A Study of the Metabolic Profiles of Penicillium dimorphosporum KMM 4689 Which Led to Its Re-Identification as Penicillium hispanicum
Abstract
1. Introduction
2. Materials and Methods
2.1. General Experimental Procedures
2.2. Fungal Strain
2.3. Cultivation of Fungus
2.4. Extraction
2.5. HPLC UV
2.6. HPLC MS
2.7. Data Analysis
2.8. Molecular Networking and Spectral Library Search
2.9. Principal Component Analysis ((PCA)
2.10. Bioasays
2.10.1. Cell Culture
2.10.2. Cell Viability Assay
2.10.3. Cell-Free DPPH Assay
2.11. DNA Extraction and Amplification
2.12. Phylogenetic Analysis
3. Results
3.1. The Effect of Culture Conditions on Metabolic Profile of Fungal Extracts and Its Bioactivity
3.1.1. HPLC UV
3.1.2. Bioactivity of Extracts
3.1.3. HPLC MS
3.2. Molecular Re-Identification of the Fungal Strain
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| N | Name | Structure | RT | Exact Mass (Measured) | Exact Mass (Calcd) | Δ, ppm | MQScore (GNPS) | Ref. |
|---|---|---|---|---|---|---|---|---|
| 1 | 16α-hydroxy-17β-methoxy-deoxydihydroisoaustamide | 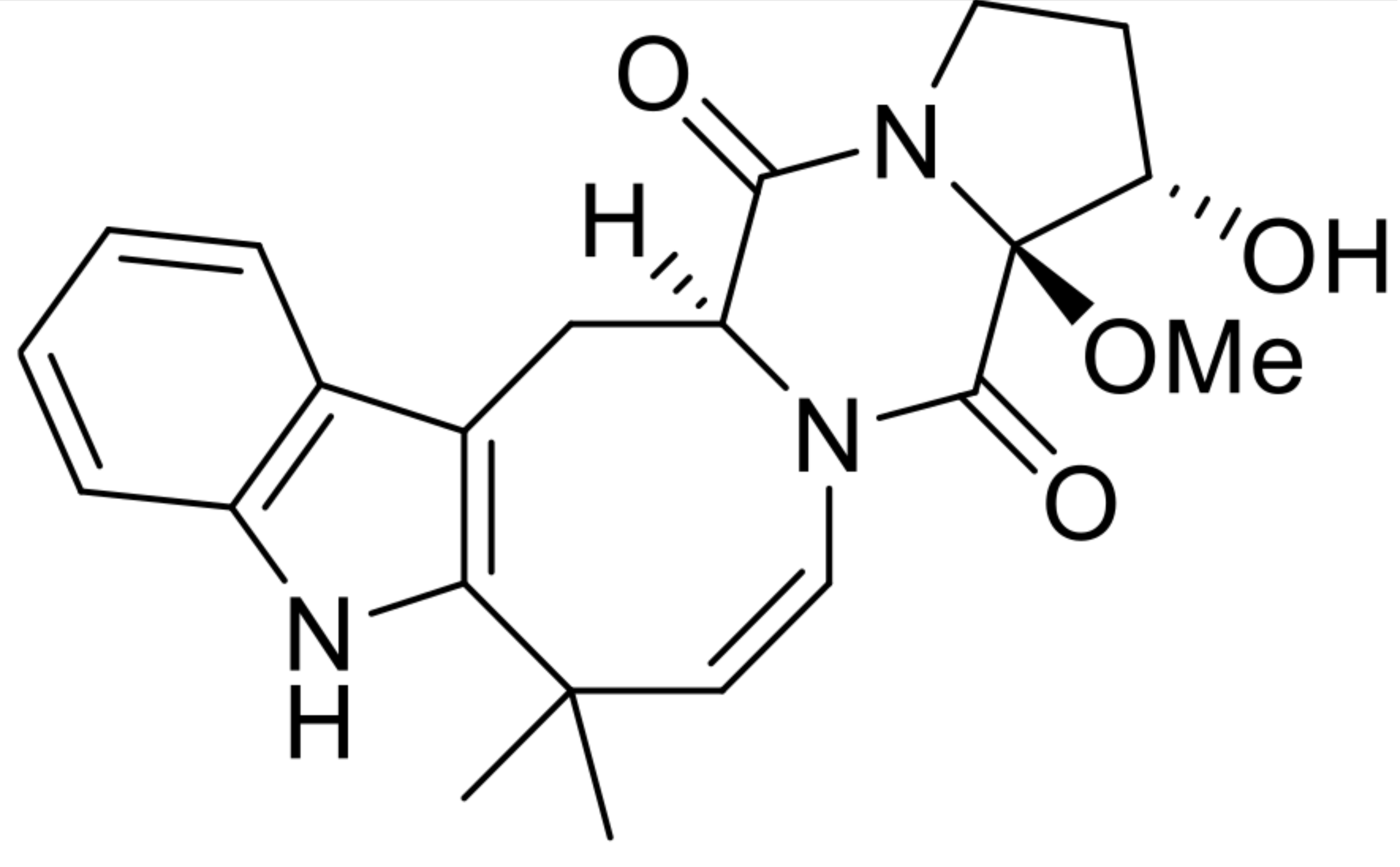 | 7.3 | 396.194 [M+H]+ | 396.1918 | −5.6 | * | [8] |
| 2 | 16β-hydroxy-17α-methoxy-deoxydihydroisoaustamide |  | 7.3 | 396.194 [M+H]+ | 396.1918 | −5.6 | * | [8] |
| 3 | 16α-hydroxy-17α-methoxy-deoxydihydroisoaustamide | 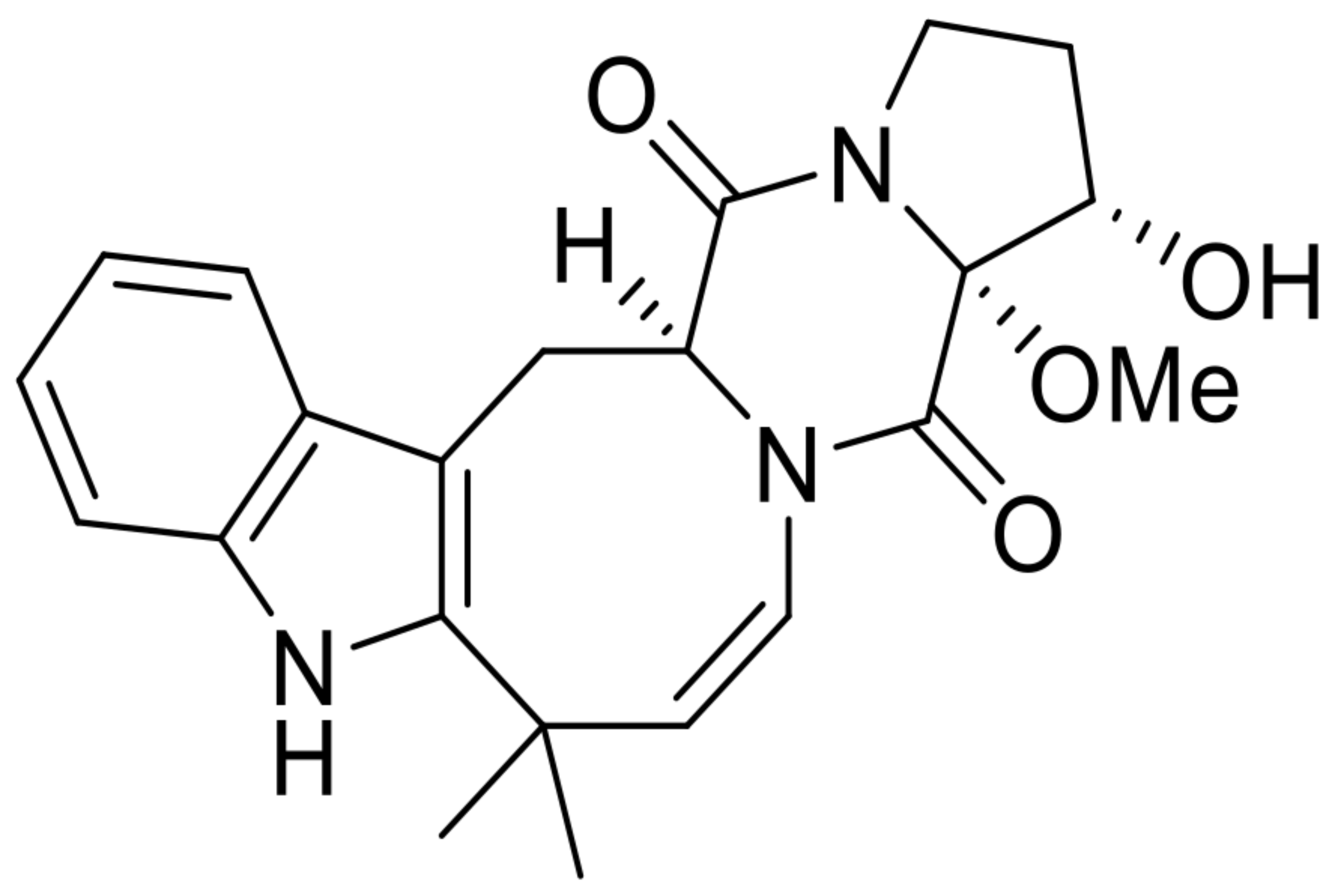 | 7.3 | 396.194 [M+H]+ | 396.1918 | −5.6 | * | [8] |
| 4 | 16,17-dihydroxy-deoxydihydroisoaustamide | 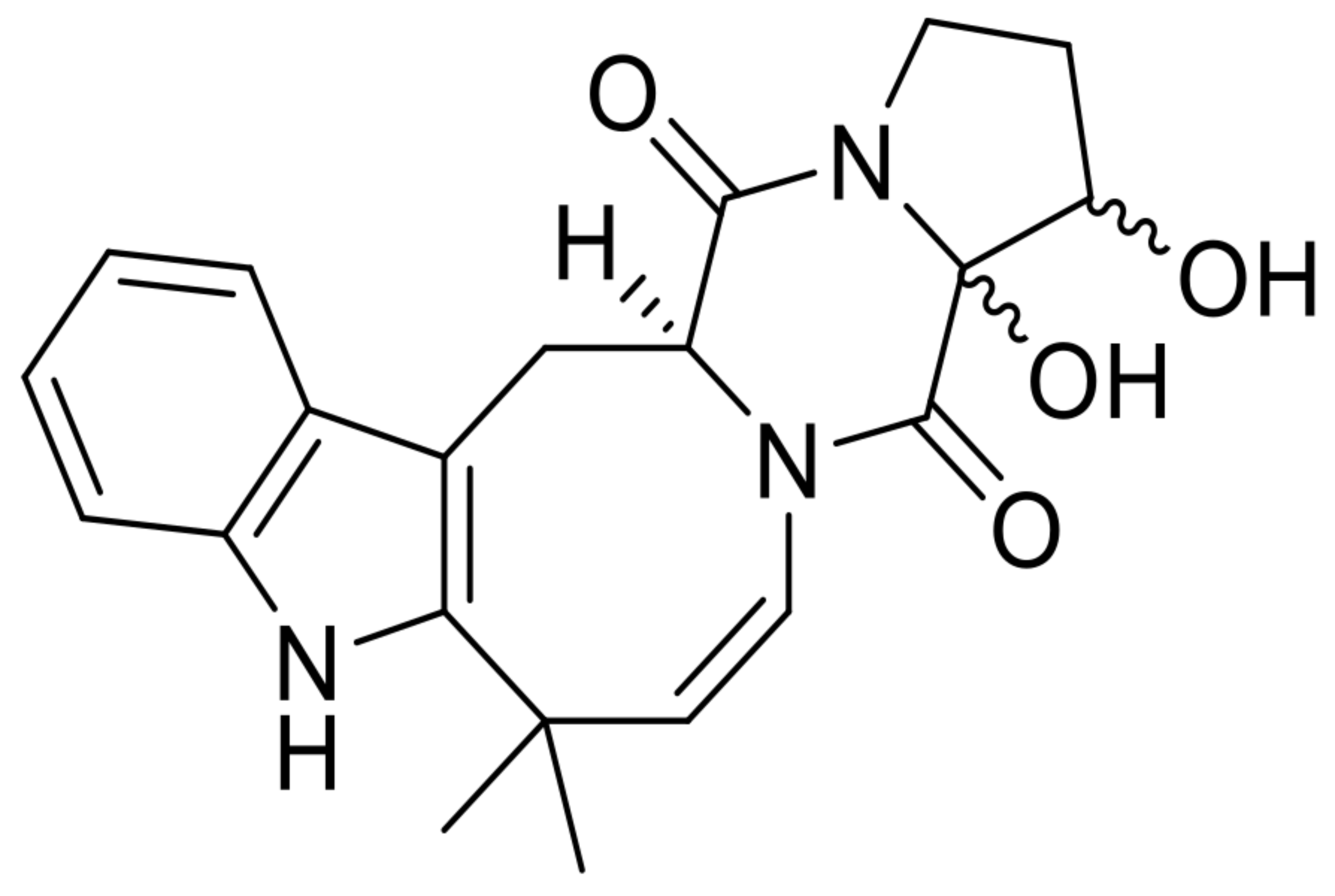 | 6.2 | 382.176 [M+H]+ | 382.1761 | 0.3 | * | [8] |
| 5 | 16β,17α-dihydroxy-deoxydihydroisoaustamide | 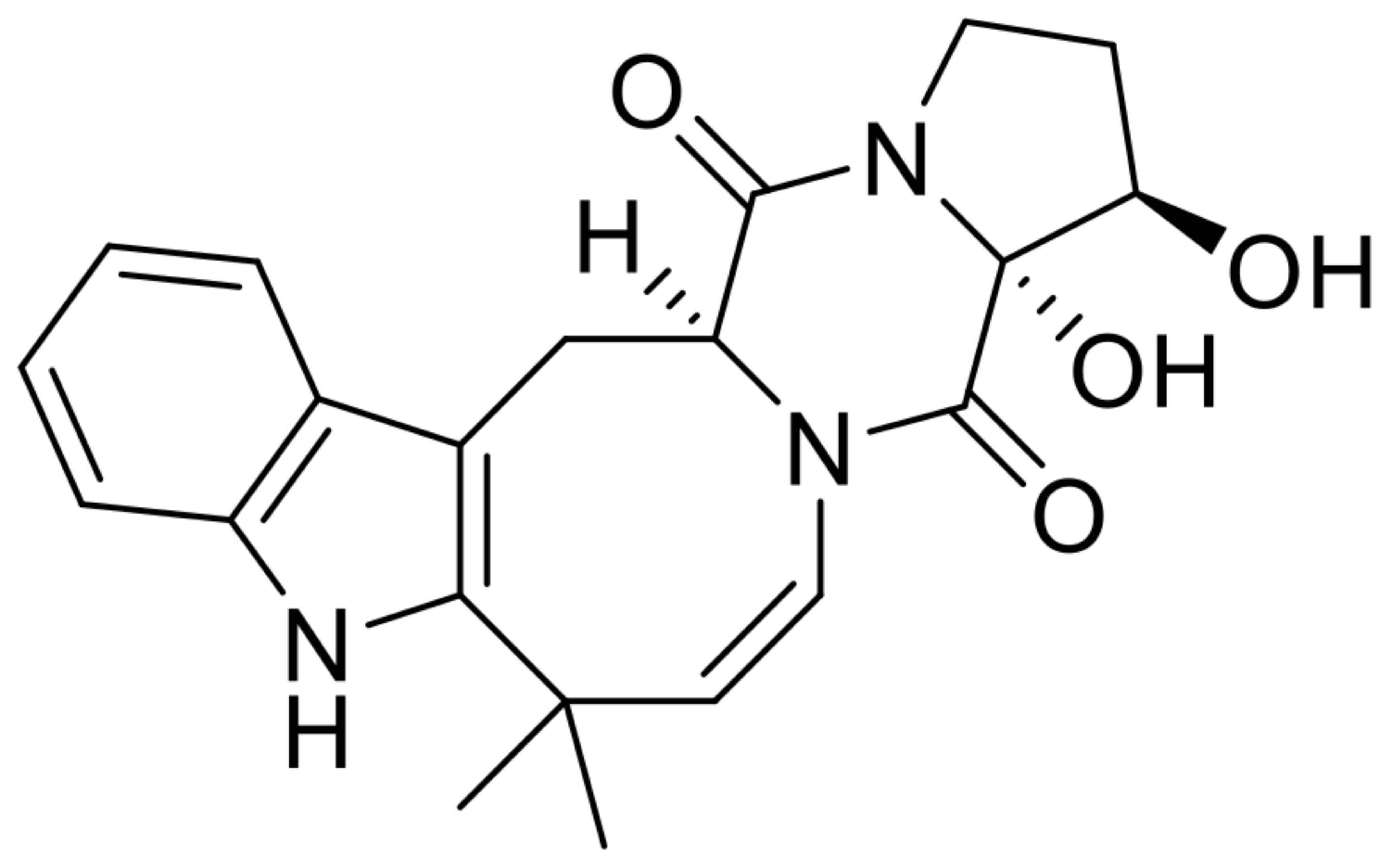 | 6.0 | 382.176 [M+H]+ | 382.1761 | 0.3 | * | [8] |
| 6 | 16α,17α-dihydroxy-deoxydihydroisoaustamide |  | 6.6 | 382.176 [M+H]+ | 382.1761 | 0.3 | * | [8] |
| 7 | 3β-hydroxydeoxyisoaustamide |  | 2.9 | 364.1645 [M+H]+ | 364.1656 | 3.0 | * | [8] |
| 8 | deoxy-14,15-dehydroisoaustamide | 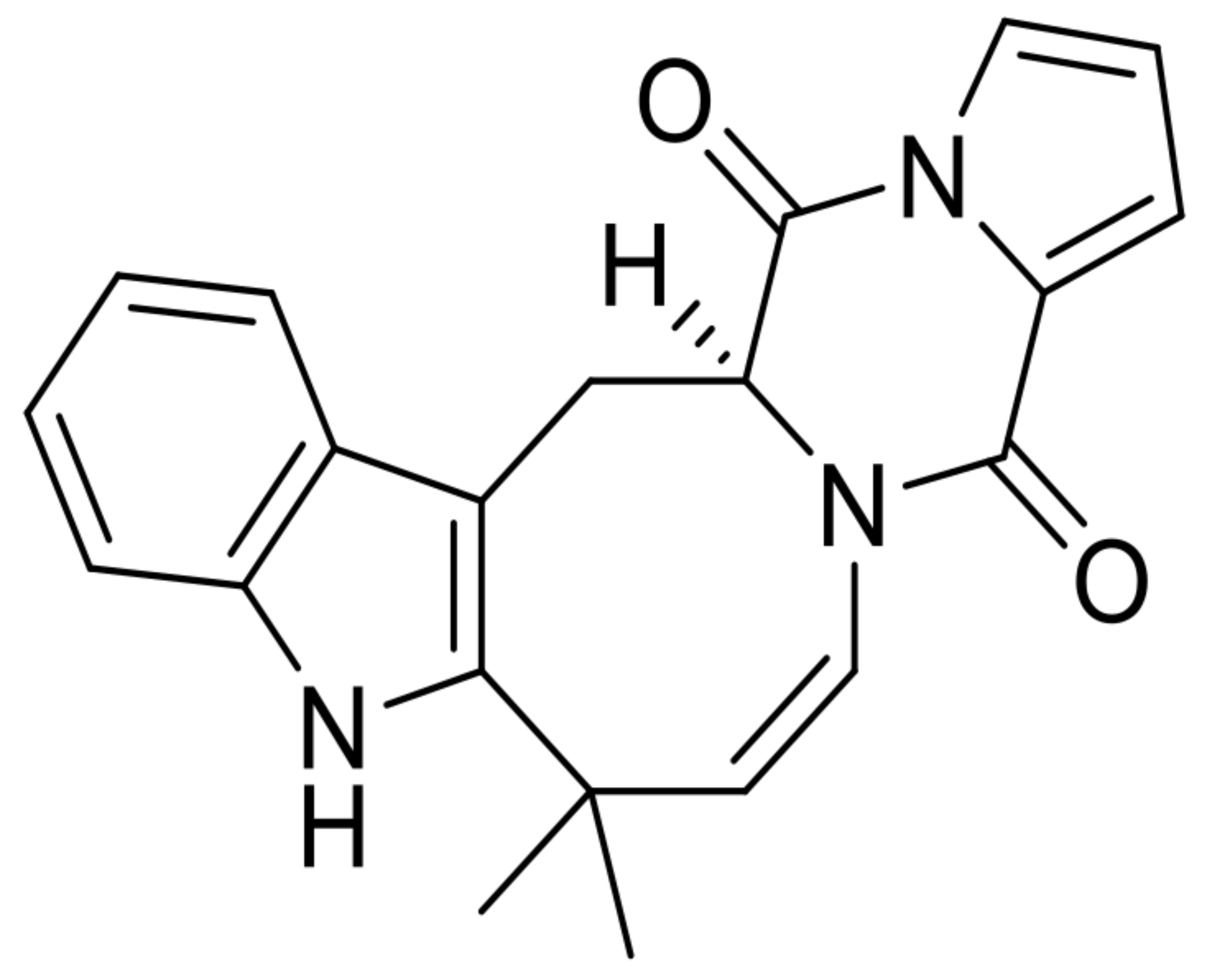 | 10.8 | 346.1562 [M+H]+ | 346.1550 | 3.4 | ** | [9] |
| 9 | (+)-deoxyisoaustamide | 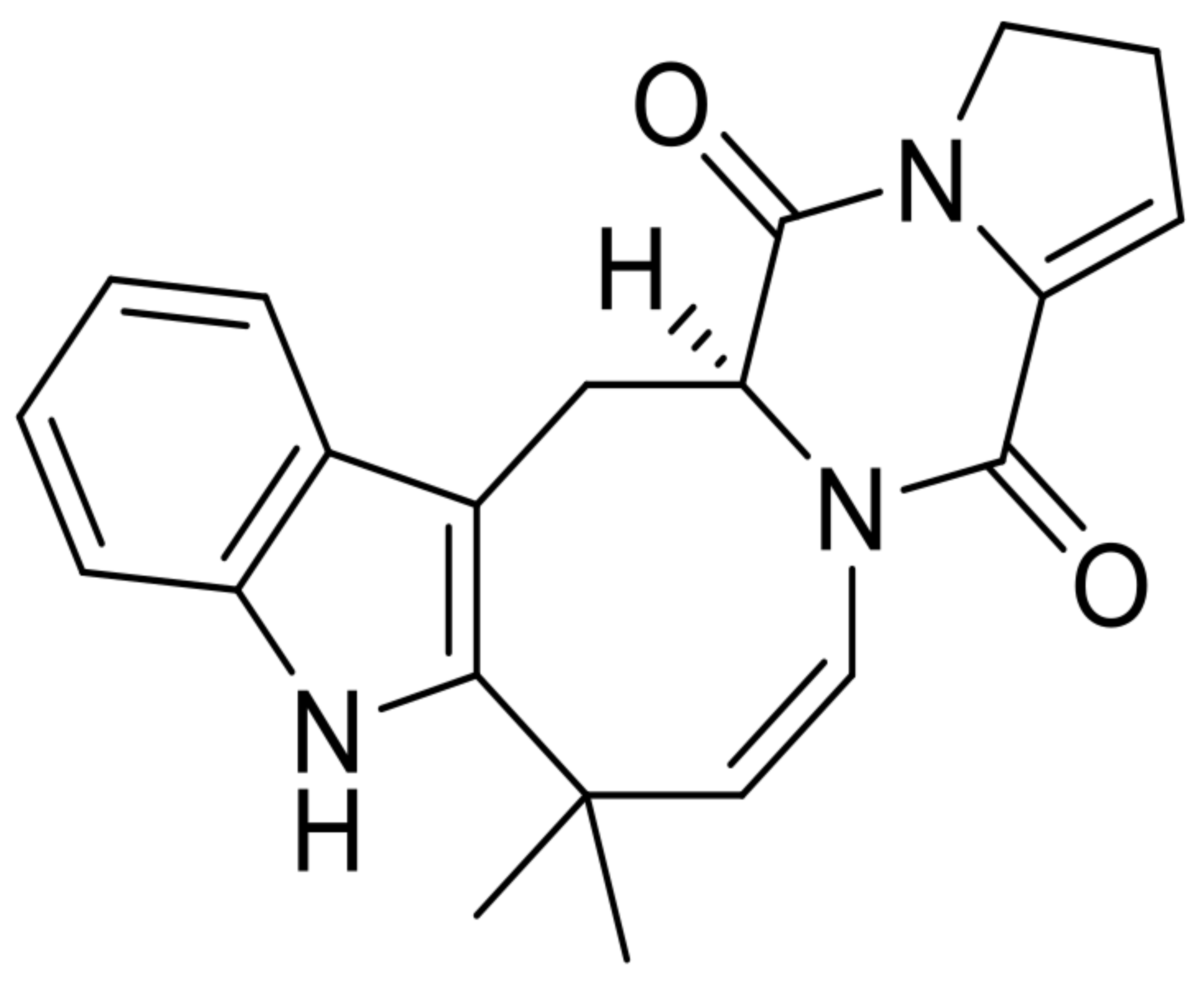 | 7.7 | 348.1697 [M+H]+ | 348.1707 | 2.9 | * | [8] |
| 10 | deoxydihydroisoaustamide | 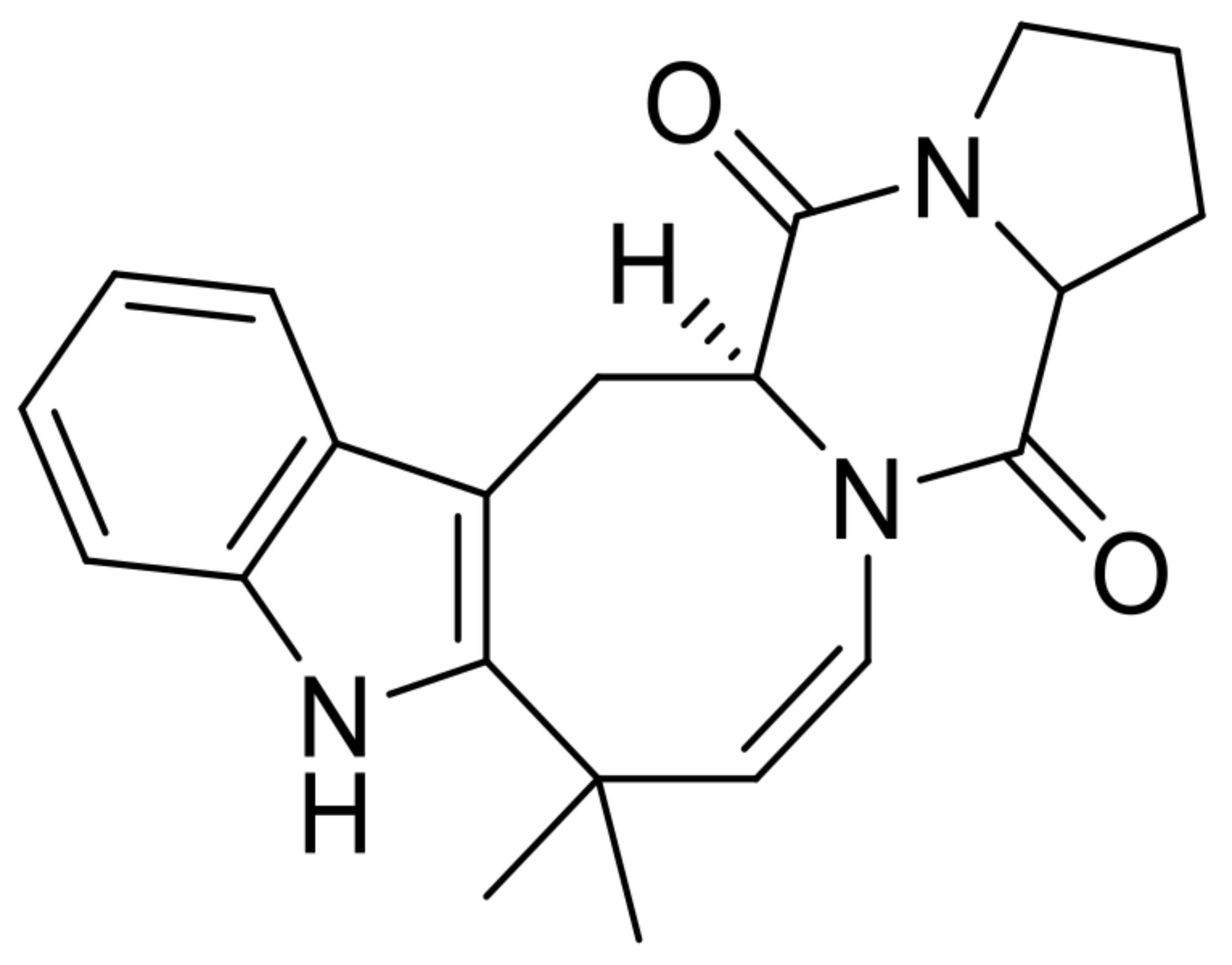 | 7.6 | 350.1870 [M+H]+ | 350.1863 | −2.0 | * | [8] |
| 11 | desoxybrevianamide E | 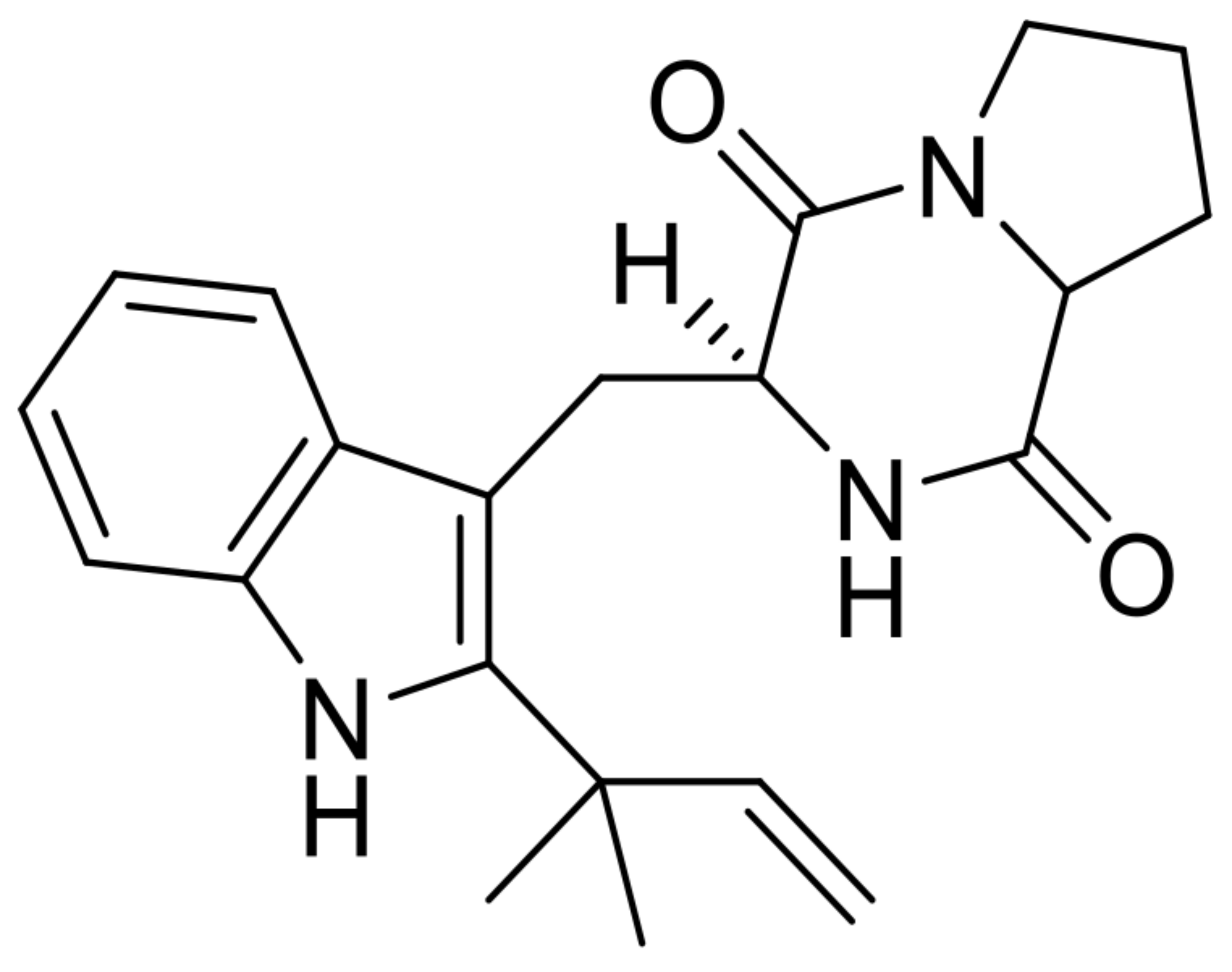 | 9.7 | 352.2010 [M+H]+ | 352.2020 | 2.7 | * | [8] |
| 12 | brevianamide F | 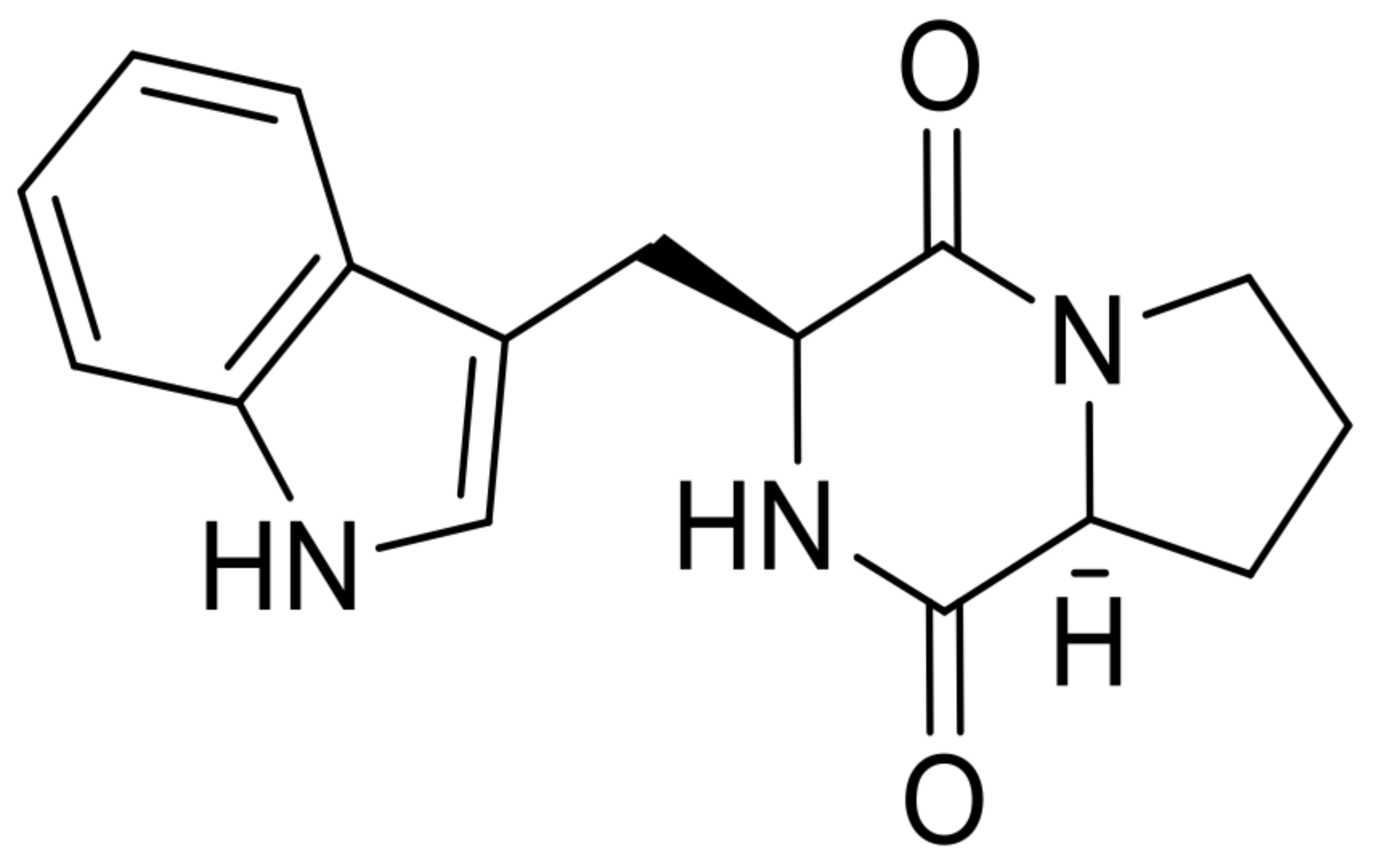 | 5.0 | 284.1386 [M+H]+ | 284.1394 | 2.7 | 0.98 | [47] |
| 13 | austamide |  | 5.2 | 364.1654 [M+H]+ | 364.1656 | 0.5 | 0.81 | [43] |
| 14 | emodine |  | 13.0 | 271.0597 [M+H]+ | 271.0601 | 1.5 | 0.94 | [14,48] |
| 15 | citreorosein |  | 8.7 | 287.0545 [M+H]+ | 287.0550 | 1.8 | 0.89 | [14,49] |
| 16 | 2-chlorocitreorosein | 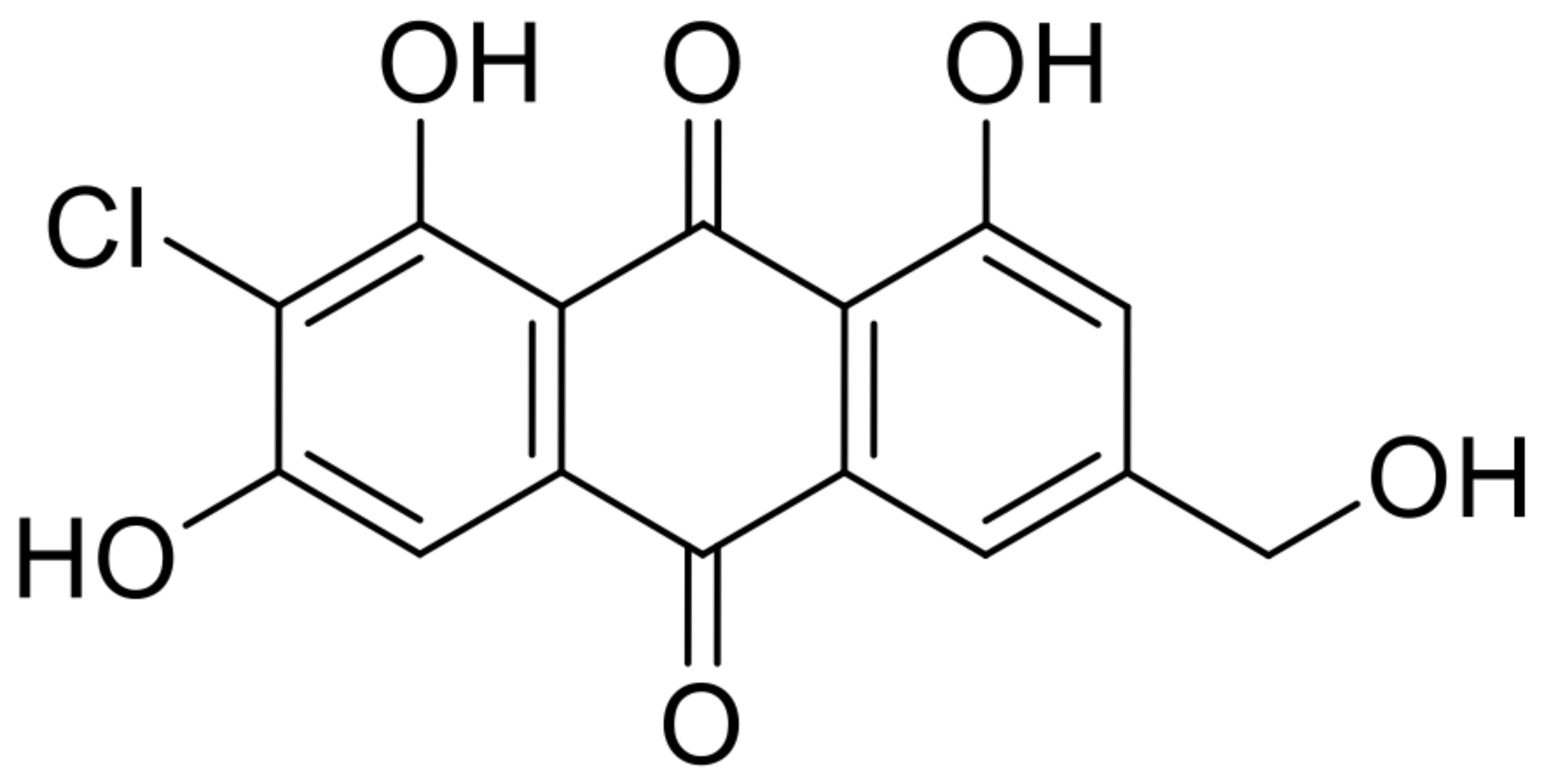 | 10.1 | 321.016 [M+H]+ | 321.0160 | 0.1 | 0.86 | [25] |
| 17 | endocrocin | 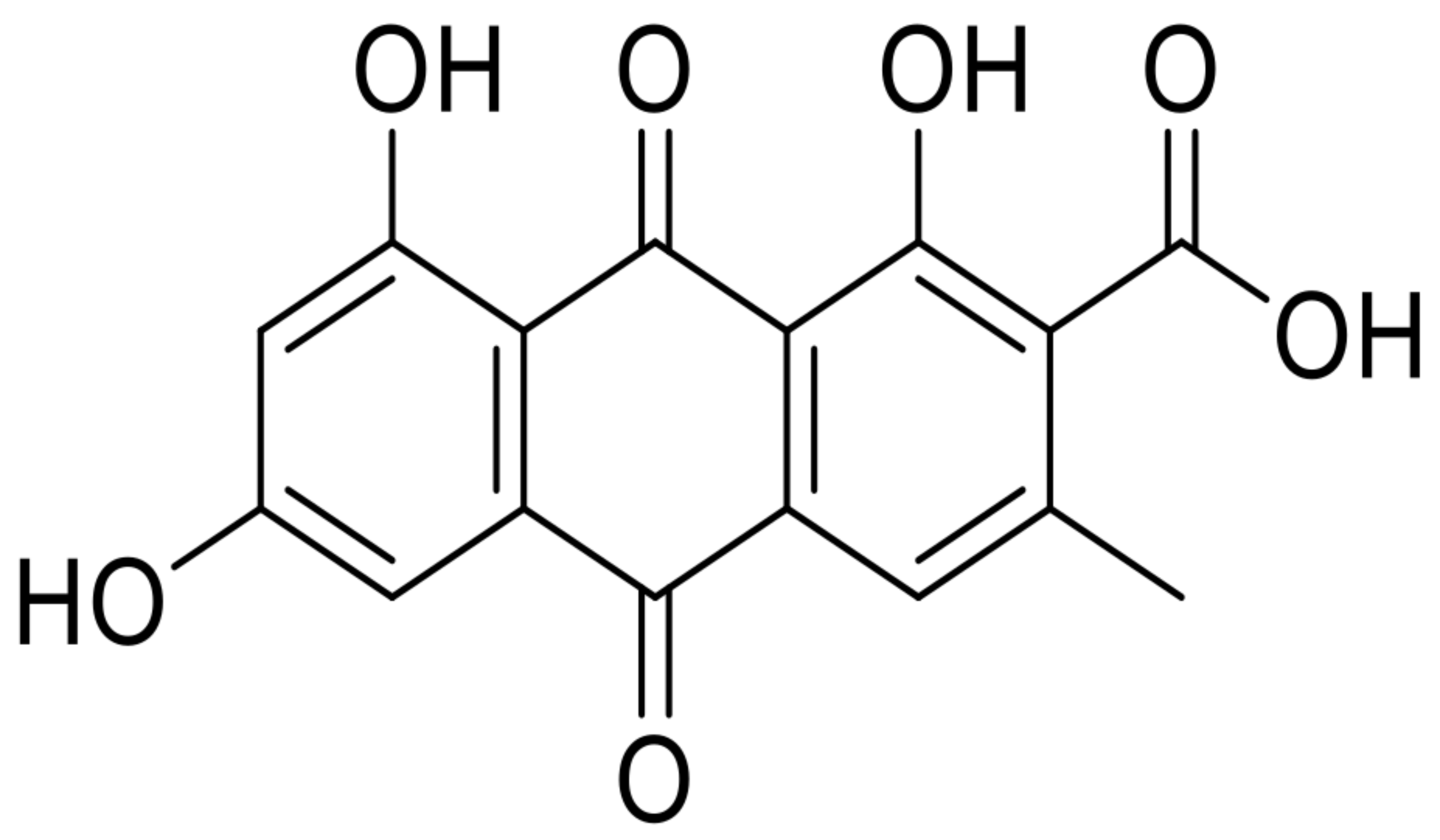 | 8.1 | 315.0490 [M+H]+ | 315.0499 | 2.9 | 0.96 | [14] |
| 18 | nephrolaevigatin A | 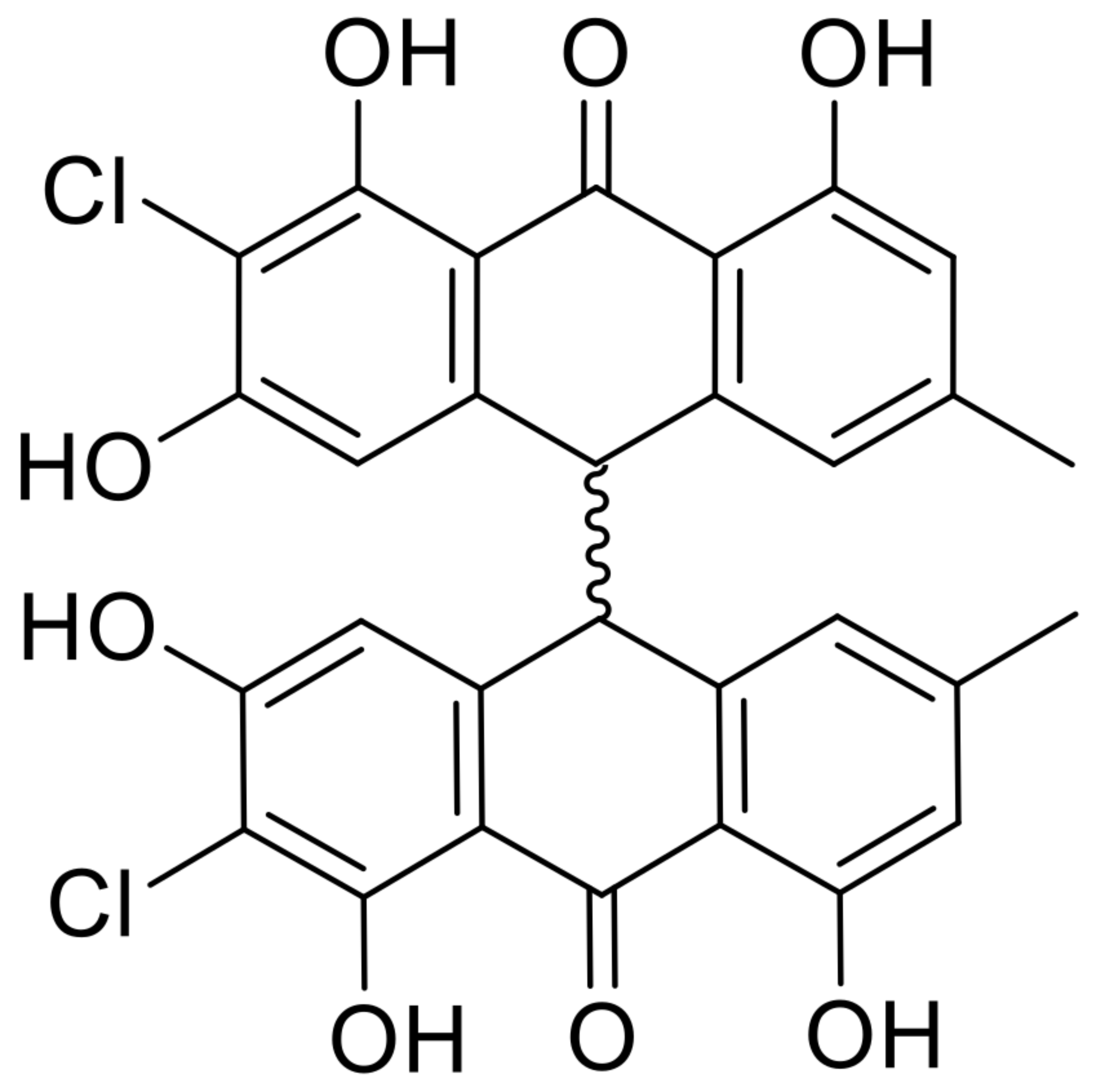 | 15.5 | 579.0609 [M+H]+ | 579.0608 | −0.2 | 0.81 | [27] |
| 19 | nephrolaevigatin B | 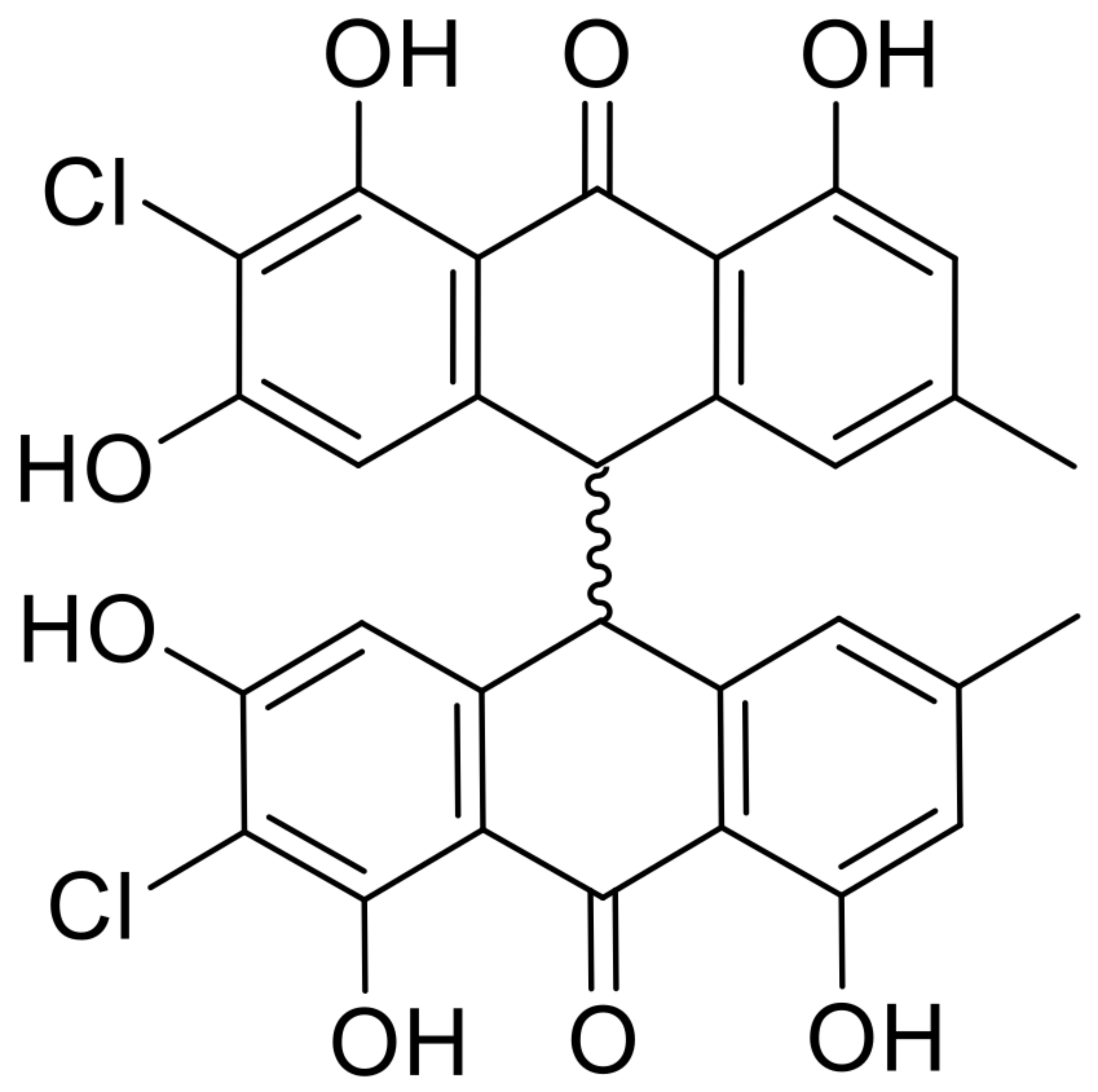 | 15.7 | 579.0614 [M+H]+ | 579.0608 | −1.04 | 0.81 | [27] |
| 20 | nephrolaevigatin C |  | 15.3 | 545.1005 [M+H]+ | 545.0998 | −1.3 | 0.89 | [27] |
| 21 | nephrolaevigatin D | 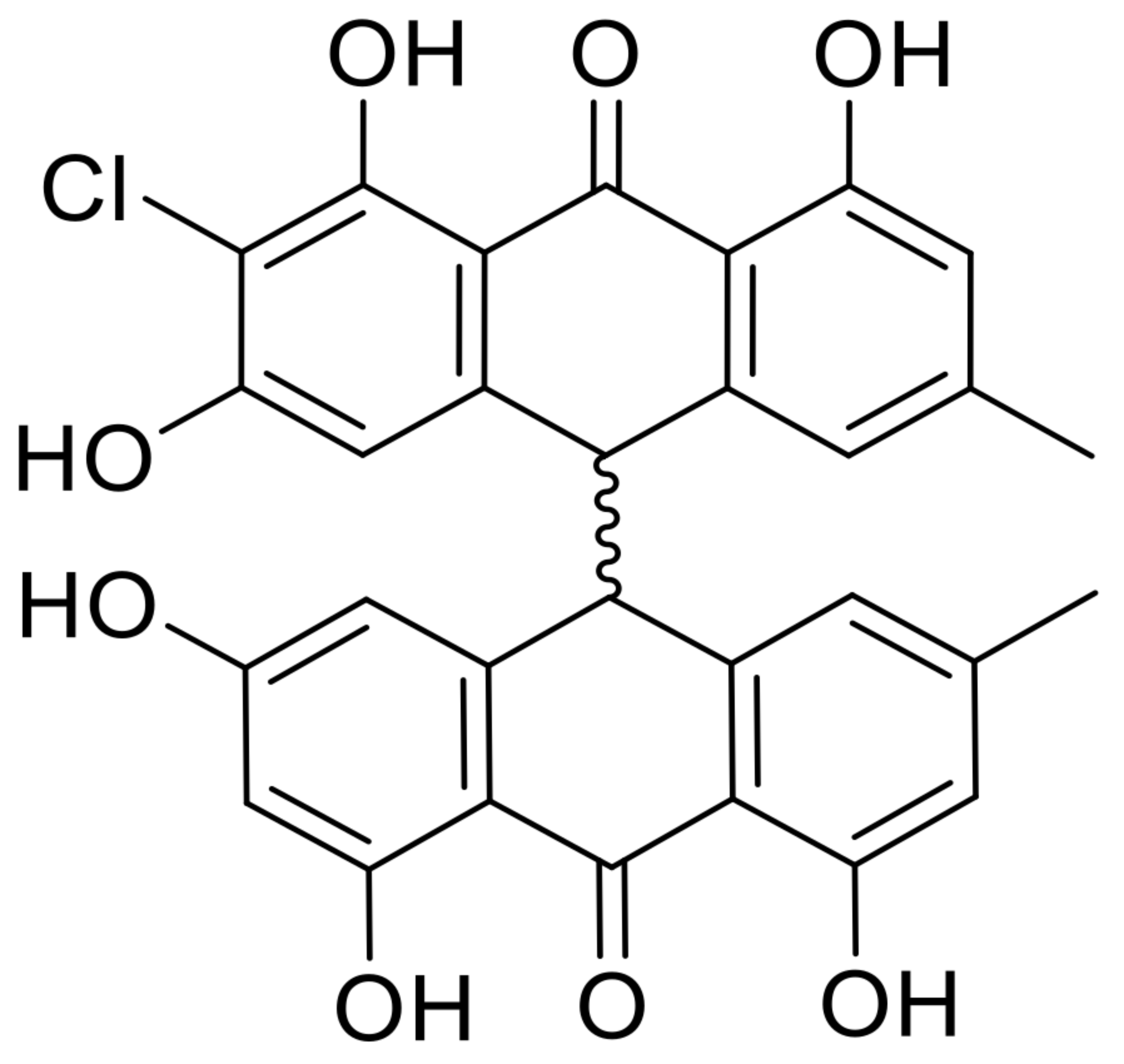 | 15.0 | 545.1012 [M+H]+ | 545.0998 | −2.6 | 0.89 | [27] |
| 22 | secalonic acid D | 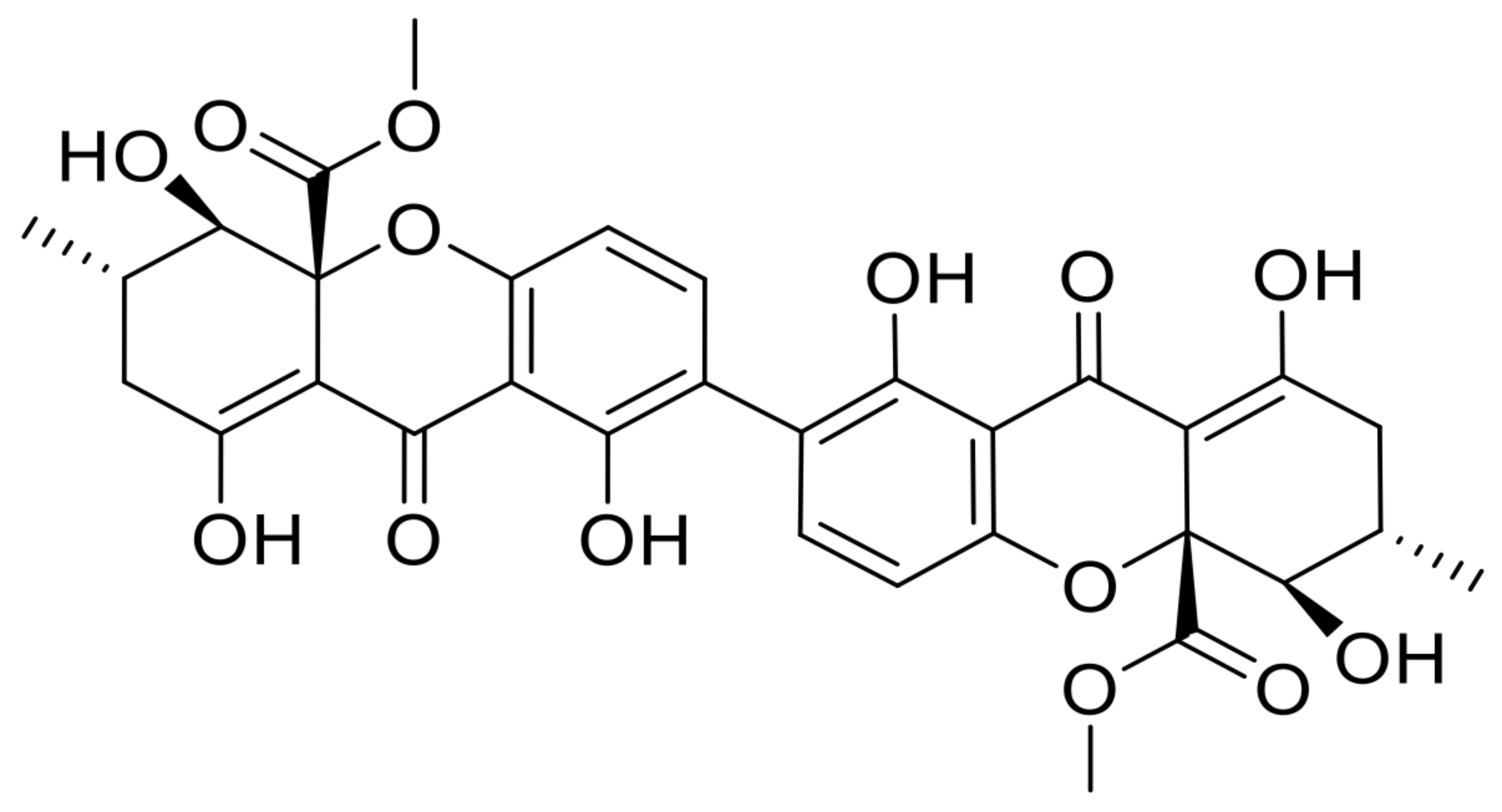 | 13.6 | 639.1747 [M+H]+ | 639.1708 | −6.1 | 0.82 | [50,51] |
| 23 | 3,4-dimethoxycinnamic acid | 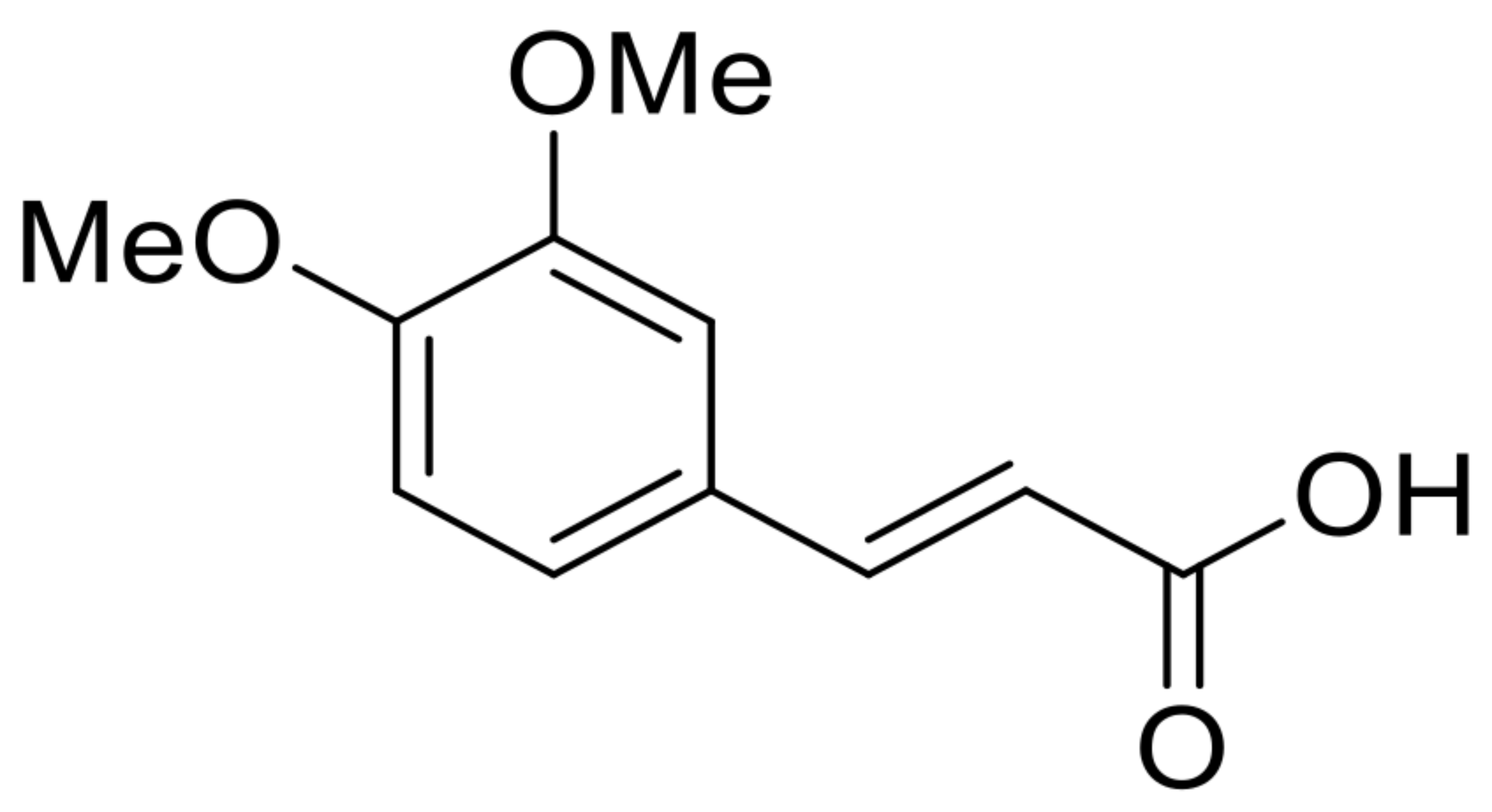 | 4.9 | 209.081 [M+H]+ | 209.0804 | −0.8 | 0.90 | [52] |
| 24 | 7-hydroxy-3-(2-hydroxypropyl)-5-methylisochromen-1-one |  | 5.1 | 235.096 [M+H]+ | 235.0965 | 2.1 | 0.86 | [53] |
| 25 | unidentified ergostane derivative | C28H42O2 | 19.9 | 411.324 [M+H]+ | 411.3258 | 4.3 |
References
- Dewapriya, P.; Kim, S.K. Marine microorganisms: An emerging avenue in modern nutraceuticals and functional foods. Food Res. Int. 2014, 56, 115–125. [Google Scholar] [CrossRef]
- Takahashi, J.A.; Barbosa, B.V.R.; Martins, B.A.; Guirlanda, C.P.; Moura, M.A.F. Use of the versatility of fungal metabolism to meet modern demands for healthy aging, functional foods, and sustainability. J. Fungi 2020, 6, 223. [Google Scholar] [CrossRef] [PubMed]
- Pinedo-Rivilla, C.; Aleu, J.; Durán-Patrón, R. Cryptic Metabolites from Marine-Derived Microorganisms Using OSMAC and Epigenetic Approaches. Mar. Drugs 2022, 20, 84. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Petersen, L.E.; Marner, M.; Labes, A.; Tasdemir, D. Rapid metabolome and bioactivity profiling of fungi associated with the leaf and rhizosphere of the baltic seagrass zostera marina. Mar. Drugs 2019, 17, 419. [Google Scholar] [CrossRef] [PubMed]
- Overy, D.; Correa, H.; Roullier, C.; Chi, W.C.; Pang, K.L.; Rateb, M.; Ebel, R.; Shang, Z.; Capon, R.; Bills, G.; et al. Does Osmotic Stress Affect Natural Product Expression in Fungi? Mar. Drugs 2017, 15, 254. [Google Scholar] [CrossRef] [PubMed]
- Igboeli, H.A.; Marchbank, D.H.; Correa, H.; Overy, D.; Kerr, R.G. Discovery of Primarolides A and B from Marine Fungus Asteromyces cruciatus Using Osmotic Stress and Treatment with Suberoylanilide Hydroxamic Acid. Mar. Drugs 2019, 17, 435. [Google Scholar] [CrossRef]
- Zhuravleva, O.I.; Antonov, A.S.; Trang, V.T.D.; Pivkin, M.V.; Khudyakova, Y.V.; Denisenko, V.A.; Popov, R.S.; Kim, N.Y.; Yurchenko, E.A.; Gerasimenko, A.V.; et al. New Deoxyisoaustamide Derivatives from the Coral-Derived Fungus Penicillium dimorphosporum KMM 4689. Mar. Drugs 2021, 19, 32. [Google Scholar] [CrossRef]
- Dyshlovoy, S.A.; Zhuravleva, O.I.; Hauschild, J.; Busenbender, T.; Pelageev, D.N.; Yurchenko, A.N.; Khudyakova, Y.V.; Antonov, A.S.; Graefen, M.; Bokemeyer, C.; et al. New Marine Fungal Deoxy-14,15-Dehydroisoaustamide Resensitizes Prostate Cancer Cells to Enzalutamide. Mar. Drugs 2023, 21, 54. [Google Scholar] [CrossRef]
- Chambers, M.C.; MacLean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Myers, O.D.; Sumner, S.J.; Li, S.; Barnes, S.; Du, X. One Step Forward for Reducing False Positive and False Negative Compound Identifications from Mass Spectrometry Metabolomics Data: New Algorithms for Constructing Extracted Ion Chromatograms and Detecting Chromatographic Peaks. Anal. Chem. 2017, 89, 8696–8703. [Google Scholar] [CrossRef] [PubMed]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, J.V.; Larsen, T.O.; Frisvad, J.C. Production of Fungal Quinones: Problems and Prospects. Biomolecules 2022, 12, 1041. [Google Scholar] [CrossRef]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F.; et al. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar] [CrossRef]
- Bedre, R. Reneshbedre/Bioinfokit: Bioinformatics Data Analysis and Visualization Toolkit, 2.0.9, Zenodo, 2022.
- Scholin, C.A.; Herzog, M.; Sogin, M.; Anderson, D.M. Identification of group- and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). II. Sequence analysis of a fragment of the LSU rRNA gene. J. Phycol. 1994, 30, 999–1011. [Google Scholar] [CrossRef]
- Elwood, H.J.; Olsen, G.J.; Sogin, M.L. The small-subunit ribosomal RNA gene sequences from the hypotrichous ciliates Oxytricha nova and Stylonychia pustulata. Mol. Biol. Evol. 1985, 2, 399–410. [Google Scholar] [PubMed]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Monisha, B.A.; Kumar, N.; Tiku, A.B. Emodin and its role in chronic diseases. Adv. Exp. Med. Biol. 2016, 928, 47–73. [Google Scholar] [PubMed]
- Graf, T.N.; Kao, D.; Rivera-Chávez, J.; Gallagher, J.M.; Raja, H.A.; Oberlies, N.H. Drug leads from endophytic fungi: Lessons learned via scaled production. Planta. Med. 2020, 86, 988–996. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Kiriyama, N.; Arahata, S. Studies on the Metabolic Products of Aspergillus fumigatus (J-4). Chemical Structure of Metabolic Products. Chem. Pharm. Bull. 1968, 16, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Kurobane, I.; Vining, L.C.; Gavin McInnes, A. Biosynthetic relationships among the secalonic acids isolation of emodin, endocrocin and secalonic acids from pyrenochaeta terrestris and aspergillus aculeatus. J. Antibiot. 1979, 32, 1256–1266. [Google Scholar] [CrossRef]
- Lagarde, A.; Mambu, L.; Mai, P.Y.; Champavier, Y.; Stigliani, J.L.; Beniddir, M.A.; Millot, M. Chlorinated bianthrones from the cyanolichen Nephroma laevigatum. Fitoterapia 2021, 149, 104811. [Google Scholar] [CrossRef]
- Wang, B.H.; Polya, G.M. The fungal teratogen secalonic acid D is an inhibitor of protein kinase C and of cyclic AMP-dependent protein kinase. Planta. Med. 1996, 62, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Swart, H.J. Penicillium dimorphosporum sp. nov. Trans. Br. Mycol. Soc. 1970, 55, 310–313. [Google Scholar] [CrossRef]
- Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Hong, S.B.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Varga, J.; Yaguchi, T.; Samson, R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef]
- Houbraken, J.; Kocsube, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R.A.; et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef]
- Hensens, O.D.; Wichmann, C.F.; Liesch, J.M.; Vanmiddlesworth, F.L.; Wilson, K.E.; Schwartz, R.E. Structure elucidation of restricticin, a novel antifungal agent from Penicillium restrictum. Tetrahedron 1991, 47, 3915–3924. [Google Scholar] [CrossRef]
- Brill, G.M.; Chen, R.H.; Rasmussen, R.R.; Whittern, D.N.; McAlpine, J.B. Calbistrins, novel antifungal agents produced by Penicillium restrictum. 2. Isolation and elucidation of structure. J. Antibiot. 1993, 46, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.; Capon, R.J.; Lacey, E.; Tennant, S.; Gill, J.H. Calbistrin E and two other new metabolites from an Australian isolate of Penicillium striatisporum. J. Nat. Prod. 2005, 68, 581–584. [Google Scholar] [CrossRef]
- Chen, X.-H.; Zhou, G.-L.; Sun, C.-X.; Zhang, X.-M.; Zhang, G.-J.; Zhu, T.-J.; Li, J.; Che, Q.; Li, D.-H. Penicacids E–G, three new mycophenolic acid derivatives from the marine-derived fungus Penicillium parvum HDN17-478. Chin. J. Nat. Med. 2020, 18, 850–854. [Google Scholar] [CrossRef] [PubMed]
- Kumla, D.; Pereira, J.A.; Dethoup, T.; Gales, L.; Freitas-Silva, J.; Costa, P.M.; Lee, M.; Silva, A.M.S.; Sekeroglu, N.; Pinto, M.M.M.; et al. Chromone derivatives and other constituents from cultures of the marine sponge-associated fungus Penicillium erubescens KUFA0220 and their antibacterial activity. Mar. Drugs 2018, 16, 289. [Google Scholar] [CrossRef]
- Asiri, I.A.M.; Badr, J.M.; Youssef, D.T.A. Penicillivinacine, antimigratory diketopiperazine alkaloid from the marine-derived fungus Penicillium vinaceum. Phytochem. Lett. 2015, 13, 53–58. [Google Scholar] [CrossRef]
- Xian, P.J.; Chen, H.Y.; Feng, Z.; Zhao, W.; Yang, X.L. Capsulactone: A new 4-hydroxy-α-pyrone derivative from an endophytic fungus Penicillium capsulatum and its antimicrobial activity. J. Asian Nat. Prod. Res. 2021, 23, 1100–1106. [Google Scholar] [CrossRef] [PubMed]
- Betina, V.; Nemec, P.; Dobias, J.; Baráth, Z. Cyanein, a New Antibiotic from Penicillium cyaneum. Folia Microbiol. 1962, 7, 353–357. [Google Scholar] [CrossRef]
- Sebak, M.; Molham, F.; Greco, C.; Tammam, M.A.; Sobeh, M.; El-Demerdash, A. Chemical diversity, medicinal potentialities, biosynthesis, and pharmacokinetics of anthraquinones and their congeners derived from marine fungi: A comprehensive update. RSC Adv. 2022, 12, 24887–24921. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Miyazawa, M. Synthesis and structure–activity relationships of phenylpropanoid amides of serotonin on tyrosinase inhibition. Bioorg. Med. Chem. Lett. 2011, 21, 1983–1986. [Google Scholar] [CrossRef]
- Choi, D.Y.; Choi, H. Natural products from marine organisms with neuroprotective activity in the experimental models of Alzheimer’s disease, Parkinson’s disease and ischemic brain stroke: Their molecular targets and action mechanisms. Arch. Pharm. Res. 2015, 38, 139–170. [Google Scholar] [CrossRef]
- Steyn, P.S. Austamide, a new toxic metabolite from Aspergillus ustus. Tetrahedron Lett. 1971, 12, 3331–3334. [Google Scholar] [CrossRef]
- Su, Y.T.; Chang, H.L.; Shyue, S.K.; Hsu, S.L. Emodin induces apoptosis in human lung adenocarcinoma cells through a reactive oxygen species-dependent mitochondrial signaling pathway. Biochem. Pharmacol. 2005, 70, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Fredenhagen, A.; Mett, H.; Meyer, T.; Buchdunger, E.; Regenass, U.; Roggo, B.E.; Petersen, F. Protein Tyrosine Kinase and Protein Kinase C Inhibition by Fungal Anthraquinones Related to Emodin. J. Antibiot. 1995, 48, 1355–1358. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Tao, L.Y.; Liang, Y.J.; Yan, Y.Y.; Dai, C.L.; Xia, X.K.; She, Z.G.; Lin, Y.C.; Fu, L.W. Secalonic acid D induced leukemia cell apoptosis and cell cycle arrest of G1 with involvement of GSK-3β/β-catenin/c-Myc pathway. Cell Cycle 2009, 8, 2444–2450. [Google Scholar] [CrossRef]
- Zhang, D.; Noviendri, D.; Nursid, M.; Yang, X.; Byeng, W.S. 12,13-Dihydroxyfumitremorgin C, fumitremorgin C, and brevianamide F, antibacterial diketopiperazine alkaloids from the marine-derived fungus Pseudallescheria sp. Nat. Prod. Sci. 2007, 13, 251–254. [Google Scholar]
- Zhang, F.; Chen, W.; Sun, L. LC-VWD-MS determination of three anthraquinones and one stilbene in the quality control of crude and prepared roots of Polygonum multiflorum Thunb. Chromatographia 2008, 67, 869–874. [Google Scholar] [CrossRef]
- Piattelli, M.; Giudici de Nicola, M. Anthraquinone pigments from Xanthoria parientina (L.). Phytochemistry 1968, 7, 1183–1187. [Google Scholar] [CrossRef]
- Steyn, P.S. The isolation, structure and absolute configuration of secalonic acid D, the toxic metabolite of Penicillium oxalicum. Tetrahedron 1970, 26, 51–57. [Google Scholar] [CrossRef]
- Santini, A.; Mikušová, P.; Sulyok, M.; Krska, R.; Labuda, R.; Šrobárová, A. Penicillium strains isolated from Slovak grape berries taxonomy assessment by secondary metabolite profile. Mycotoxin Res. 2014, 30, 213–220. [Google Scholar] [CrossRef]
- Kovács, Z.; Dinya, Z.; Antus, S. LC-SSI-MS Techniques as Efficient Tools for Characterization of Nonvolatile Phenolic Compounds of a Special Hungarian Wine. J. Chromatogr. Sci. 2004, 42, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.X.; Bao, L.; Yang, X.L.; Guo, H.; Yang, R.N.; Ren, B.; Zhang, L.X.; Dai, H.Q.; Guo, L.D.; Liu, H.W. Polyketides with antimicrobial activity from the solid culture of an endolichenic fungus Ulocladium sp. Fitoterapia 2012, 83, 209–214. [Google Scholar] [CrossRef] [PubMed]
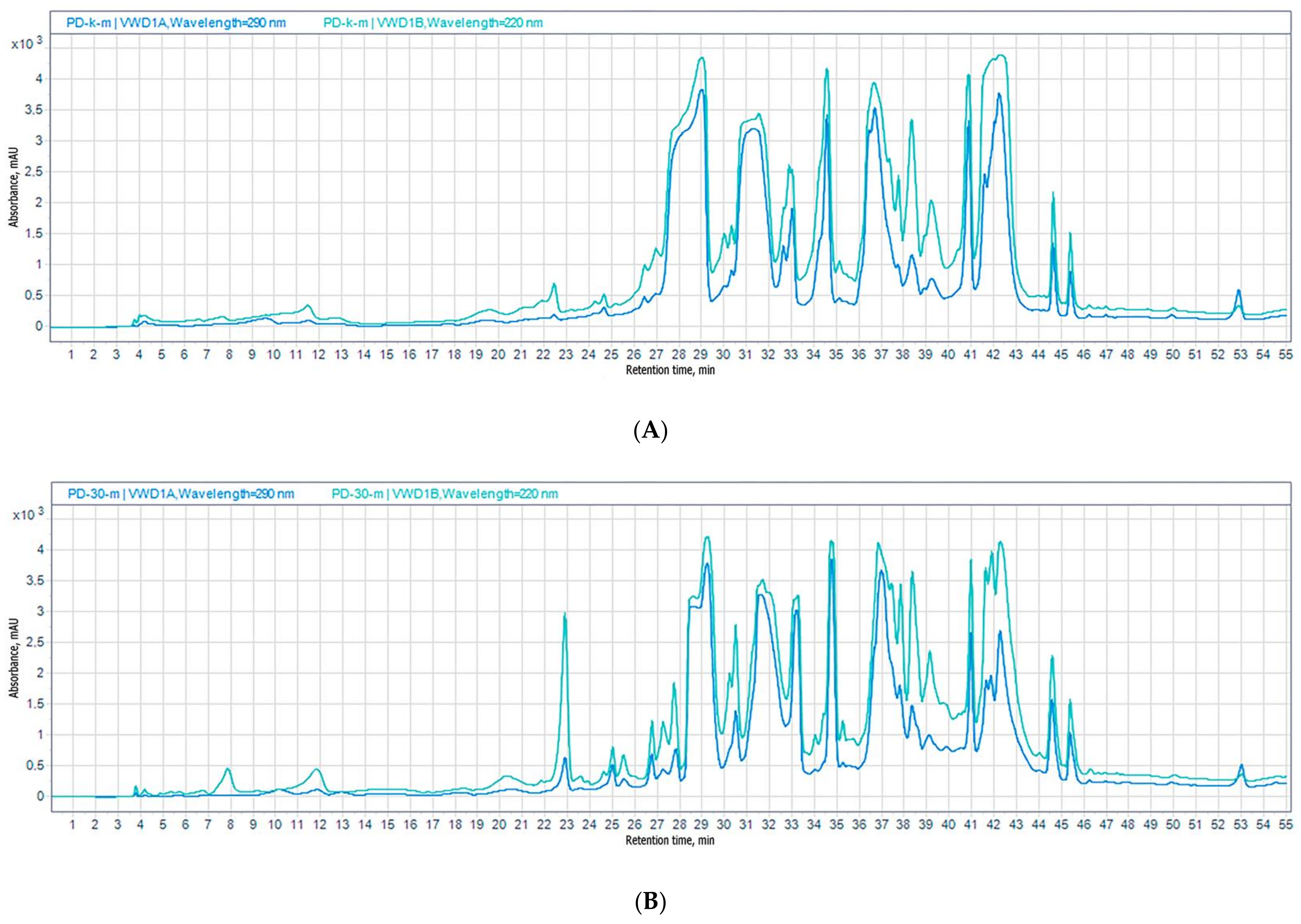
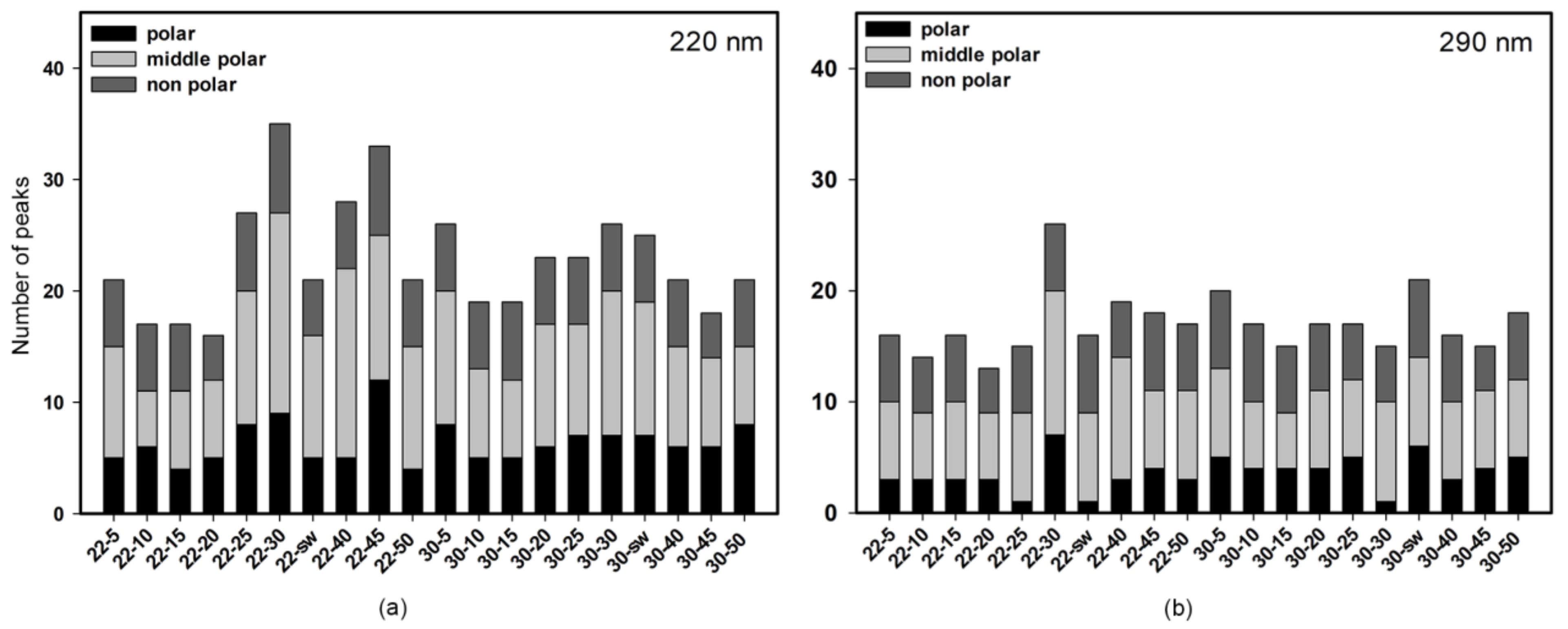




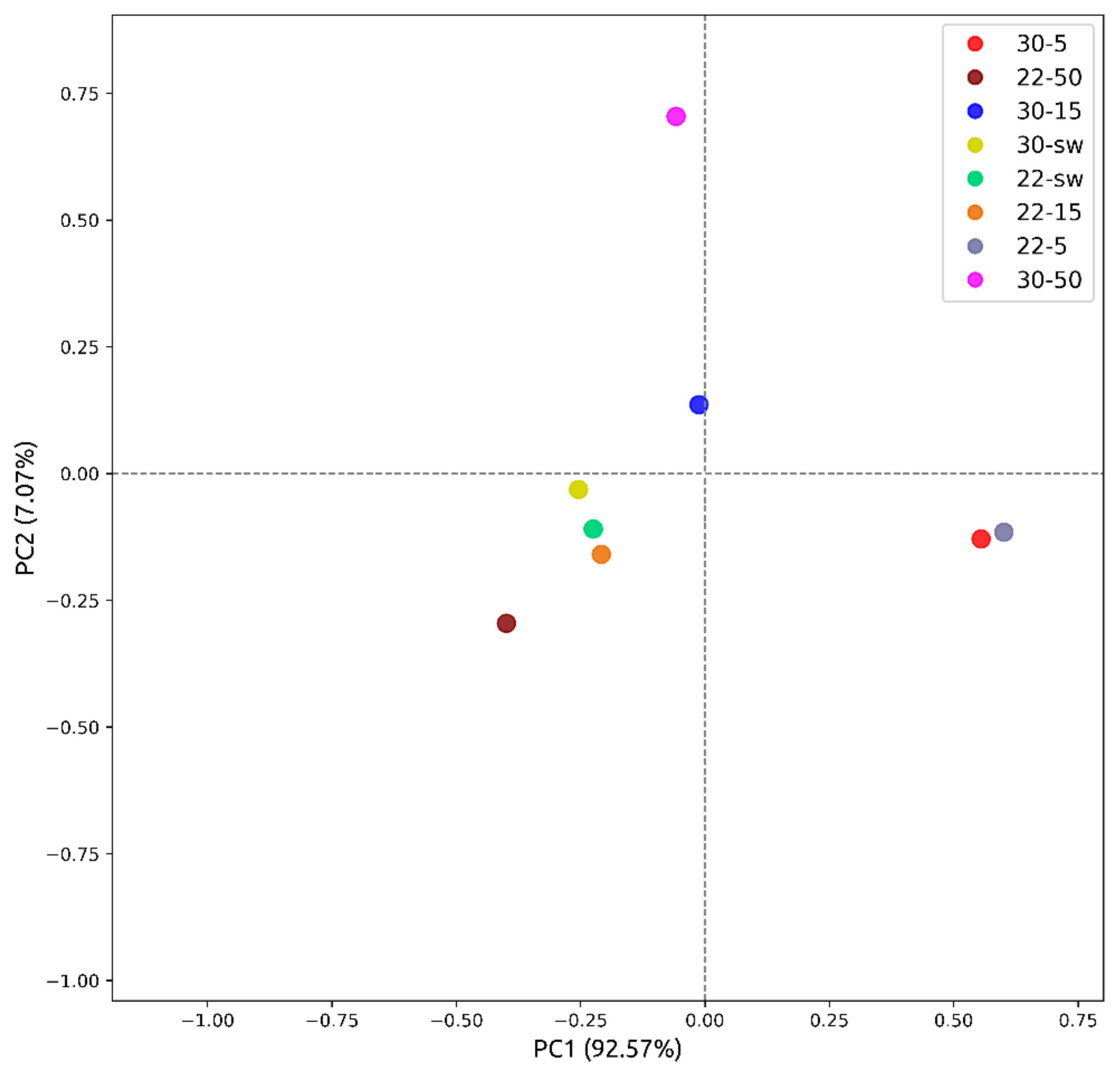

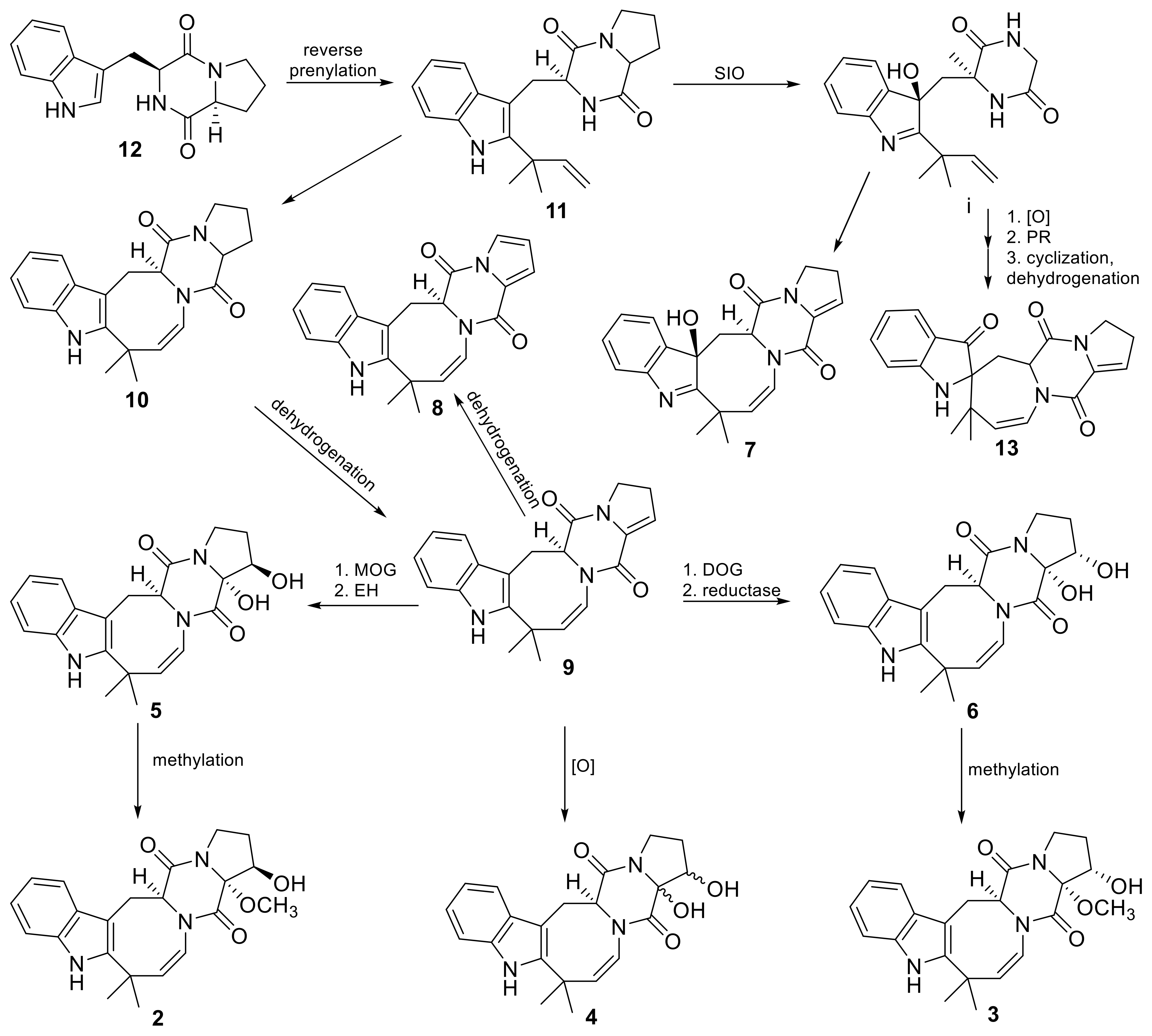
| No | Cultivation Conditions | Sample Code | EtOAc Extract Amount, mg | Purified Extract Amount, mg |
|---|---|---|---|---|
| 1 | 22 °C, sea water | 22-sw | 452.3 | 178.1 |
| 2 | 22 °C, sea salt (5 mg/mL) | 22-5 | 536.9 | 211.4 |
| 3 | 22 °C, sea salt (10 mg/mL) | 22-10 | 246.2 | 96.6 |
| 4 | 22 °C, sea salt (15 mg/mL) | 22-15 | 259.5 | 34.8 |
| 5 | 22 °C, sea salt (20 mg/mL) | 22-20 | 374.3 | 47.0 |
| 6 | 22 °C, sea salt (25 mg/mL) | 22-25 | 344.3 | 32.3 |
| 7 | 22 °C, sea salt (30 mg/mL) | 22-30 | 343.6 | 23.3 |
| 8 | 22 °C, sea salt (40 mg/mL) | 22-40 | 294.5 | 31.7 |
| 9 | 22 °C, sea salt (45 mg/mL) | 22-45 | 151.8 | 22.2 |
| 10 | 22 °C, sea salt (50 mg/mL) | 22-50 | 391.1 | 50.6 |
| 11 | 30 °C, sea water | 30-sw | 409.4 | 161.2 |
| 12 | 30 °C, sea salt (5 mg/mL) | 30-5 | 415.8 | 163.7 |
| 13 | 30 °C, sea salt (10 mg/mL) | 30-10 | 294.3 | 114.4 |
| 14 | 30 °C, sea salt (15 mg/mL) | 30-15 | 408.4 | 53.6 |
| 15 | 30 °C, sea salt (20 mg/mL) | 30-20 | 337.5 | 44.3 |
| 16 | 30 °C, sea salt (25 mg/mL) | 30-25 | 311.5 | 47.8 |
| 17 | 30 °C, sea salt (30 mg/mL) | 30-30 | 358.0 | 32.8 |
| 18 | 30 °C, sea salt (40 mg/mL) | 30-40 | 268.1 | 30.5 |
| 19 | 30 °C, sea salt (45 mg/mL) | 30-45 | 155.2 | 22.7 |
| 20 | 30 °C, sea salt (50 mg/mL) | 30-50 | 142.1 | 29.4 |
| Taxon | Collection Number | GenBank Accession Number | |
|---|---|---|---|
| ITS | β-Tubulin | ||
| Penicillium canis S.W. Peterson | NRRL 62,798 T | KJ511291 | |
| Penicillium capsulatum Raper et Fennell | CBS 301.48 T | AF033429 | MN969375 |
| Penicillium catenatum D.B. Scott | CBS 352.67 T | KC411754 | |
| Penicillium cyaneum (Bainier et Sartory) Biourge | CBS 315.48 T | AF033427 | JX091552 |
| Penicillium dimorphosporum H.J. Swart | CBS 456.70 T | AF081804 | |
| Penicillium erubescens D.B. Scott | CBS 318.67 T | AF033464 | |
| Penicillium georgiense S.W. Peterson et B.W. Horn | CBS 132826 T | EF422852 | EF506223 |
| Penicillium guttulosum J.C. Gilman et E.V. Abbott | NRRL 907 T | HQ646592 | |
| Penicillium hermansii Houbraken, Seifert et Samson | CBS 124,296 T | MG333472 | |
| Penicillium hispanicum C. Ramírez, A.T. Martínez et Ferrer | KMM 4689 CBS 691.77 T | MW325972 JX841247 | OP407668 KJ834456 |
| Penicillium laeve (K. Ando et Manoch) Houbraken et Samson | CBS 136,665 T | KF667369 | |
| Penicillium menonorum S.W. Peterson | NRRL 50,410 T | HQ646591 | |
| Penicillium nepalense Takada et Udagawa | CBS 203.84 T | KC411692 | |
| Penicillium ornatum Udagawa | CBS 190.68 T | KC411687 | KJ834479 |
| Penicillium ovatum (K. Ando et Nawawi) Houbraken et Samson | CBS 136,664 T | KF667370 | |
| Penicillium parvofructum Guevara-Suarez, Cano-Canals, Cano et Stchigel | CBS 141,690 T | AF033460 | |
| Penicillium parvum Raper et Fennell | CBS 359.48 T | AF033460 | |
| Penicillium pimiteouiense S.W. Peterson | CBS 102,479 T | AF037431 | |
| Penicillium ramusculum Bat. et H. Maia | CBS 251.56 T | EF433765 | EU427269 |
| Penicillium rubidurum Udagawa et Y. Horie | CBS 609.73 T | AF033462 | |
| Penicillium striatisporum Stolk | CBS 705.68 T | AF038938 | |
| Penicillium vinaceum J.C. Gilman et E.V. Abbott | CBS 389.48 T | AF033461 | |
| Talaromyces marneffei (Segretain, Capponi et Sureau) Samson, N. Yilmaz, Frisvad et Seifert | CBS 388.87 T | JN899344 | JX091389 |
| Extract Code | Cell Viability 1, % of Control | DPPH Radicals 2, % | |
|---|---|---|---|
| PC-3 | H9c2 | ||
| 22-5 | 85.4 ± 0.8 | 93.5 ± 3.8 | 19.9 ± 0.9 |
| 22-10 | 56.6 ± 7.6 | 70.1 ± 10.2 | 20.4 ± 2.3 |
| 22-15 | 56.8 ± 9.6 | 46.6 ± 0.7 | 25.8 ± 1.8 |
| 22-20 | 64.8 ± 6.5 | 67.2 ± 5.6 | 26.9 ± 4.1 |
| 22-25 | 46.0 ± 8.1 | 67.1 ± 8.6 | 29.5 ± 3.1 |
| 22-30 | 56.4 ± 4.7 | 57.9 ± 4.1 | 40.7 ± 8.0 |
| 22-sw | 44.2 ± 6.6 | 53.2 ± 2.2 | 39.6 ± 2.5 |
| 22-40 | 60.3 ± 4.4 | 57.9 ± 6.2 | 28.4 ± 2.4 |
| 22-45 | 68.3 ± 5.0 | 64.8 ± 4.7 | 35.3 ± 5.0 |
| 22-50 | 27.7 ± 4.0 | 64.4 ± 4.1 | 19.1 ± 1.8 |
| 30-5 | 67.6 ± 9.5 | 70.5 ± 3.1 | 16.2 ± 4.0 |
| 30-10 | 62.6 ± 6.4 | 65.4 ± 2.1 | 23.1 ± 2.4 |
| 30-15 | 70.2 ± 2.6 | 67.0 ± 2.5 | 40.1 ± 4.6 |
| 30-20 | 77.4 ± 12.2 | 70.9 ± 4.4 | 29.1 ± 2.2 |
| 30-25 | 74.4 ± 5.4 | 75.1 ± 0.8 | 31.9 ± 1.9 |
| 30-30 | 48.1 ± 6.6 | 73.5 ± 5.2 | 33.1 ± 5.2 |
| 30-sw | 70.9 ± 1.2 | 62.7 ± 3.6 | 21.4 ± 5.2 |
| 30-40 | 50.5 ± 10.7 | 82.3 ± 6.4 | 31.0 ± 8.3 |
| 30-45 | 53.4 ± 1.0 | 69.8 ± 1.0 | 36.9 ± 9.3 |
| 30-50 | 86.9 ± 2.8 | 72.2 ± 2.2 | 42.9 ± 3.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nesterenko, L.E.; Popov, R.S.; Zhuravleva, O.I.; Kirichuk, N.N.; Chausova, V.E.; Krasnov, K.S.; Pivkin, M.V.; Yurchenko, E.A.; Isaeva, M.P.; Yurchenko, A.N. A Study of the Metabolic Profiles of Penicillium dimorphosporum KMM 4689 Which Led to Its Re-Identification as Penicillium hispanicum. Fermentation 2023, 9, 337. https://doi.org/10.3390/fermentation9040337
Nesterenko LE, Popov RS, Zhuravleva OI, Kirichuk NN, Chausova VE, Krasnov KS, Pivkin MV, Yurchenko EA, Isaeva MP, Yurchenko AN. A Study of the Metabolic Profiles of Penicillium dimorphosporum KMM 4689 Which Led to Its Re-Identification as Penicillium hispanicum. Fermentation. 2023; 9(4):337. https://doi.org/10.3390/fermentation9040337
Chicago/Turabian StyleNesterenko, Liliana E., Roman S. Popov, Olesya I. Zhuravleva, Natalya N. Kirichuk, Viktoria E. Chausova, Kirill S. Krasnov, Mikhail V. Pivkin, Ekaterina A. Yurchenko, Marina P. Isaeva, and Anton N. Yurchenko. 2023. "A Study of the Metabolic Profiles of Penicillium dimorphosporum KMM 4689 Which Led to Its Re-Identification as Penicillium hispanicum" Fermentation 9, no. 4: 337. https://doi.org/10.3390/fermentation9040337
APA StyleNesterenko, L. E., Popov, R. S., Zhuravleva, O. I., Kirichuk, N. N., Chausova, V. E., Krasnov, K. S., Pivkin, M. V., Yurchenko, E. A., Isaeva, M. P., & Yurchenko, A. N. (2023). A Study of the Metabolic Profiles of Penicillium dimorphosporum KMM 4689 Which Led to Its Re-Identification as Penicillium hispanicum. Fermentation, 9(4), 337. https://doi.org/10.3390/fermentation9040337








