [58], and Mycoplasma mycoides, which leads to Contagious Bovine Pleuropneumonia (CBPP) [59], are known to be pathogenic in humans and animals. In this study, we identified four Lactobacillus spp. candidates that may have inhibitory effects on Mycoplasma.
Additionally, our results demonstrated that four probiotic strains, including Rhodobacter sphaeroides 2.4.1, Phaeobacter inhibens DSM 17395, Megasphaera elsdenii DSM 20460, and Bacillus coagulans ATCC 7050, were negatively associated with both Salmonella spp. and Escherichia coli. Moreover, nine probiotic strains were found to be negatively linked with Salmonella spp. only. Among these strains, four probiotics associated with antimicrobial efficacy in aquaculture (Phaeobacter inhibens DSM 17395, Micrococcus luteus NCTC 2665, Pseudomonas fluorescens A506) were shown to be negatively associated with one or both of Salmonella spp. and E. coli. These findings suggest that these antimicrobial probiotics may also exert antibacterial effects in humans or livestock. The identification of probiotic strains with potential inhibitory effects on pathogenic bacteria is crucial for the development of effective probiotic interventions in both human and animal health. Our findings provide valuable insights into the associations between probiotics and pathogens and serve as a foundation for further research on the specific roles and mechanisms of action of these probiotic strains in inhibiting pathogenic bacteria. Understanding these relationships will help optimize the potential applications and benefits of probiotics for improving the health of various host species.
Nonetheless, these findings require further validation, particularly at the strain level, and the variations in the effectiveness of different strains against pathogenic bacteria warrant more in-depth examination and discussion.
The scope of strains utilized in this study is somewhat limited, which presents the issue of potentially combining similar strains (but not representative genomes) into analogous categories. To conduct a more comprehensive and representative investigation, the establishment of a more extensive database of probiotic strains is essential. Furthermore, achieving quantitative accuracy at the strain level continues to be a challenging scientific conundrum that is yet to be fully addressed, and the development of improved methods remains a valuable area of exploration [60].
5. Conclusions
The isolation of highly efficient probiotic strains is of the utmost importance. In this study, we generated an expression profile of probiotic strains and found multiple differentially expressed (overexpression or suppressed expression) probiotics in the gut/rumen of hosts. This study provided a useful reference for indigenous probiotic isolation and animal model selection in the future, and suggest new candidate probiotic strains for human and animals. In addition, the network analysis identified several probiotic strains, such as Rhodobacter sphaeroides 2.4.1, Phaeobacter inhibens DSM 17395, Megasphaera elsdenii DSM 20460, Bacillus coagulans ATCC 7050, and Lactobacillus spp., negatively linked with pathogenic bacteria, suggesting the potential antibacterial properties of these probiotics in humans and animals.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation9050417/s1, Table S1: Information of the probiotic strains used in this study; Table S2: The significantly upregulated or downregulated probiotic strains in human and animals.
Author Contributions
Conceptualization, F.D. and Y.L.; methodology, Y.P., R.C. and Z.Z.; formal analysis, Y.P., R.C., X.L. and Z.Z.; data curation, R.J.; writing—original draft preparation, Y.P. and R.C.; writing—review and editing, S.H., J.Z., F.D. and J.C.; visualization, Z.Z., R.J., T.X. and X.L.; funding acquisition, Y.L. and F.D. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Youth project of Guangdong Foshan joint fund of the Guangdong Natural Science Foundation (2022A1515110819), National Natural Science Foundation of China (No 32170430), Guangdong Provincial Key Laboratory of Animal Molecular Design and Precise Breeding (2019B030301010), and Key Laboratory of Animal Molecular Design and Precise Breeding of Guangdong Higher Education Institutes (2019KSYS011).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rianda, D.; Agustina, R.; Setiawan, E.A.; Manikam, N.R.M. Effect of probiotic supplementation on cognitive function in children and adolescents: A systematic review of randomised trials. Benef. Microbes 2019, 10, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Galdeano, C.M.; Cazorla, S.I.; Dumit, J.M.L.; Vélez, E.; Perdigón, G. Beneficial effects of probiotic consumption on the immune system. Ann. Nutr. Metab. 2019, 74, 115–124. [Google Scholar]
- Jha, R.; Das, R.; Oak, S.; Mishra, P. Probiotics (direct-fed microbials) in poultry nutrition and their effects on nutrient utilization, growth and laying performance, and gut health: A systematic review. Animals 2020, 10, 1863. [Google Scholar] [CrossRef] [PubMed]
- Kambale, R.M.; Nancy, F.I.; Ngaboyeka, G.A.; Kasengi, J.B.; Bindels, L.B.; Van der Linden, D. Effects of probiotics and synbiotics on diarrhea in undernourished children: Systematic review with meta-analysis. Clin. Nutr. 2021, 40, 3158–3169. [Google Scholar] [CrossRef]
- Curciarello, R.; Canziani, K.E.; Salto, I.; Barbiera Romero, E.; Rocca, A.; Doldan, I.; Peton, E.; Brayer, S.; Sambuelli, A.M.; Goncalves, S.; et al. Probiotic Lactobacilli isolated from kefir promote down-regulation of inflammatory lamina propria t cells from patients with active IBD. Front. Pharmacol. 2021, 12, 658026. [Google Scholar] [CrossRef]
- Ebell, M.H. Probiotic Ineffective for Treatment of Acute Gastroenteritis in Young Children. Am. Fam. Physician 2019, 99, 717. [Google Scholar]
- Kothari, D.; Patel, S.; Kim, S.-K. Probiotic supplements might not be universally-effective and safe: A review. Biomed. Pharmacother. 2019, 111, 537–547. [Google Scholar] [CrossRef]
- Betancur, C.; Martínez, Y.; Tellez-Isaias, G.; Avellaneda, M.C.; Velázquez-Martí, B. In vitro characterization of indigenous probiotic strains isolated from Colombian creole pigs. Animals 2020, 10, 1204. [Google Scholar] [CrossRef]
- Chiang, M.-L.; Chen, H.-C.; Chen, K.-N.; Lin, Y.-C.; Lin, Y.-T.; Chen, M.-J. Optimizing production of two potential probiotic Lactobacilli strains isolated from piglet feces as feed additives for weaned piglets. Asian-Australas. J. Anim. Sci. 2015, 28, 1163. [Google Scholar] [CrossRef]
- Ouwehand, A.C.; Salminen, S.; Isolauri, E. Probiotics: An overview of beneficial effects. In Proceedings of the Lactic Acid Bacteria: Genetics, Metabolism and Applications: Proceedings of the Seventh Symposium on Lactic acid Bacteria: Genetics, Metabolism and Applications, Egmond aan Zee, The Netherlands, 1–5 September 2002; Springer: Dordrecht, The Netherlands, 2002; pp. 279–289. [Google Scholar]
- Wang, Y.; Hu, Y.; Liu, F.; Cao, J.; Lv, N.; Zhu, B.; Zhang, G.; Gao, G.F. Integrated metagenomic and metatranscriptomic profiling reveals differentially expressed resistomes in human, chicken, and pig gut microbiomes. Environ. Int. 2020, 138, 105649. [Google Scholar] [CrossRef]
- Li, F.; Hitch, T.C.; Chen, Y.; Creevey, C.J.; Guan, L.L. Comparative metagenomic and metatranscriptomic analyses reveal the breed effect on the rumen microbiome and its associations with feed efficiency in beef cattle. Microbiome 2019, 7, 6. [Google Scholar] [CrossRef]
- Kamke, J.; Kittelmann, S.; Soni, P.; Li, Y.; Tavendale, M.; Ganesh, S.; Janssen, P.H.; Shi, W.; Froula, J.; Rubin, E.M.; et al. Rumen metagenome and metatranscriptome analyses of low methane yield sheep reveals a Sharpea-enriched microbiome characterised by lactic acid formation and utilisation. Microbiome 2016, 4, 56. [Google Scholar] [CrossRef]
- Chung, Y.W.; Gwak, H.-J.; Moon, S.; Rho, M.; Ryu, J.-H. Functional dynamics of bacterial species in the mouse gut microbiome revealed by metagenomic and metatranscriptomic analyses. PLoS ONE 2020, 15, e0227886. [Google Scholar] [CrossRef]
- Tisza, M.J.; Buck, C.B. A catalog of tens of thousands of viruses from human metagenomes reveals hidden associations with chronic diseases. Proc. Natl. Acad. Sci. USA 2021, 118, e2023202118. [Google Scholar] [CrossRef]
- Sun, Y.; Li, H.; Zheng, L.; Li, J.; Hong, Y.; Liang, P.; Kwok, L.-Y.; Zuo, Y.; Zhang, W.; Zhang, H. iProbiotics: A machine learning platform for rapid identification of probiotic properties from whole-genome primary sequences. Brief. Bioinform. 2021, 23, bbab477. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Rosen, B.D.; Bickhart, D.M.; Schnabel, R.D.; Koren, S.; Elsik, C.G.; Tseng, E.; Rowan, T.N.; Low, W.Y.; Zimin, A.; Couldrey, C.; et al. De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience 2020, 9, giaa021. [Google Scholar] [CrossRef]
- The International Sheep Genomics Consortium; Archibald, A.; Cockett, N.; Dalrymple, B.; Faraut, T.; Kijas, J.; Maddox, J.; McEwan, J.; Hutton Oddy, V.; Raadsma, H.; et al. The sheep genome reference sequence: A work in progress. Anim. Genet. 2010, 41, 449–453. [Google Scholar]
- Wallis, J.W.; Aerts, J.; Groenen, M.A.; Crooijmans, R.P.; Layman, D.; Graves, T.A.; Scheer, D.E.; Kremitzki, C.; Fedele, M.J.; Mudd, N.K.; et al. A physical map of the chicken genome. Nature 2004, 432, 761–764. [Google Scholar] [CrossRef]
- Groenen, M.A.; Archibald, A.L.; Uenishi, H.; Tuggle, C.K.; Takeuchi, Y.; Rothschild, M.F.; Rogel-Gaillard, C.; Park, C.; Milan, D.; Megens, H.-J.; et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 2012, 491, 393–398. [Google Scholar] [CrossRef]
- Keane, T.M.; Goodstadt, L.; Danecek, P.; White, M.A.; Wong, K.; Yalcin, B.; Heger, A.; Agam, A.; Slater, G.; Goodson, M.; et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 2011, 477, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Koren, S.; Rhie, A.; Rautiainen, M.; Bzikadze, A.V.; Mikheenko, A.; Vollger, M.R.; Altemose, N.; Uralsky, L.; Gershman, A.; et al. The complete sequence of a human genome. Science 2022, 376, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Rotmistrovsky, K.; Agarwala, R. BMTagger: Best Match Tagger for removing human reads from metagenomics datasets. 2011; preprint. [Google Scholar]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Chuvochina, M.; Chaumeil, P.-A.; Rinke, C.; Mussig, A.J.; Hugenholtz, P. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 2020, 38, 1079–1086. [Google Scholar] [CrossRef]
- Youngblut, N.D.; Ley, R.E. Struo2: Efficient metagenome profiling database construction for ever-expanding microbial genome datasets. PeerJ 2021, 9, e12198. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Watts, S.C.; Ritchie, S.C.; Inouye, M.; Holt, K.E. FastSpar: Rapid and scalable correlation estimation for compositional data. Bioinformatics 2019, 35, 1064–1066. [Google Scholar] [CrossRef]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.-L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Grotkjær, T.; Bentzon-Tilia, M.; D’Alvise, P.; Dierckens, K.; Bossier, P.; Gram, L. Phaeobacter inhibens as probiotic bacteria in non-axenic Artemia and algae cultures. Aquaculture 2016, 462, 64–69. [Google Scholar] [CrossRef]
- García-Galán, A.; De la Fe, C.; Gomis, J.; Bataller, E.; Sánchez, A.; Quereda, J.; García-Roselló, E.; Gómez-Martín, A. The addition of Lactobacillus spp. negatively affects Mycoplasma bovis viability in bovine cervical mucus. BMC Vet. Res. 2020, 16, 251. [Google Scholar] [CrossRef]
- Ma, C.; Zhang, C.; Chen, D.; Jiang, S.; Shen, S.; Huo, D.; Huang, S.; Zhai, Q.; Zhang, J. Probiotic consumption influences universal adaptive mutations in indigenous human and mouse gut microbiota. Commun. Biol. 2021, 4, 1198. [Google Scholar] [CrossRef]
- Aziz, G.; Tariq, M.; Zaidi, A.H. Mining indigenous honeybee gut microbiota for Lactobacillus with probiotic potential. Microbiology 2021, 167, 001032. [Google Scholar] [CrossRef]
- Lecocq, A.; Natsopoulou, M.; Berggreen, I.; Eilenberg, J.; Heckmann, L.-H.L.; Nielsen, H.; Stensvold, C.; Jensen, A. Probiotic properties of an indigenous Pediococcus pentosaceus strain on Tenebrio molitor larval growth and survival. J. Insects Food Feed 2021, 7, 975–986. [Google Scholar] [CrossRef]
- Rebollar, E.A.; Antwis, R.E.; Becker, M.H.; Belden, L.K.; Bletz, M.C.; Brucker, R.M.; Harrison, X.A.; Hughey, M.C.; Kueneman, J.G.; Loudon, A.H.; et al. Using “omics” and integrated multi-omics approaches to guide probiotic selection to mitigate chytridiomycosis and other emerging infectious diseases. Front. Microbiol. 2016, 7, 68. [Google Scholar] [CrossRef]
- Yan, W.; Sun, C.; Yuan, J.; Yang, N. Gut metagenomic analysis reveals prominent roles of Lactobacillus and cecal microbiota in chicken feed efficiency. Sci. Rep. 2017, 7, 45308. [Google Scholar] [CrossRef]
- Fesseha, H.; Demlie, T.; Mathewos, M.; Eshetu, E. Effect of Lactobacillus species probiotics on growth performance of dual-purpose chicken. Vet. Med. Res. Rep. 2021, 12, 75–83. [Google Scholar] [CrossRef]
- Forte, C.; Manuali, E.; Abbate, Y.; Papa, P.; Vieceli, L.; Tentellini, M.; Trabalza-Marinucci, M.; Moscati, L. Dietary Lactobacillus acidophilus positively influences growth performance, gut morphology, and gut microbiology in rurally reared chickens. Poult. Sci. 2018, 97, 930–936. [Google Scholar] [CrossRef]
- Peng, Q.; Zeng, X.; Zhu, J.; Wang, S.; Liu, X.; Hou, C.; Thacker, P.; Qiao, S. Effects of dietary Lactobacillus plantarum B1 on growth performance, intestinal microbiota, and short chain fatty acid profiles in broiler chickens. Poult. Sci. 2016, 95, 893–900. [Google Scholar] [CrossRef]
- Nakphaichit, M.; Sobanbua, S.; Siemuang, S.; Vongsangnak, W.; Nakayama, J.; Nitisinprasert, S. Protective effect of Lactobacillus reuteri KUB-AC5 against Salmonella Enteritidis challenge in chickens. Benef. Microbes 2019, 10, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Tian, Y.; Cao, Y.; Li, J.; Guo, H.; Su, Y.; Tian, Y.; Wang, C.; Wang, T.; Zhang, L. Probiotic properties of Lactobacillus paracasei subsp. paracasei L1 and its growth performance-promotion in chicken by improving the intestinal microflora. Front. Physiol. 2019, 10, 937. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Ni, X.; Zeng, D.; Wang, H.; Jing, B.; Yin, Z.; Pan, K. Effect of a dietary probiotic, Lactobacillus johnsonii BS15, on growth performance, quality traits, antioxidant ability, and nutritional and flavour substances of chicken meat. Anim. Prod. Sci. 2016, 57, 920–926. [Google Scholar] [CrossRef]
- Olnood, C.G.; Beski, S.S.; Choct, M.; Iji, P.A. Novel probiotics: Their effects on growth performance, gut development, microbial community and activity of broiler chickens. Anim. Nutr. 2015, 1, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Fatufe, A.; Matanmi, O. The effect of probiotics supplementation on the growth performance of two strains of cockerels. J. Cent. Eur. Agric. 2008, 9. [Google Scholar] [CrossRef]
- Yasueda, A.; Mizushima, T.; Nezu, R.; Sumi, R.; Tanaka, M.; Nishimura, J.; Kai, Y.; Hirota, M.; Osawa, H.; Nakajima, K.; et al. The effect of Clostridium butyricum MIYAIRI on the prevention of pouchitis and alteration of the microbiota profile in patients with ulcerative colitis. Surg. Today 2016, 46, 939–949. [Google Scholar] [CrossRef]
- Chen, L.; Li, S.; Zheng, J.; Li, W.; Jiang, X.; Zhao, X.; Li, J.; Che, L.; Lin, Y.; Xu, S.; et al. Effects of dietary Clostridium butyricum supplementation on growth performance, intestinal development, and immune response of weaned piglets challenged with lipopolysaccharide. J. Anim. Sci. Biotechnol. 2018, 9, 62. [Google Scholar] [CrossRef]
- Yang, C.; Cao, G.; Ferket, P.; Liu, T.; Zhou, L.; Zhang, L.; Xiao, Y.; Chen, A. Effects of probiotic, Clostridium butyricum, on growth performance, immune function, and cecal microflora in broiler chickens. Poult. Sci. 2012, 91, 2121–2129. [Google Scholar] [CrossRef]
- Kohiruimaki, M.; Ohtsuka, H.; Tanami, E.; Kitagawa, M.; Masui, M.; Ando, T.; Kawamura, S. Effects of active egg white product/Clostridium butyricum Miyairi 588 additive on peripheral leukocyte populations in periparturient dairy cows. J. Vet. Med. Sci. 2008, 70, 321–323. [Google Scholar] [CrossRef]
- Cai, L.; Yu, J.; Hartanto, R.; Qi, D. Dietary supplementation with Saccharomyces cerevisiae, Clostridium butyricum and their combination ameliorate rumen fermentation and growth performance of heat-stressed goats. Animals 2021, 11, 2116. [Google Scholar] [CrossRef]
- Ariyoshi, T.; Hagihara, M.; Takahashi, M.; Mikamo, H. Effect of Clostridium butyricum on gastrointestinal infections. Biomedicines 2022, 10, 483. [Google Scholar] [CrossRef]
- Tran, N.T.; Li, Z.; Ma, H.; Zhang, Y.; Zheng, H.; Gong, Y.; Li, S. Clostridium butyricum: A promising probiotic confers positive health benefits in aquatic animals. Rev. Aquac. 2020, 12, 2573–2589. [Google Scholar] [CrossRef]
- Sato, Y.; Kuroki, Y.; Oka, K.; Takahashi, M.; Rao, S.; Sukegawa, S.; Fujimura, T. Effects of dietary supplementation with Enterococcus faecium and Clostridium butyricum, either alone or in combination, on growth and fecal microbiota composition of post-weaning pigs at a commercial farm. Front. Vet. 2019, 6, 26. [Google Scholar] [CrossRef]
- Wang, J.; Ishfaq, M.; Guo, Y.; Chen, C.; Li, J. Assessment of probiotic properties of Lactobacillus salivarius isolated from chickens as feed additives. Front. Vet. Sci. 2020, 7, 415. [Google Scholar] [CrossRef]
- Waites, K.B.; Talkington, D.F. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 2004, 17, 697–728. [Google Scholar] [CrossRef]
- Mackie, D.; Ball, H.; Logan, E. Isolation of Mycoplasma californicum from an outbreak of bovine mastitis and the experimental reproduction of the disease. Vet. Rec. 1982, 110, 578–580. [Google Scholar] [CrossRef]
- Krasteva, I.; Liljander, A.; Fischer, A.; Smith, D.G.; Inglis, N.F.; Scacchia, M.; Pini, A.; Jores, J.; Sacchini, F. Characterization of the in vitro core surface proteome of Mycoplasma mycoides subsp. mycoides, the causative agent of contagious bovine pleuropneumonia. Vet. Microbiol. 2014, 168, 116–123. [Google Scholar] [CrossRef]
- Anyansi, C.; Straub, T.J.; Manson, A.L.; Earl, A.M.; Abeel, T. Computational methods for strain-level microbial detection in colony and metagenome sequencing data. Front. Microbiol. 2020, 11, 1925. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).