Isolation and Characterization of Lignocellulolytic Bacteria from Municipal Solid Waste Landfill for Identification of Potential Hydrolytic Enzyme
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Physico-Chemical Characterization of Waste Samples
2.2. Isolation and Identification of Bacteria
2.2.1. Isolation of Potential Bacteria
2.2.2. Preliminary Identification of Potential Bacteria
2.3. Screening for Ligninolytic and Cellulolytic Ability
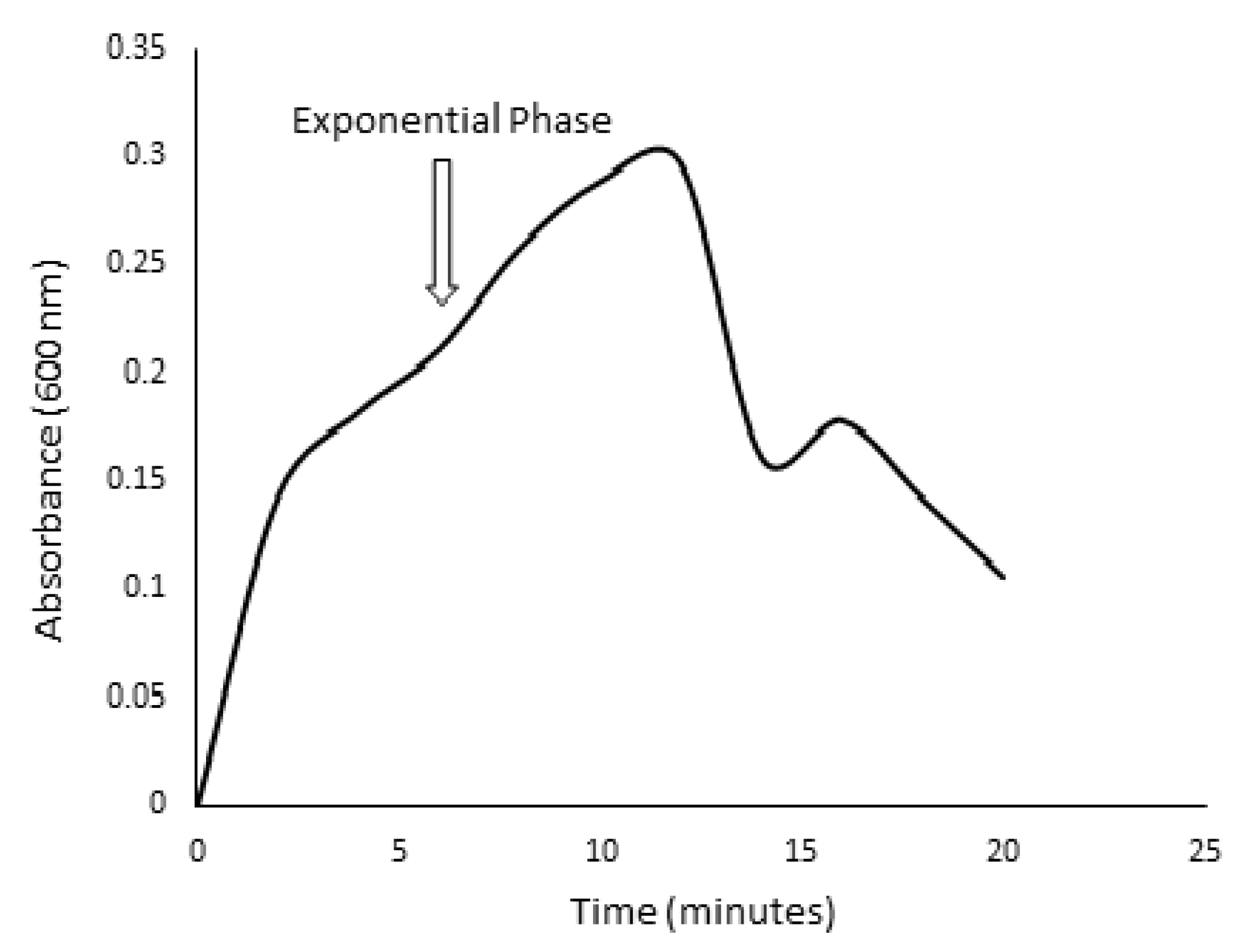
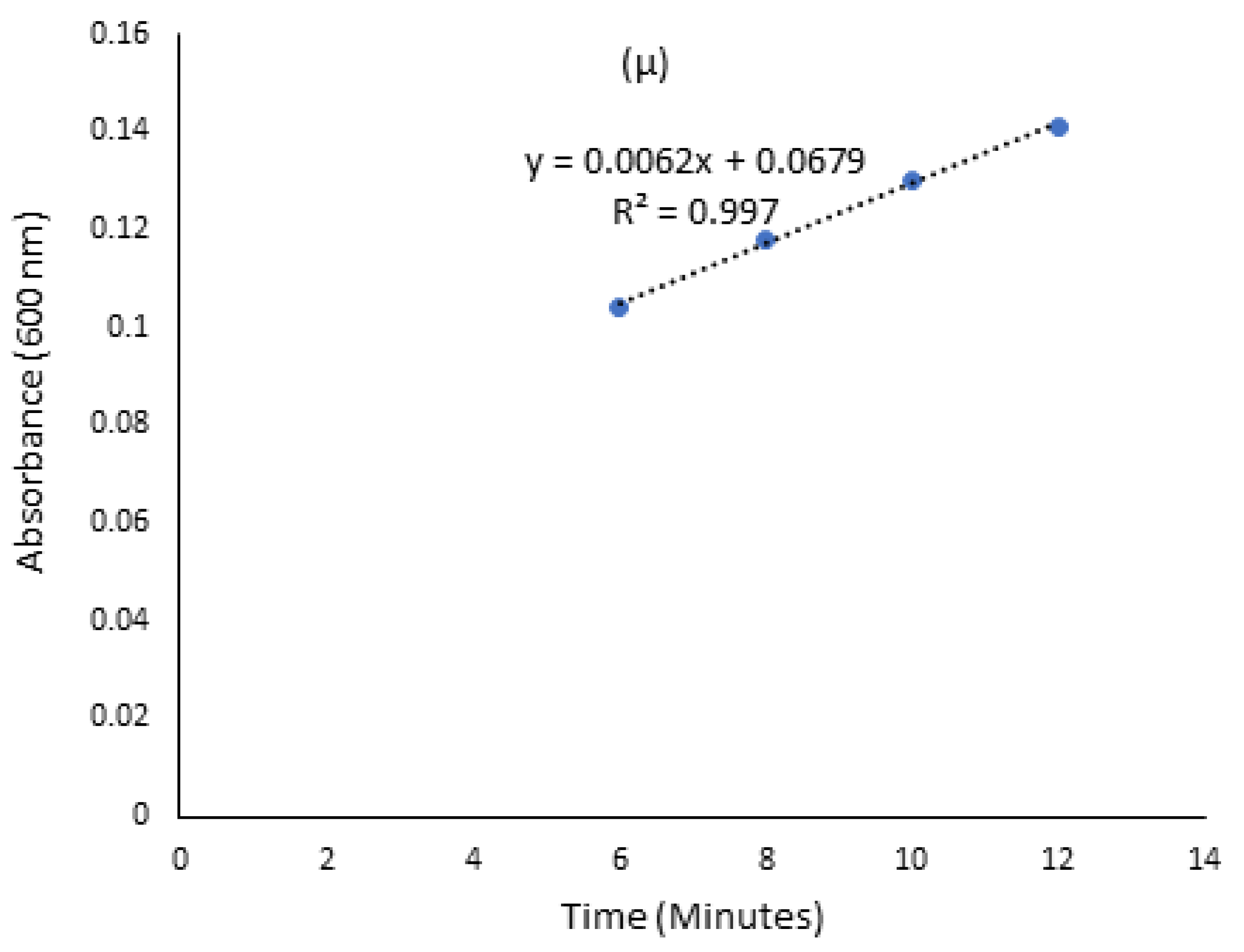
2.4. Kinetic Growth Studies for Ligninolytic and Cellulolytic Ability of Isolated Bacteria
2.5. Enzyme Assays
2.5.1. Qualitative Screening for Hydrolytic Enzyme Production
2.5.2. Quantitative Screening for Cellulolytic Ability of Microorganisms
2.6. DNA Extraction and Molecular Characterization
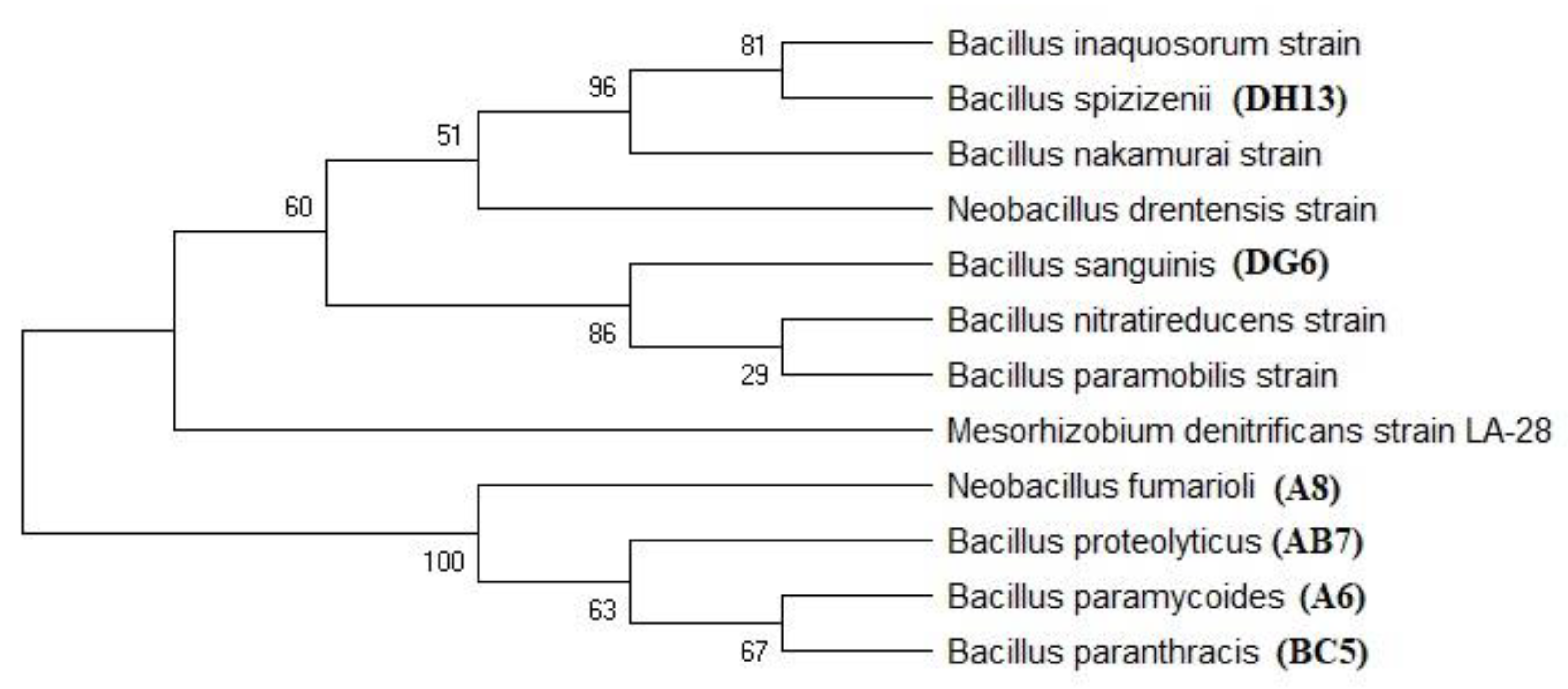
Phylogenetic Analysis
2.7. Statistical Analysis
3. Results and Discussion
3.1. Identification and Isolation of Bacteria
3.2. Screening for Lignocellulolytic Ability and Kinetic Studies
| Starch | Lignin | ||||||
|---|---|---|---|---|---|---|---|
| Strain | Growth Rate (µ) | Doubling Time (min) | R2 | Strain | Growth Rate (µ) | Doubling Time (min) | R2 |
| A1 | 0.0041 | 169.0603 | 0.8229 | A1 | 0.0048 | 144.4057 | 0.7293 |
| A17 | 0.0039 | 177.73 | 0.6297 | A17 | 0.0121 | 57.28489 | 0.6275 |
| A19 | 0.0415 | 16.70234 | 0.6025 | A19 | 0.0184 | 37.67104 | 0.8382 |
| A2 | 0.0027 | 256.7212 | 0.7946 | A2 | 0.0024 | 288.8113 | 0.6095 |
| A4 | 0.0092 | 75.34208 | 0.9605 | A4 | 0.0075 | 92.41962 | 0.6279 |
| A6 | 0.0075 | 92.41962 | 0.6614 | A6 | 0.0095 | 72.96286 | 0.8324 |
| A8 | 0.008 | 86.6434 | 0.9046 | A8 | 0.0099 | 70.01487 | 0.8266 |
| AB1 | 0.0053 | 130.7825 | 0.9891 | AB1 | 0.0017 | 407.7336 | 0.9928 |
| AB11 | 0.0036 | 192.5409 | 0.5615 | AB11 | 0.0046 | 150.6842 | 0.8631 |
| AB16 | 0.0027 | 256.7212 | 0.6173 | AB16 | 0.0238 | 29.12383 | 0.7028 |
| AB18 | 0.0075 | 92.41962 | 0.7002 | AB18 | 0.004 | 173.2868 | 0.4408 |
| AB2 | 0.004 | 173.2868 | 0.9231 | AB2 | 0.0053 | 130.7825 | 0.8207 |
| AB4 | 0.0082 | 84.53014 | 0.8176 | AB4 | 0.0044 | 157.5335 | 0.8001 |
| AB7 | 0.013 | 53.31901 | 0.9337 | AB7 | 0.0078 | 88.86502 | 0.5844 |
| BC5 | 0.0068 | 101.9334 | 0.6162 | BC5 | 0.0538 | 12.88378 | 0.7952 |
| CE10 | 0.0047 | 147.4781 | 0.828 | CE10 | 0.0011 | 630.1338 | 0.5326 |
| CE11 | 0.0098 | 70.7293 | 0.8073 | CE11 | 0.0015 | 462.0981 | 0.8242 |
| CE15 | 0.0145 | 47.80325 | 0.839 | CE15 | 0.0032 | 216.6085 | 0.7068 |
| CE16 | 0.0062 | 111.7979 | 0.78 | CE16 | 0.0077 | 90.01911 | 0.7832 |
| CE2 | 0.0223 | 31.08283 | 0.8646 | CE2 | 0.0064 | 108.3042 | 0.86 |
| CE3 | 0.01 | 69.31472 | 0.9494 | CE3 | 0.0052 | 133.2975 | 0.9643 |
| CE4 | 0.0093 | 74.53195 | 0.7904 | CE4 | 0.0011 | 630.1338 | 0.8701 |
| CE5 | 0.0014 | 495.1051 | 0.6426 | CE5 | 0.0042 | 165.035 | 0.972 |
| CE6A | 0.0038 | 182.4072 | 0.7998 | CE6A | 0.0017 | 407.7336 | 0.9966 |
| CE7 | 0.0025 | 277.2589 | 0.8929 | CE7 | 0.0077 | 90.01911 | 0.5235 |
| CE8 | 0.0062 | 111.7979 | 0.997 | CE8 | 0.0042 | 165.035 | 0.6056 |
| CE9 | 0.0108 | 64.18029 | 0.9912 | CE9 | 0.0033 | 210.0446 | 0.6926 |
| CEX5 | 0.0066 | 105.0223 | 0.7638 | CEX5 | 0.0034 | 203.8668 | 0.8187 |
| CF1 | 0.0031 | 223.5959 | 0.9907 | CF1 | 0.0125 | 55.45177 | 0.7057 |
| CF13 | 0.0076 | 91.20358 | 0.9516 | CF13 | 0.0046 | 150.6842 | 0.9648 |
| CF14 | 0.0053 | 130.7825 | 0.1813 | CF14 | 0.0036 | 192.5409 | 0.6019 |
| CF16 | 0.0026 | 266.5951 | 0.7536 | CF16 | 0.0035 | 198.0421 | 0.9336 |
| CF5 | 0.0029 | 239.0163 | 0.7325 | CF5 | 0.0158 | 43.87007 | 0.7799 |
| CF7A | 0.0018 | 385.0818 | 0.8547 | CF7A | 0.0123 | 56.35343 | 0.8978 |
| CF7B | 0.0014 | 495.1051 | 0.5178 | CF7B | 0.0036 | 192.5409 | 0.8947 |
| CF8 | 0.0032 | 216.6085 | 0.9119 | CF8 | 0.0093 | 74.53195 | 0.8128 |
| CFM4A | 0.0043 | 161.197 | 0.9572 | CFM4A | 0.0037 | 187.3371 | 0.4752 |
| CFM4B | 0.0034 | 203.8668 | 0.8847 | CFM4B | 0.0022 | 315.0669 | 0.8039 |
| DG1 | 0.004 | 173.2868 | 0.8421 | DG1 | 0.0083 | 83.51171 | 0.6603 |
| DG12 | 0.0076 | 91.20358 | 0.6823 | DG12 | 0.003 | 231.0491 | 0.6246 |
| DG13 | 0.0033 | 210.0446 | 0.9287 | DG13 | 0.0085 | 81.54673 | 0.8483 |
| DG15 | 0.0203 | 34.14518 | 0.8143 | DG15 | 0.0051 | 135.9112 | 0.8992 |
| DG16 | 0.0064 | 108.3042 | 0.6816 | DG16 | 0.0016 | 433.217 | 0.3942 |
| DG18 | 0.002 | 346.5736 | 0.9231 | DG18 | 0.0092 | 75.34208 | 0.9148 |
| DG20 | 0.0043 | 161.197 | 0.9198 | DG20 | 0.0027 | 256.7212 | 0.8144 |
| DG21 | 0.0077 | 90.01911 | 0.9434 | DG21 | 0.0133 | 52.11633 | 0.9905 |
| DG3 | 0.0006 | 1155.245 | 0.89 | DG3 | 0.0022 | 315.0669 | 0.5738 |
| DG5 | 0.0049 | 141.4586 | 0.63 | DG5 | 0.0042 | 165.035 | 0.784 |
| DG6 | 0.0115 | 60.27367 | 0.9083 | DG6 | 0.0116 | 59.75407 | 0.8613 |
| DGM1 | 0.0037 | 187.3371 | 0.7002 | DGM1 | 0.0103 | 67.29584 | 0.8878 |
| DH13 | 0.0112 | 61.88814 | 0.9173 | DH13 | 0.0089 | 77.88171 | 0.9231 |
| DH15 | 0.0051 | 135.9112 | 0.9261 | DH15 | 0.0043 | 161.197 | 0.6154 |
| DH18 | 0.0025 | 277.2589 | 0.8929 | DH18 | 0.0078 | 88.86502 | 0.5787 |
| DH2 | 0.0021 | 330.0701 | 0.9303 | DH2 | 0.0043 | 161.197 | 0.6789 |
| DH23 | 0.0103 | 67.29584 | 0.9842 | DH23 | 0.0068 | 101.9334 | 0.7872 |
| DH28 | 0.014 | 49.51051 | 0.7901 | DH28 | 0.0032 | 216.6085 | 0.7977 |
| DH29 | 0.005 | 138.6294 | 0.9328 | DH29 | 0.0039 | 177.73 | 0.6145 |
| DH3 | 0.0221 | 31.36413 | 0.8494 | DH3 | 0.0042 | 165.035 | 0.9012 |
| DH31 | 0.0533 | 13.00464 | 0.9603 | DH31 | 0.0058 | 119.5081 | 0.9878 |
| DH8 | 0.0068 | 101.9334 | 0.8647 | DH8 | 0.0057 | 121.6048 | 0.8454 |
| DH9 | 0.0046 | 150.6842 | 0.7145 | DH9 | 0.0058 | 119.5081 | 0.8814 |
3.3. Enzyme Assays
3.4. Molecular Characterization and Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhat, R.A.; Singh, D.V.; Qadri, H.; Dar, G.H.; Dervash, M.A.; Bhat, S.A.; Unal, B.T.; Ozturk, M.; Hakeem, K.R.; Yousaf, B. Vulnerability of municipal solid waste: An emerging threat to aquatic ecosystems. Chemosphere 2022, 287, 132223. [Google Scholar] [CrossRef]
- Salazar-Adams, A. The efficiency of municipal solid waste collection in Mexico. Waste Manag. 2021, 133, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Gautam, M.; Agrawal, M. Greenhouse Gas Emissions from Municipal Solid Waste Management: A Review of Global Scenario. In Carbon Footprint Case Studies; Springer: Singapore, 2021; pp. 123–160. [Google Scholar]
- Hameed, M.; Bhat, R.A.; Pandit, B.A.; Ramzan, S.; Dijoo, Z.K.; Wani, M.A. Qualitative assessment of compost engendered from municipal solid waste and green waste by indexing method. J. Air Waste Manage. Assoc. 2022, 72, 210–219. [Google Scholar] [CrossRef]
- Luo, H.; Zeng, Y.; Cheng, Y.; He, D.; Pan, X. Recent advances in municipal landfill leachate: A review focusing on its characteristics, treatment, and toxicity assessment. Sci. Total Environ. 2020, 703, 135468. [Google Scholar] [CrossRef] [PubMed]
- Antony, J.; Niveditha, S.V.; Gandhimathi, R.; Ramesh, S.T.; Nidheesh, P.V. Stabilized landfill leachate treatment by zero valent aluminium-acid system combined with hydrogen peroxide and persulfate based advanced oxidation process. Waste Manag. 2020, 106, 1–11. [Google Scholar] [CrossRef]
- Tulebayeva, N.; Yergobek, D.; Pestunova, G.; Mottaeva, A.; Sapakova, Z. Green economy: Waste management and recycling methods. E3S Web Conf. 2020, 159, 01012. [Google Scholar] [CrossRef]
- Yu, X.; Sui, Q.; Lyu, S.; Zhao, W.; Liu, J.; Cai, Z.; Yu, G.; Barcelo, D. Municipal solid waste landfills: An underestimated source of pharmaceutical and personal care products in the water environment. Environ. Sci. Technol. 2020, 54, 9757–9768. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Feng, K.; Li, Y.; Li, Q. Microbial characteristics of the leachate contaminated soil of an informal landfill site. Chemosphere 2022, 287, 132155. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Liang, G.; Alex, J.; Zhang, T.; Ma, C. Research progress of energy utilization of agricultural waste in China: Bibliometric analysis by citespace. Sustainability 2020, 12, 812. [Google Scholar] [CrossRef]
- De La Peña, L.; Guo, R.; Cao, X.; Ni, X.; Zhang, W. Accelerating the energy transition to achieve carbon neutrality. Resour. Conserv. Recycl. 2022, 177, 105957. [Google Scholar] [CrossRef]
- Zhang, L.; Chung, J.; Jiang, Q.; Sun, R.; Zhang, J.; Zhong, Y.; Ren, N. Characteristics of rumen microorganisms involved in anaerobic degradation of cellulose at various pH values. RSC Adv. 2017, 7, 40303–40310. [Google Scholar] [CrossRef]
- Paul, S.; Dutta, A. Challenges and opportunities of lignocellulosic biomass for anaerobic digestion. Resour. Conserv. Recycl. 2018, 130, 164–174. [Google Scholar] [CrossRef]
- De la Cruz, F.B.; Chanton, J.P.; Barlaz, M.A. Measurement of carbon storage in landfills from the biogenic carbon content of excavated waste samples. Waste Manag. 2013, 33, 2001–2005. [Google Scholar] [CrossRef]
- Bendeddouche, W.; Bedrane, S.; Zitouni, A.; Bachir, R. Highly efficient catalytic one-pot biofuel production from lignocellulosic biomass derivatives. Int. J. Energy Res. 2021, 45, 2148–2159. [Google Scholar] [CrossRef]
- Kaloudas, D.; Pavlova, N.; Penchovsky, R. Phycoremediation of wastewater by microalgae: A review. Environ. Chem. Lett. 2021, 19, 2905–2920. [Google Scholar] [CrossRef]
- Dar, M.A.; Syed, R.; Pawar, K.D.; Dhole, N.P.; Xie, R.; Pandit, R.S.; Sun, J. Evaluation and characterization of the cellulolytic bacterium, Bacillus pumilus SL8 isolated from the gut of oriental leafworm Spodoptera litura: An assessment of its potential value for lignocellulose bioconversion. Environ. Technol. Innov. 2022, 27, 102459. [Google Scholar] [CrossRef]
- Vasco-Correa, J.; Ge, X.; Li, Y. Biological pretreatment of lignocellulosic biomass. In Biomass Fractionation Technologies for a Lignocellulosic Feedstock Based Biorefinery; Elsevier: Amsterdam, The Netherlands, 2016; pp. 561–585. [Google Scholar]
- Poszytek, K.; Ciężkowska, M.; Skłodowska, A.; Drewniak, Ł. Microbial consortium with high cellulolytic activity (mchca) for enhanced biogas production. Front. Microbiol. 2016, 7, 324. [Google Scholar] [CrossRef]
- Ferdeș, M.; Dincă, M.N.; Moiceanu, G.; Zăbavă, B. Ștefania; Paraschiv, G. Microorganisms and enzymes used in the biological pretreatment of the substrate to enhance biogas production: A review. Sustainability 2020, 12, 7205. [Google Scholar] [CrossRef]
- Güllert, S.; Fischer, M.A.; Turaev, D.; Noebauer, B.; Ilmberger, N.; Wemheuer, B.; Alawi, M.; Rattei, T.; Daniel, R.; Schmitz, R.A.; et al. Deep metagenome and metatranscriptome analyses of microbial communities affiliated with an industrial biogas fermenter, a cow rumen, and elephant feces reveal major differences in carbohydrate hydrolysis strategies. Biotechnol. Biofuels. 2016, 9, 121. [Google Scholar] [CrossRef] [PubMed]
- Ransom-Jones, E.; McCarthy, A.J.; Haldenby, S.; Doonan, J.; McDonald, J.E. Lignocellulose-degrading microbial communities in landfill sites represent a repository of unexplored biomass-degrading diversity. mSphere 2017, 2, e00300-17. [Google Scholar] [CrossRef]
- Al-Yaqout, A.; Hamoda, M.F. Long-term temporal variations in characteristics of leachates from a closed landfill in an arid region. Water Air Soil Pollut. 2020, 231, 319. [Google Scholar] [CrossRef]
- Kundu, A.; Majumdar, B. Optimization of the cellulase free xylanase production by immobilized Bacillus pumilus. Iran. J. Biotechnol. 2018, 16, 273–278. [Google Scholar] [CrossRef]
- Ezeilo, C.A.; Okonkwo, S.I.; Onuora, C.C.; Linu-Chibuezeh, A.; Ugwunnadi, N.E. Determination of heavy metals in some fruits and vegetables from selected market’s in Anambra state. ACTA Sci. Nutr. Heal. 2020, 4, 163–171. [Google Scholar]
- Malik, W.A.; Javed, S. Biochemical characterization of cellulase from Bacillus subtilis strain and its effect on digestibility and structural modifications of lignocellulose rich biomass. Front. Bioeng. Biotechnol. 2021, 9, 800265. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Wang, X.; Zhou, J.; Lü, X. Degradation of switchgrass by Bacillus subtilis 1AJ3 and expression of a beta-glycoside hydrolase. Front. Microbiol. 2022, 13, 922371. [Google Scholar] [CrossRef]
- Shajahan, S.; Moorthy, I.G.; Sivakumar, N.; Selvakumar, G. Statistical modeling and optimization of cellulase production by Bacillus licheniformis NCIM 5556 isolated from the hot spring, Maharashtra, India. J. King Saud Univ. Sci. 2017, 29, 302–310. [Google Scholar] [CrossRef]
- Islam, M.; Sarkar, P.K.; Mohiuddin, A.K.M.; Suzauddula, M. Optimization of fermentation condition for cellulase enzyme production from Bacillus sp. Malays. J. Halal Res. 2019, 2, 19–24. [Google Scholar] [CrossRef]
- Aulitto, M.; Fusco, S.; Bartolucci, S.; Franzén, C.J.; Contursi, P. Bacillus coagulans MA-13: A promising thermophilic and cellulolytic strain for the production of lactic acid from lignocellulosic hydrolysate. Biotechnol. Biofuels 2017, 10, 210. [Google Scholar] [CrossRef]
- Balla, A.; Silini, A.; Cherif-Silini, H.; Bouket, A.C.; Boudechicha, A.; Luptakova, L.; Alenezi, F.N.; Belbahri, L. Screening of cellulolytic bacteria from various ecosystems and their cellulases production under multi-stress conditions. Catalysts 2022, 12, 769. [Google Scholar] [CrossRef]
- Bhagat, S.A.; Kokitkar, S.S. Isolation and identification of bacteria with cellulose-degrading potential from soil and optimization of cellulase production. J. Appl. Biol. Biotechnol. 2021, 9, 154–161. [Google Scholar] [CrossRef]
- Liang, Y.-L.; Zhang, Z.; Wu, M.; Wu, Y.; Feng, J.-X. Isolation, screening, and identification of cellulolytic bacteria from natural reserves in the subtropical region of china and optimization of cellulase production by Paenibacillus terrae ME27-1. Biomed Res. Int. 2014, 2014, 512497. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Li, L.; Peng, X.; Zhang, R.; Song, L. Methanogen community dynamics and methanogenic function response to solid waste decomposition. Front. Microbiol. 2021, 12, 743847. [Google Scholar] [CrossRef] [PubMed]
- Yong, Z.J.; Bashir, M.J.K.; Ng, C.A.; Sethupathi, S.; Lim, J.W.; Show, P.L. Sustainable waste-to-energy development in Malaysia: Appraisal of environmental, financial, and public issues related with energy recovery from municipal solid waste. Processes 2019, 7, 676. [Google Scholar] [CrossRef]
- Forster, J.C. Soil Sampling, Handling, Storage and Analysis. In Methods in Applied Soil Microbiology and Biochemistry; Elsevier: Amsterdam, The Netherlands, 1995; pp. 49–121. [Google Scholar]
- Radojević, M.; Bashki, V. Soil, Sediment, Sludge and Dust analysis. In Practical Environmental Analysis; Radojevic, M., Bashkin, V.N., Eds.; Royal Society of Chemistry: Cambridge, UK, 1999; pp. 274–377. ISBN 978-0-85404-594-5. [Google Scholar]
- Reynolds, J. Serial Dilution Protocols; American Society for Microbiology: Washington, DC, USA, 2005; pp. 1–7. [Google Scholar]
- Bergey, D.H.; Holt, J.G. Bergey’s Manual of Determinative Bacteriology, 9th ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2000. [Google Scholar]
- Smith, A.C.; Hussey, M.A. Gram Stain Protocols; American Society for Microbiology: Washington, DC, USA, 2016; pp. 1–9. [Google Scholar]
- Dalgaard, P.; Ross, T.; Kamperman, L.; Neumeyer, K.; McMeekin, T.A. Estimation of bacterial growth rates from turbidimetric and viable count data. Int. J. Food Microbiol. 1994, 23, 391–404. [Google Scholar] [CrossRef] [PubMed]
- Muloiwa, M.; Nyende-Byakika, S.; Dinka, M. Comparison of unstructured kinetic bacterial growth models. S. Afr. J. Chem. Eng. 2020, 33, 141–150. [Google Scholar] [CrossRef]
- Breidt, F.; Romick, T.L.; Fleming, H.P. A rapid method for the determination of bacterial growth kinetics. J. Rapid Methods Autom. Microbiol. 1994, 3, 59–68. [Google Scholar] [CrossRef]
- Lal, A.; Cheeptham, N. ASM ATLAS Protocol: Starch Agar; American Society for Microbiology: Washington, DC, USA, 2012; p. 11. [Google Scholar]
- Masi, C.; Gemechu, G.; Tafesse, M. Isolation, screening, characterization, and identification of alkaline protease-producing bacteria from leather industry effluent. Ann. Microbiol. 2021, 71, 24. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Yi, S.; Tay, J.-H.; Maszenan, A.M.; Tay, S.T.-L. A culture-independent approach for studying microbial diversity in aerobic granules. Water Sci. Technol. 2003, 47, 283–290. [Google Scholar] [CrossRef]
- Farhana Zakaria, S.N.; Abdul Aziz, H. Characteristic of leachate at Alor Pongsu Landfill Site, Perak, Malaysia: A comparative study. IOP Conf. Ser. Earth Environ. Sci. 2018, 140, 012013. [Google Scholar] [CrossRef]
- Ma, J.; Wu, S.; Shekhar, N.V.R.; Biswas, S.; Sahu, A.K. Determination of physicochemical parameters and levels of heavy metals in food wastewater with environmental effects. Bioinorg. Chem. Appl. 2020, 2020, 8886093. [Google Scholar] [CrossRef]
- Saha, C.K.; Ammon, C.; Berg, W.; Loebsin, C.; Fiedler, M.; Brunsch, R.; von Bobrutzki, K. The effect of external wind speed and direction on sampling point concentrations, air change rate and emissions from a naturally ventilated dairy building. Biosyst. Eng. 2013, 114, 267–278. [Google Scholar] [CrossRef]
- Zhai, Y.; Li, X.; Wang, T.; Wang, B.; Li, C.; Zeng, G. A review on airborne microorganisms in particulate matters: Composition, characteristics and influence factors. Environ. Int. 2018, 113, 74–90. [Google Scholar] [CrossRef]
- Delignette-Muller, M.L. Relation between the generation time and the lag time of bacterial growth kinetics. Int. J. Food Microbiol. 1998, 43, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.; Bokka, V.; Sen, S. Dependence of bacterial growth rate on dynamic temperature changes. IET Syst. Biol. 2020, 14, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Chukwuma, O.B.; Rafatullah, M.; Tajarudin, H.A.; Ismail, N. Bacterial diversity and community structure of a municipal solid waste landfill: A source of lignocellulolytic potential. Life 2021, 11, 493. [Google Scholar] [CrossRef]
- Guder, D.G.; Krishna, M.S.R. Isolation and characterization of potential cellulose degrading bacteria from sheep rumen. J. Pure Appl. Microbiol. 2019, 13, 1831–1839. [Google Scholar] [CrossRef]
- Alrumman, S.; Mostafa, Y.S.M.; Al-Qahtani, S.; Taha, T.H.T. Hydrolytic enzyme production by thermophilic bacteria isolated from Saudi hot springs. Open Life Sci. 2018, 13, 470–480. [Google Scholar] [CrossRef]
- Logan, N.A.; Lebbe, L.; Hoste, B.; Goris, J.; Forsyth, G.; Heyndrickx, M.; Murray, B.L.; Syme, N.; Wynn-Williams, D.D.; De Vos, P. Aerobic endospore-forming bacteria from geothermal environments in northern Victoria Land, Antarctica, and Candlemas Island, South Sandwich archipelago, with the proposal of Bacillus fumarioli sp. nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 1741–1753. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.K.; Yadav, R.C.; Choudhary, P.; Sharma, S.K.; Bhagat, N. Mitigation of drought stress in wheat (Triticum aestivum L.) by inoculation of drought tolerant Bacillus paramycoides DT-85 and Bacillus paranthracis DT-97. J. Appl. Biol. Biotechnol. 2022, 10, 59–69. [Google Scholar] [CrossRef]
- Bhaskar, N.; Sudeepa, E.; Rashmi, H.; Tamilselvi, A. Partial purification and characterization of protease of Bacillus proteolyticus CFR3001 isolated from fish processing waste and its antibacterial activities. Bioresour. Technol. 2007, 98, 2758–2764. [Google Scholar] [CrossRef]
- Hallol, M.; Helmy, O.; Shawky, A.-E.; El-Batal, A.; Ramadan, M. Optimization of alpha-amylase production by a local Bacillus paramycoides isolate and immobilization on chitosan-loaded barium ferrite nanoparticles. Fermentation 2022, 8, 241. [Google Scholar] [CrossRef]
- Omeiri, M.; Khnayzer, R.; Yusef, H.; Tokajian, S.; Salloum, T.; Mokh, S. Bacillus spp. isolated from soil in Lebanon can simultaneously degrade methomyl in contaminated soils and enhance plant growth. Biocatal. Agric. Biotechnol. 2022, 39, 102280. [Google Scholar] [CrossRef]
- Akpor, O.B.; Akinwusi, O.D.; Ogunnusi, T.A. Production, characterization and pesticidal potential of Bacillus species metabolites against sugar ant (Camponotus consobrinus). Heliyon 2021, 7, e08447. [Google Scholar] [CrossRef] [PubMed]
- Thomas, N.N.; Archana, V.; Shibina, S.; Edwin, B.T. Isolation and characterization of a protease from Bacillus sps. Mater. Today Proc. 2021, 41, 685–691. [Google Scholar] [CrossRef]

| Deionized Water (mL) | Sole Carbon Source (g) | |
|---|---|---|
| Lignin | 10 | 0.1 |
| Starch | 10 | 0.1 |
| Sampling Point | Latitude | Longitude | pH | Physical Appearance of Sediment | Temperature, °C |
|---|---|---|---|---|---|
| A | N5°12′6.9″ | E100°25′24.7″ | 5.97 | black | 32 |
| B | N5°12′14.8″ | E100°25′33.8″ | 6.32 | red | 35 |
| C | N5°12′7.6″ | E100°25′26.3″ | 6.3 | brown | 36 |
| D | N5°11′57.7″ | E100°25′36.0″ | 6.9 | loamy | 28 |
| Isolate | Amylase | Cellulase | Xylanase | Protease |
|---|---|---|---|---|
| A3 | ++ | + | + | + |
| A4 | + | − | − | − |
| A6 | + | − | − | + |
| A8 | + | − | − | − |
| DH31 | ++ | +++ | +++ | − |
| AB7 | + | + | + | + |
| BC5 | + | + | + | − |
| DG6 | + | ++ | ++ | + |
| DG21 | +++ | − | − | − |
| BD25 | − | +++ | +++ | − |
| DH13 | + | +++ | +++ | + |
| Sample ID | Coverage | Similarity | BP | Accession | Matched Bacteria from NCBI |
|---|---|---|---|---|---|
| A6 | 98 | 99.91 | 1160 | OQ288926 | Bacillus paramycoides |
| DG6 | 96 | 98.81 | 2322 | OQ288927 | Bacillus Sanguinis |
| A8 | 91 | 97.11 | 1270 | OQ288921 | Neobacillus fumarioli |
| DG21 | 92 | 99.75 | 1315 | OQ288922 | Bacillus paramycoides |
| DH13 | 97 | 98.96 | 1277 | OQ288871 | Bacillus spizizenii |
| AB7 | 97 | 98.78 | 1185 | OQ288869 | Bacillus proteolyticus |
| BC5 | 95 | 98.92 | 1260 | OQ288870 | Bacillus paranthracis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chukwuma, O.B.; Rafatullah, M.; Kapoor, R.T.; Tajarudin, H.A.; Ismail, N.; Siddiqui, M.R.; Alam, M. Isolation and Characterization of Lignocellulolytic Bacteria from Municipal Solid Waste Landfill for Identification of Potential Hydrolytic Enzyme. Fermentation 2023, 9, 298. https://doi.org/10.3390/fermentation9030298
Chukwuma OB, Rafatullah M, Kapoor RT, Tajarudin HA, Ismail N, Siddiqui MR, Alam M. Isolation and Characterization of Lignocellulolytic Bacteria from Municipal Solid Waste Landfill for Identification of Potential Hydrolytic Enzyme. Fermentation. 2023; 9(3):298. https://doi.org/10.3390/fermentation9030298
Chicago/Turabian StyleChukwuma, Ogechukwu Bose, Mohd Rafatullah, Riti Thapar Kapoor, Husnul Azan Tajarudin, Norli Ismail, Masoom Raza Siddiqui, and Mahboob Alam. 2023. "Isolation and Characterization of Lignocellulolytic Bacteria from Municipal Solid Waste Landfill for Identification of Potential Hydrolytic Enzyme" Fermentation 9, no. 3: 298. https://doi.org/10.3390/fermentation9030298
APA StyleChukwuma, O. B., Rafatullah, M., Kapoor, R. T., Tajarudin, H. A., Ismail, N., Siddiqui, M. R., & Alam, M. (2023). Isolation and Characterization of Lignocellulolytic Bacteria from Municipal Solid Waste Landfill for Identification of Potential Hydrolytic Enzyme. Fermentation, 9(3), 298. https://doi.org/10.3390/fermentation9030298










