Abstract
The skin microbiome is composed of a complex association of bacteria, fungi, and viruses. The maintenance of skin commensal microbes is essential for preventing the overgrowth of pathogenic microorganisms or already present opportunistic pathogens. Thus, the development of bioactive compounds capable of modulating skin microbiome has become an important topic for both researchers and the cosmetic industry. Increasingly, scientific evidence highlights that metabolites derived from probiotics have a great potential to prevent diseases affecting the skin. These compounds have recently been called postbiotics and are defined as a “preparation of inanimate microorganisms and/or their components that confers a health benefit on the host”. Postbiotics are obtained from fermentations performed almost exclusively by lactic acid bacteria and yeast. Short-chain fatty acids, bacteriocins, and organic acids are some examples of postbiotics. These compounds exhibit antimicrobial, immunomodulatory, antioxidant, and anti-inflammatory activities. In addition, postbiotic production possesses technological advantages, including high stability and increased safety, compared to viable probiotics. In this article, a model for the large-scale production of postbiotics and their uses in cosmetic formulations are reviewed. In addition, results obtained from in vivo tests for the treatment of alopecia, acne, atopic dermatitis, and wound healing are discussed. Finally, technological advances are shown based on a survey of the main patents filed in the area of postbiotics.
1. Introduction
In the last decade, high-throughput sequencing has helped scientists unravel the relationships between microorganisms and human health. Dedicated studies are mainly related to human intestinal microbiota, while only recently, skin microbiome has been explored [1,2,3]. These recent studies have helped in understanding the interactions and relationships between bacteria, fungi, and viruses associated with skin diseases, such as atopic dermatitis, alopecia, acne vulgaris, and psoriasis [4,5]. It has changed our perception of the importance of microorganisms for skin health, stimulating the development of new products that protect, respect, or rebalance skin microbiome [6]. The development of dermo-cosmetics derived from probiotics seems to be a promising alternative mainly due to their beneficial health effects. However, the application of viable microorganisms to the skin still presents major challenges in technology and regulations [6,7].
Interestingly, since 2002, there has been evidence that some of the bioactive properties of probiotics are not necessarily related to their viability, as dead cells can confer biological responses equal or superior to their live counterparts [8]. Therefore, these compounds/molecules derived from inactivated probiotics have been termed postbiotics, which are defined as a “preparation of inanimate microorganisms and/or their components that confers a health benefit on the host” [9]. Postbiotics can be obtained either from metabolites generated during microbial growth or from intact (i.e., inanimate) dead strains after cell disruption and fragmentation. In general, the bioactive properties of postbiotics responsible for the beneficial effects on host health are associated with several components that include bacteriocins, lipoteichoic acids, surface layer proteins, peptides, polysaccharides, and organic acids. Furthermore, postbiotics must be obtained from defined and characterized microbial strains using a well-delineated and reliable technological process for production and inactivation that exhibits high reproducibility [10].
This review aims to present the importance of microorganisms in maintaining skin homeostasis and to provide an overview of the production, formulations, and potential therapeutic applications of postbiotics for the treatment of skin diseases and injuries.
2. Skin Microbiome
Skin is a complex organ with a symbiotic relationship between microbial communities and the host tissue. Recent studies have revealed how resident microbial communities play a crucial role in maintaining the normal healthy function of the skin and the immune system. An abundant and diverse community of bacteria, viruses, eukaryotes, such as fungi and arthropods [4,5], composes the human skin microbiota. The diversity and abundance vary considerably between individuals and between different sites on the skin, due to our different genetics, diet, lifestyle, gender, age, ethnicity, and habitat [11,12,13].
Unlike the gut and stool microbiome which has been studied and described extensively for years, research on the skin and scalp microbiome is still scarce [14]. The evolution of studies on microbial diversity of human skin started at the end of 1950s by using improved cell culture-methods and identification according to their morphological or biochemical characteristics [15]. Thenceforth, culture-dependent based studies reported the genera Staphylococci, Micrococci, Corynebacteria, Brevibacteria, Propionibacteria, and Acinetobacter from normal skin [16]. At the species level, Staphylococcus aureus, Streptococcus pyogenes, and Pseudomonas aeruginosa were identified by culture methods, such as colonizers in abnormal conditions [17]. However, cultivation provides an inaccurate assessment and underestimates the total diversity of the community [18].
In 2007, the National Institute of Health funded the Human Microbiome Project (HMP) with the aim to characterize the microbial communities present at specific body sites and to further the understanding of how microbiome impacts human health and disease [19]. In recent decades, culture-independent methodologies were developed to overcome the limitations of conventional microbiology testing through DNA analyses without any culturing step [20]. Initially, the applied techniques included denaturing gradient gel electrophoresis (DGGE), single-stranded conformation polymorphism (SSCP), real-time quantitative PCR (qPCR), automated PCR-based techniques (PCR-ARDRA, ARISA-PCR, AP-PCR, and AFLP), and terminal restriction fragment length polymorphism (T-RFLP) [14,15,16,17,18,19,20,21]. Quantitative PCR is used to identify and quantify microbial communities using specific DNA markers. Hammoudi et al. [22] detected a Mycobacterium ulcerans-specific marker from an asymptomatic Buruli skin ulcer by using the abovementioned technique. However, many of the microorganism-specific primers that are employed for qPCR are, in fact, not specific or do not cover all microbiome. Furthermore, microbial qPCR methods typically target the rRNA genetic markers, and because species differ markedly in number of rRNA operon copies per genome, the performance of qPCR is limited [18].
Researchers in microbiology and dermatology started using metagenomic approaches based on DNA sequencing for more accurate analysis, including characterization of the complete diversity of the skin microbiome as well their relative abundance, and link how microbial diversity may contribute to skin health and dermatological conditions [20,21,22,23]. At the beginning, Sanger sequencing technology using 16S rRNA gene profiling revealed the bacterial diversity that inhabits the human skin [24]. At least 19 phyla were observed to be part of the bacterial skin microbiome. The major microbial phyla are Actinobacteria (51.8%), Firmicutes (24.4%), Proteobacteria (16.5%), and Bacteroidetes (6.3%). The majority of the identified genera are Corynebacterium (22.8%; Actinobacteria), Propionibacterium (23.0%; Actinobacteria), and Staphylococcus (16.8%; Firmicutes) [25]. Swab-scrubbed forehead skin samples of five healthy volunteers were analyzed based on 16S ribosomal (rRNA) and Sanger sequencing as well as by conventional culture methods in order to provide a profile of the cutaneous microbiota. Seventeen species were detected, including Cutibacterium acnes, P. granulosum, S. warneri, S. mitis/salivarius, S. epidermidis, Acinetobacter johnsonii, Acinetobacter calcoaceticus, Acinetobacter junii, Finegoldia magna, Streptococcus cristatus, Stenotrophomonas maltophilia, Corynebacterium pseudogenitalium, Acidovorax temperans, Dietzia maris, Bacillus licheniformis, B. subtilis, and Neisseria subflava [24].
Currently, high-throughput DNA sequencing (HTS) methodologies primarily use second generation (Illumina and 454 pyrosequencing) short-read sequencing for studying microbial communities [14]. Two main approaches associated with HTS are used: marker gene studies [5,11,13,18,26,27,28,29] and whole-genome shotgun metagenomic (WGS) approaches [23,30,31]. Marker gene analyses rely on the sequencing of a gene-specific region to reveal the diversity and composition of specific taxonomic groups present in an environmental sample. The principal marker genes used in microbial studies for amplicon sequencing are the 16S ribosomal (rRNA) gene to analyse the presence of archaea and bacteria; the internal transcribed spacer (ITS) region to characterize the composition of the fungal community; and the 18S rRNA to report the occurrence of eukaryotes [18]. These original sequencing approaches exploit sequence variation in conserved taxonomic markers as molecular fingerprints to identify members of microbial communities [21].
The opposite of the marker gene method is the WGS approach, which captures the entire complement of genetic material in a sample to analyse the biodiversity and the functional capabilities of the microbial community studied without a targeted amplification step [18,20,32]. As the entire genetic material of a sample is recovered, it simultaneously captures all genetic material in a sample, including human, bacterial, fungal, archaeal, and viral, thus allowing relative kingdom abundances to be inferred. An additional advantage is that the obtained datasets provide sufficient resolution to differentiate species and even strains within a species [23]. The ability to distinguish strains is important as more studies reveal the functional differences that exist between strains within a species [19,23,27,33].
In sequencing surveys of healthy adults, the composition of microbiome is highly dependent on characteristics based on skin type and location, with changes in the relative abundance of bacterial taxa associated with three main physiological skin sites. The first is the moist site, including the axilla, inner elbow, or inguinal fold. The moist regions are colonized mostly by Staphylococcus and Corynebacteria species. The axillar area consists mainly of Gram-positive bacteria of the genera Staphylococcus, Micrococcus, Corynebacterium, and Propionibacterium. The second is the dry sites host, predominantly Staphylococcus, Propionibacterium, Micrococcus, Corynebacterium, Enhydrobacter, and Streptococcus. The drier sites include the upper buttock area. The third, called sebaceous areas, include the forehead, the alar crease (side of the nostril), the retro auricularcrease (behind the ear), and the back. Sebaceous sites have a higher density of particularly lipophilic species, mainly Propionibacterium, which has adapted to this lipid-rich, anaerobic environment [17,34].
To date, the marker genes and WGS studies coupled to HTS methodology have established new milestones in skin microbiome, revealing a diverse community with operational taxonomic units (OTUs) represented by Bacteroides, Porphyromonas, Aerococcus, Rikenellaceae, Clostridiales, Lachnospiraceae, Shuttleworthia, Dorea, Ruminococcaceae Oscillospira Eikenella Bacillales, Lachnospiraceae, Agrobacterium, Veillonella, Rhizobium, Delftia, Comamonadaceae, Tepidimonas, Variovorax, Legionella, Dermacoccus, and Deinococcus [23,35]. Over the past few years, metagenomic analyses with HTS technologies performed from healthy and diseased human skin unveiled the presence of over 65 genera and species of bacteria, fungi, and viruses. From these, 290 species were reported for the first time when compared to previous works using traditional culture-dependent methods. A complete list of these new skin microbiota identified by culture-independent methods is shown in the Supplementary Materials (Table S1).
Most microbiome studies concentrate on identifying bacterial composition, but the fungi community are also a crucial part of the skin microbiota [4]. Culture-based studies have reported Malassezia, Rhodotorula, Debaromyces, Cryptococcus, and, in some sites, Candida as fungal skin commensals [36]. Relative abundance of fungal genera from different areas of the skin revealed by HTS methodology include Arthrodermataceae, Aspergillus, Rhodotorula, Cryptococcus, Chrysosporium, Candida, Penicillium, Leptosphaerulina, Phoma, Saccharomyces, Ustilago, and Epicoccum [31,37,38]. The predominant fungi detected using phylogenetic markers such as 18S rRNA belonged to the genera Malassezia, including the most frequent species Malassezia globosa, Malassezia restricta, and Malassezia sympodialis [27]. Demodex spp. are tiny mites living in the pilosebaceous follicles and were also identified by a culture-independent approach in rosacea pathogenesis [39].
In addition, healthy human skin is also colonized by a remarkable diversity of viruses [40]. Many eukaryotic viruses from the families Polyomaviridae, Papillomaviridae, Circoviridae, Caudovirales Adenoviridae, Anelloviridae, Cavemovirus, Luteoviridae, Parvoviridae, Poxviridae, Reoviridae, Retroviridae, and Herpesviridae are commonly detected [23,40,41]. In the Papillomaviridae family, sequences are related to the beta-, alpha-, mupa-, and gammapapilloma viruses, whereas Polyomaviridae related sequences corresponded to Merkel cell polyomavirus (MCPyV), human polyomavirus 6 (HPyV6), human polyomavirus 7 (HPyV7), and human polyomavirus 9 (HPyV9) [23,41]. The interactions of the virome with other members of the microbiota have barely been studied [40]. However, the skin virome can also contribute to the health and disease status of the host through the suppressive actions of bacteriophages [42]. The majority of the phages belong to the Siphoviridae, Podoviridae, and Myoviridae [43]. Among phage species with abundant host bacteria, two significantly differential abundant phage species, Acinetobacter phage Presley and Pseudomonas phage O4, are identified in psoriasis lesional and healthy skin [42]. Although viruses are generally considered as pathogen agents, complex viral flora are present on the surface of human skin that appears healthy in various individuals [23].
The update of the major skin metataxonomic studies at different sites is valuable for elucidating the etiology of common skin disorders [17]. Some of these diseases are noninfectious but can be influenced by shifts and imbalances in skin microbiota. Organisms that cause cutaneous infections can also be studied via metagenomics, which could be particularly useful in those infections associated with a wide range of clinical features and wide geographic and host variability [14,18,26]. Over the past years, many studies were conducted on the lesional skin of common skin diseases to identify which species are most abundant and whether this microbial composition differs from healthy skin. Under healthy skin conditions, most of the microbes living on that organ behave as commensal or mutualistic organisms. The commensal skin microbiota inhibits the spread of opportunistic parasites. In case of skin barrier breach or immunosuppression, these carefully balanced relationships may shift from commensalism to pathogenicity [12,18,32], developing skin conditions such as acne [32,44], atopic dermatitis (AD) [45], and psoriasis [46].
The microbial community structure in psoriasis patients were investigated using high throughput 16S rRNA and WGS sequencing [28,42]. The four major skin genera, Corynebacterium, Propionibacterium, Staphylococcus, and Streptococcus are not different between psoriasis lesion, unaffected, and healthy control skin. In contrast, a significant univariate decrease in relative abundances of Cupriavidus, Flavisolibacter, Methylobacterium, and Schlegelella genera was observed in psoriasis-associated skin when compared to the healthy control [47]. Another study did claim that Staphylococcus species were overrepresented in psoriasis skin, but, overall, the microbiomes of psoriatic and unaffected sites display few discriminative features at the species level [46].
AD pathology is a skin disease associated by Staphylococcus aureus colonization on both lesional and non-lesional skin [12]. Chng et al. [27], using the two profiling approaches, 16S rRNA and WGS, identified distinct non-flare, baseline skin microbiome signatures enriched for Veillonella and Haemophilus and the potential pathobionts belonging to the genera Streptococcus and Gemella but depleted for Dermacoccus, Deinococcus and Methylobacterium in AD versus normal healthy skin. At the species level, nine bacteria, including Corynebacterium matruchotii, Gemella haemolysans Haemophilus parainfluenzae, Streptococcus cristatus, S. mitis, C. pseudogenitalium, Veillonella parvula, S. sanguinis, and C. tuberculostearicum were identified as having significant AD-associated microbiome differences. S. epidermidis, a commensal present on non-inflamed skin, appears to the best antagonist of S. aureus. Fyhrquist et al. [28] showed an increased abundance of S. aureus with a depletion of S. epidermidis and Corynebacterium spp. among AD patients when compared to the healthy controls. WGS data revealed, for the first time, a decrease in Malassezia relative abundance in AD-associated microbiomes. However, an enrichment of M. dermatis and M. sympodalis species in AD-susceptible skin were detected, suggesting the biological relevance of species switching in AD pathology [27].
Acne has been largely associated with Propionibacterium acnes (synonymous Cutibacterium acnes) proliferation [44]. However, studies using metagenomics found that the abundance and bacterial load of C. acnes do not differ significantly between acne and healthy skins [12]. Barnard et al. [32], using the WGS approach, reported that the healthy individuals had a slightly higher relative abundance of C. acnes than the acne patients (93.8% vs. 88.5%). Other cutaneous Propionibacterium species, including P. humerusii, P. granulosum, and P. avidum, were also discovered in healthy and acne patients. At the strain level, C. acnes populations in older healthy individuals are similar to those of the younger healthy cohort, consisting of mostly RT1, RT2, and RT3 strains, with little or no RT4, RT5, and RT8 strains spotted [23,32]. Using SLST markers targeting the tuf2 gene, staphylococcal strains were found to selectively exclude acne-associated C. acnes and co-exist with healthy skin-associated phylotypes through regulation of the antimicrobial activity. Overall, these findings highlight the importance of skin-resident staphylococci and suggest that selective microbial interference is a contributor to healthy skin homeostasis [30].
Evidence of rosacea-associated microorganisms was shown for Demodex folliculorum, S. epidermidis, B. oleronius, Helicobacter pylori, and Chlamydia pneumonia, but the results have been inconsistent [48]. 16S rRNA gene sequencing showed different subtypes of rosacea harbor Demodex mites with different microbiota [42]. Rosacea severity increased with age and was associated with a decrease in the relative abundance of C. acnes and an increase in Snodgrassella alvi [48].
Bacterial taxa can also be readily associated with younger or older skin [13]. Lately, a number of studies focused on changes related to the skin microbiome in relation with its aging and therapeutic interventions [12]. Alkema et al. [29], in which 16S rRNA gene sequencing data observed an association of Corynebacterium, Chryseobacterium, and Veillonella with the older (59–68 years) age group. In a study reported by Kim et al. [35], an overrepresentation of Alistipes, Prevotella, Porphyromonas, Sphingobacterium, Lactobacillus, Aerococcus, Oscillospira, and Ruminococcus was found in the test group representing 25–35- -year-old healthy female volunteers and an overrepresentation of Micrococcus, Corynebacterium, Dermacoccus, Actinomyces Streptococcus, Lysinibacillus, and Bacillus in a group constituted by 56–63-year-old females.
Studies of the skin microbiota through its main milestones has been coupled with the development of sequencing technology. Indeed, the research that applied culture-independent approaches and HTS technology produced the main progress in our knowledge of the composition and function of this microbial community. The advances in third generation sequencing (Oxford Nanopore Technologies MinION and Pacific Biosciences Sequel platforms) long-read technologies provide opportunities to overcome some of the limitations of short-read sequencing provided by the second-generation sequencing methods [20,21]. Until now, third generation sequencing has not yet been used for generating the microbiome of human skin. This is essential for identifying members of the different genus, which are difficult to classify to the species level with most amplicon sequencing approaches. The ability to differentiate strains is important as more studies reveal the functional differences that exist between strains within a species [14,31]. Such methods enable the identification of potential causal relationships between microbial communities and clinical outcomes [29]. Obtaining insights on the skin microbiome composition to species level in skin health and disease will help to develop more effective postbiotic, prebiotic, probiotic, or drug therapies to treat skin diseases associated with microbiome dysbiosis.
3. Overview of Postbiotics
Although the beneficial health effects promoted by probiotics are often associated with cell viability, current knowledge allows us to state that not all mechanisms are directly related to this characteristic [49]. Recently, it has been shown that non-viable microorganisms (postbiotics) and the by-product of microbial metabolism can also confer anti-inflammatory, antimicrobial, antioxidant, and immunomodulatory effects on the host [7]. Therefore, postbiotics represent new groups of compounds that exhibit biological activities [50], and are shown in (Figure 1).
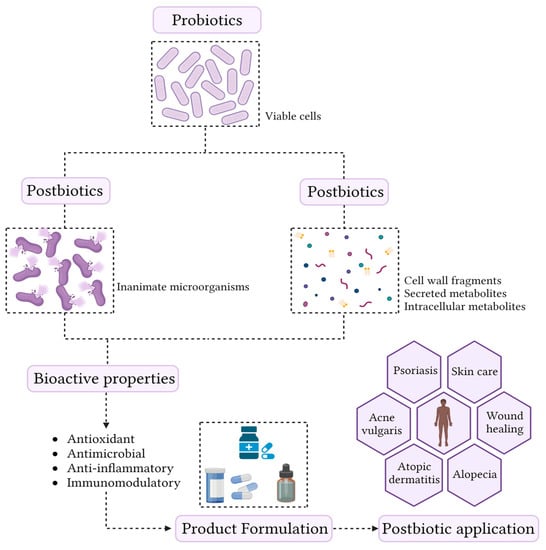
Figure 1.
Conceptualization of the term postbiotic and schematic representation of the skin health benefits.
The most important and well-known postbiotics are bacteriocins, short-chain fatty acids (SCFA), organic acids, and tryptophan (Trp) [50,51]. The mechanisms of action of these compounds are similar to those of probiotics and can be classified as direct or indirect. The direct mechanisms result in an action on the host cell, while the indirect action can stimulate the growth of microorganisms beneficial to health and inhibit the growth of pathogenic and opportunistic microorganisms [51,52]. However, these effects have been described primarily in the gut, but the impact of these compounds on epithelial cells and the skin microbiome have not yet been fully elucidated.
From a technological point of view, postbiotics have several advantages over probiotics, which include longer shelf life, stability over a wide temperature and pH range, defined chemical composition, no ability to transfer antibiotic resistances, and the fact that they can be used in immunosuppressed people [51]. In addition, postbiotics eliminate the need to maintain viable cells. These characteristics allow postbiotics to be incorporated into formulations, which makes their application innovative in the cosmetic ingredient market. Although this is a relatively new area, postbiotics are an emerging trend. Currently, there are about 17 companies that commercialize probiotics for cosmetic purposes, and most of these compounds are derived from bacteria belonging to the Lactobacillus genus. Among eukaryotes, these products are obtained almost exclusively from the yeast Saccharomyces cerevisiae [7]. There is a great prospect for the growth and development of the postbiotics market. However, this new area of science still lacks research, and new efforts should be made to identify and characterize new postbiotics from different microbial strains, whether probiotic or not.
4. Postbiotic Production Process
Using the information available in the literature, a flowsheet to produce postbiotics has been proposed (Figure 2). It is highlighted to show the main differences in the downstream steps to assist researchers and companies in decision measures. Initially, the proper probiotic strain is inserted in an inoculum tank bioreactor (IT-01) with small-scale volumes (1 to 10 L). After initial growth and establishing the exponential phase, the probiotic cells are transferred to a larger scale bioreactor (over 10 L), usually a stirring tank reactor operating in batches without aeration, as both Lactobacillus and Saccharomyces are favored by anaerobic conditions [53]. After reaching adequate biomass concentration, the fermentation broth is removed from the bioreactor and can be destined to three main postbiotic downstream processes: (i) conventional biomass removal; (ii) direct cell lysis and subsequent cell debris removal; and (iii) probiotic-derived secondary metabolite extraction. The conventional postbiotic production pathway begins with a centrifuge (C-01) that separates the biomass from the cell-free supernatant. The supernatant undergoes a filtration (F-01) with a rotating drum filter, and it is directed to a homogenizing tank (HT-01).
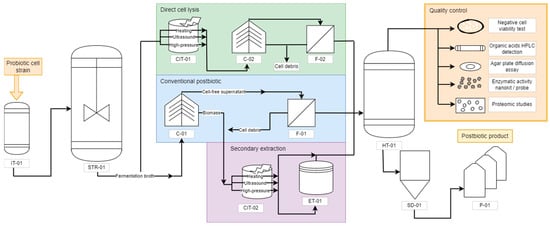
Figure 2.
Flowchart of postbiotic production. IT = inoculum tank; STR = stirred tank reactor; C = centrifuge; F = filter; CIT = cell inactivation tank; ET = extraction tank; HT = homogenizing tank; SD = spray dryer; P = packaging.
The fermentation broth can also be directly destined to cell lysis without previous concentration. The inactivation methods include ultrasound, high-pressure breakage, and enzymatic treatment, performed in the cell inactivation tank (CIT-01) [54,55]. This will liberate both intracellular and cell membrane/wall molecules into the solution, requiring further centrifugation (C-02) and filtration (F-02) to remove large cell debris before being directed to a homogenizing tank (HT-01). Another possibility is to perform cell lysis of the centrifuged biomass in a secondary pathway. The resulting biomass of the C-01 centrifuge can be coupled with the cell debris of other unit operations, such as the filtration F-01, and inactivated, resulting in other postbiotic product. The same inactivation methods can be applied to the C-01 biomass in a cell inactivation tank (CIT-02). Whenever necessary, the inactivation can be followed by specific extraction methods in an extraction tank (ET-01), where solvents can remove specific molecules from the original solution due to their stronger solubilizing properties—for instance, using ethanol or ethyl acetate to extract carbenicillin, cephalexin, cephalothin, and tetracycline [56]. After the inactivation, with or without extraction, the metabolites are destined to a homogenizing tank (HT-01).
The homogenizing tank is positioned at the end of each strategy to guarantee that the product will present a similar composition in all fractions. The most promising alternative when the desired product is in a solid particulate form is the spray dryer (SD-01) followed by packaging (P-01). Although the spray dryer uses heated air, it is less aggressive than the inactivation methods and generally presents protective compounds that are added before drying to soluble starch, such as glucose, sucrose, lactose, and maltodextrin [57].
Before being released into the market, the postbiotic should undergo quality control for its composition. Beyond the traditional beneficial features characterization, which can be performed similarly to the probiotic tests (non-toxicity, antipathogenic activity, and functional properties) [53], a negative cell viability test must be performed to guarantee the absence of viable cells. The most recommended test is direct plate cultivation without the use of several dilutions. Alternatives also include microscopies or real-time PCR, which may present obstacles due to the presence of cell debris and remaining genetic material in the solution [58]. Additionally, further characterization of the postbiotic is recommended, such as evaluation of the saccharides, organic acids, antibiotics, bacteriocins, proteins, and enzymes. Monosaccharides, N-acetylglucosamine, alcohols, and organic acids can be detected through chromatographic methods, such as HPLC and thin layer [59,60]. For the antibiotics and bacteriocins, although they can be identified through chromatography and spectrometry, their activity is better tested with agar diffusion well tests against classical pathogens to verify if the existing biocides remain active [61,62]. As for the proteins, an interesting strategy is to characterize a proteomic to find unnamed proteins and enzymes as well, other than the regularly known ones [63]. Enzymatic activity is difficult to detect in a general solution, yet it is possible to detect specific enzymes using nanokits or probes developed for the exclusive detection of their activity, such as the detection of sphingomyelinase through fluorescent nanoliposome [64].
However, it is important to note that the industrial production of postbiotics should be carried out with caution. There is not enough data in the literature demonstrating the effect of scale-up on their biological functions, since all studies were performed on a laboratory scale. Therefore, it is still necessary to evaluate the influence of each step on the stability and biological activity of postbiotics.
5. Postbiotics Formulation
The composition of skin cosmetic products contains up to 20 different ingredients necessary to provide the intended efficacy, safety, and commercial acceptability. Water, surfactants, preservatives, barrier agents, enhancers, humectants, and active ingredients are the main key ingredients, and the categorization of the final product (e.g., lotion, cream, moisturizing) depends on the concentrations of these chemical compounds [65]. Ultra-pure water is the basis of skin products, acting as the solvent basis to achieve the final consistency and contributing to the improvement of skin moisturizing levels [66]. Surfactants are amphiphilic substances with the capability to reduce the surface between liquids with different polarities due to the presence of both hydrophobic and hydrophilic moieties in their chemical structure [67], providing a homogeneous and uniform texture to the final product. Preservatives, such as parabens and formaldehyde, are substances able to prevent the growth of spoilage and harmful microorganisms, counteract the generation of reactive oxygen species (ROS), and impede the oxidation of cosmetics [68]. Barrier agents reduce the water loss and direct contact to sensitizer, irritant, or allergen compounds that may be present on the formulae through the creation of a thin, hydrophobic layer over the skin. Enhancers, such as denatured alcohol, glycols, and esters, are added in order to improve the penetration of active ingredients through lipid fluidization, lipid extraction, and lipid ordering mechanisms [69]. Humectants, as suggested, improve the hydration of the skin surface through the attraction of water from lower layers and posterior fixation in the stratum corneum via formation of hydrogen bounds [65].
In addition to this ‘basic’ composition, synthetic-active ingredients—encompassing anti-inflammatory, corticosteroids, immunosuppressive, and antimicrobial agents—are also added to the formulation of skin products directed towards the treatment of skin disorders, such as acne vulgaris, psoriasis, atopic dermatitis, and wound healing [70,71,72]. However, recent studies highlighted that prolonged exposition to these synthetic compounds as a sole treatment can result in numerous adverse events, including skin irritation, dryness, exfoliation, erythema, and striae [73,74]. As previously discussed, these reactions lead to a change in the structure of the microbial community, comprising the biological functions of the skin and severity increase in atopic dermatitis [75,76], acne vulgaris [77,78], and psoriasis [79,80]. This fact, allied to a market change to address the superior demand for green cosmetics, lead to the development of biocosmetics [66].
Among the biotechnological solutions presented, the addition of postbiotics as active ingredients proved to be one of the most promising due to the absence of bacteremia and fungaemia risks, besides the inherent stability during industrial processes and shelf life [9]. The formulation of cosmetic products containing postbiotics also have proven to be less costly, once it is unnecessary to maintain cell viability in the final product during transportation and storage. Golkar et al. [81] proposed the formulation of cold creams for wound healing in animal models by the direct addition (0.01% [w/w]) of lyophilized supernatant obtained after centrifugation of Lactobacillus fermentum, L. reuteri, and Bacillus subtilis ferments in the synthetic medium. Direct addition of LactoSporin® (2% [w/w]), an extracellular, low-weight protein metabolite produced by the patented probiotic B. coagulans MTCC 5856 [82], have also been evaluated by Majeed et al. [83] from the formulation of topical creams to the treatment of mild-to-moderate acne lesions.
Although cost-effective, the direct addition of postbiotics expose these substances to external conditions (e.g., UV light, temperature, pH, oxidation) in topical formulations, which may result in the loss of stability and biological activity. In this sense, the encapsulation with microparticles and nanoparticles is a viable option, providing the necessary protection and allowing the active ingredients to be retained in the top of epidermis [84]. A recent study conducted by Ashoori et al. [85] investigated the characterization and application of topical formulations, each containing 1 g of probiotic lysates from L. reuteri, L. fermentum, and B. subtilis sp. Natto in 100 g of chitosan nanogel (1% w/w), for wound healing in vivo. The characterization of the nanocapsules revealed spherical and uniform capsules with sizes ranging from 10 to 50 nm and appropriate physical stability, thus supporting the chitosan nanogel as an appropriate carrier for postbiotics in cosmetic formulae. Although it is a field recently investigated by science, cosmetic products containing postbiotics in their formulae are already available in the market with alleged claims and benefits, such as ProRenew Complex CLR™ NP (Onlystar Bio-Technology, Beijing, China), Bioptimized™ Guava (Innovacos, Mt. Arlington, NJ, USA), and PREBIOME™ (Radiant) [7].
6. Applications and Effects of Postbiotics in the Treatment of Skin Condition
Table 1 shows the main advances related to in vivo evaluation of the efficacy of postbiotics associated with skin health. The biological properties of postbiotics have been explored by researchers to develop alternative treatments for alopecia, acne vulgaris, atopic dermatitis, and wound healing. The postbiotics are derived from well-defined microorganisms or a combination of microorganisms of the Lactobacillus and Bacillus genera (Table 1). However, other bacterial (e.g., Bifidobacterium, Lactococcus, Leuconostoc, Pediococcus, Weissella, Enterococcus, and Fructobacillus) and fungal (e.g., Citeromyce, Cystofilobasidium, Hanseniaspora, Issatchenki, Pichia, and Saccharomyces) have already been described and characterized as probiotic microorganisms, and can still be explored as safe sources of postbiotics [53]. In general, the application of postbiotics occurs by incorporating these molecules in the formulation of creams for topical use or by encapsulation and oral administration [86,87]. Therefore, in the next topics, we will address the advances and in vivo applications of postbiotics, and the possible mechanisms of action related to skin health benefits.

Table 1.
In vivo evaluation of the efficacy of postbiotics for the treatment of diseases affecting the skin.
6.1. Alopecia
Alopecia areata (AA) is an autoimmune condition that affects the scalp, causing hair loss. In some more severe cases, AA can spread to the entire scalp (alopecia totalis) or even the body (alopecia universalis) [90]. Due to these characteristics, AA is often associated with mental health problems, especially social anxiety [91]. To date, there is no cure for AA [92]. Treatment for AA includes topical or intralesional application of corticosteroids for mild cases to immunotherapy with diphenylcyclopropenone or squaric acid dibutylester for severe cases [93]. However, during treatment, patients may experience some side effects, such as lymph node enlargement, blisters, headache, intense itching, weight gain, and hyperpigmentation [92,93,94]. In addition, the long-term application can lead to a process of skin atrophy [92].
Due to these conditions, some alternatives for the treatment of AA have been proposed and are under evaluation, such as low-level light therapy [95], Janus Kinase (JAK) inhibitors [96], and platelet-rich plasma (PRP) treatment, which is known as an ‘elixir’ for hair growth [97]. PRP is usually obtained by centrifuging the patient’s own blood and used to formulate a gel for topical application. However, the mechanism of action of PRP treatment has not been fully elucidated, but it is speculated that several growth factors that are present in platelets, mainly polypeptides, act by stimulating hair follicle cell differentiation and proliferation as well as inhibiting the process of apoptosis [98,99,100]. Although the use of PRP has shown great potential in several studies [101,102,103], the treatment still has many limitations, as there is no standardization of the production process and platelet concentration, besides presenting low stability, which makes topical applications difficult [88,98].
To overcome these barriers, scientists have sought modern alternatives, which include the use of bioactive peptides obtained from biotechnological tools combined with postbiotics [88,98]. This approach aims to mimic the growth factors found in PRP treatment and thus opens avenues for new therapeutic approaches. For instance, a double-blind randomized study of 160 people was conducted to evaluate the efficacy of a gel formulation containing postbiotics for topical application for the treatment of AA. These volunteers were randomly divided into two groups: (i) the treatment group (received the gel TR-PRP plus-Cel) and (ii) the control group (control received placebo). Among the treated patients, about 47.50% showed a complete regression of hair loss, while 13.75% exhibited a partial regression and only 6.25% of treated patients reported no response. In the control group, only 5% of individuals reported complete regression [88]. The positive effect of TR-PRP plus-Cels was associated with the postbiotic plantaricin A (PlnA) and Lactobacillus kunkeei-fermented bee bread (bee bread). PlnA is a peptide that presents several biological properties, among them the ability to induce key mediators of epithelial cell proliferation, migration, and differentiation, while bee bread is known for its immunomodulatory role. In addition, both compounds exhibit high antioxidant and antimicrobial activity [88,104,105,106]. Rinaldi et al. [88] speculated that the postbiotics present in TR-PRP plus-Cels may also positively modulate the structure of the scalp microbial community. However, further studies are still needed to evaluate the effect of microbial metabolites on the scalp and how these compounds can be used as an alternative method for and treatment of AA.
6.2. Acne Vulgaris
Acne vulgaris is considered a chronic inflammatory disease affecting the pilosebaceous unit (hair follicles in the skin associated with a sebaceous gland) present on the face, neck, chest, and coast [107]. Acne is characterized by papules, nodules, comedones, pustules, cysts, and often scars on the affected area [108,109]. The pathogenesis of acne is not fully known, but some studies indicate that factors such as androgen-induced excessive sebum production, hyperkeratinization, obstruction of the sebaceous follicles, bacterial colonization of the hair follicles by C. acnes, and inflammation are the main factors behind this disease [107,110]. Although acne is not a life-threatening disease, it can trigger a range of psychological problems that include anxiety, social isolation, and even suicidal intent [109,111,112]. In general, there is no ideal treatment for acne, but topical therapies with retinoids, benzoyl peroxide, and antibiotics, when used in combination, show efficacy in mild and moderate cases. On the other hand, in severe cases, oral antibiotics combined with the topical use of benzoyl peroxide are generally recommended [107,113,114].
Benzoyl peroxide has been used for the treatment of acne since 1934 and its mechanism consists of reducing the aerobic bacteria population by strong oxidation processes [27]. Furthermore, benzoyl peroxide does not allow C. acnes to develop resistance to antibiotics. Retinoids, on the other hand, can reduce the obstruction within the follicle, and are mainly used in cases where the patient presents comedonal and inflammatory acne. However, treatment with these compounds is usually associated with redness, irritation, dryness, and excessive flaking of the skin [113,115,116].
Postbiotics have been shown to be a potential alternative in the treatment of acne [117]. For instance, a study conducted by Majeed et al. [83] compared the effect of LactoSporin® (a filtered extract obtained from a fermented Bacillus coagulans MTCC 5856) with benzoyl peroxide in 64 individuals diagnosed with mild and moderate acne. Both treatments showed significant improvements in dermatological assessment of closed and open comedones and papule count. However, the effect of postbiotics on closed comedones appeared earlier compared to benzoyl peroxide, showing an advantage over the standard treatment. Furthermore, the application of LactoSporin resulted in a significant decrease in sebum secretion, leading to a reduction in oiliness, spots, and redness around acne. This effect may be associated with the ability of the postbiotic to inhibit the enzyme 5-alpha reductase, as shown in in vitro tests [118], as 5-alpha reductase plays an important role during the production of hormones that stimulate sebaceous gland secretion favoring the growth of C. acnes [119]. In addition, LactoSporin has been shown to have microbial activities against P. aeruginosa, S. aureus, S. epidermidis, and, most importantly, it was found to be effective against C. acnes. In addition, it was able to inhibit the formation of microbial biofilms [83,118].
Further studies involving the characterization and application of novel postbiotics for the treatment of acne vulgaris are still under development. An interesting approach was recently suggested by Chung et al. [120] and consists of the development of a postbiotic complex (PC), i.e., the use of two or more strains. This strategy has great potential, because it is difficult for a single strain to present all the desired biological properties. As the biological activities of various probiotics are known, it is possible to obtain postbiotics from these different strains and formulate new products with the desired characteristics, thus increasing their range of application. For example, Chung et al. [120] evaluated in vitro the biological activity of a CP produced from Lactobacillus helveticus HY7801 Lactobacillus lactis HY449. The results showed that the CP derived from these strains showed an antibacterial effect against S. aureus and C. acnes, in addition to anti-inflammatory activity, regulating the levels of inflammatory cytokines and hyaluronic acid in keratinocytes. The authors also evaluated the main metabolites present in CP and attributed these biological properties to the molecule’s hypoxanthine, 2-hydroxyisocaproic acid, succinic acid, ornithine, and GABA. However, it is still necessary to formulate products containing these potential postbiotics and to prove their efficacy through in vivo tests.
6.3. Atopic Dermatitis
Atopic dermatitis (AD) is a chronic inflammatory skin disease characterized by the presence of pruritic eczema. Although this disease is more common in children, adults are also affected [121,122,123]. Several studies have shown that the pathophysiology of AD is mainly associated with a process of immune system dysregulation, loss of skin barrier efficiency, microbial dysbiosis of the skin, and the itch/scratch cycle [122,124,125,126]. However, the literature suggests that problems in the epidermal barrier are the main factor responsible for AD. It is speculated that these alterations may be associated mainly with genetic/epigenetic and environmental factors [127,128]. Treatment of mild to moderate AD usually includes the use of corticosteroids, topical emollients, and calcineurin inhibitors. About 20% of patients present moderate to severe symptoms, making topical treatments not very effective due to the marked inflammation process [129,130]. Generally, these patients undergo phototherapy and treatment with systemic immune modulators [130]. However, these treatments are associated with significant adverse effects and do not offer long-term “cures”, while other patients may show resistance to these standard treatments [90,128].
Therefore, the use of probiotics and postbiotics has been proposed as an alternative treatment for AD [87,89,131]. For example, a double-blind placebo-controlled study showed that administration of heat-treated milk fermented by L. acidophilus L-92 exhibited immunoregulatory properties, as it was able to attenuate the symptoms of Japanese cedar pollen allergy [132]. Later, Hong et al. [89] and Inoue et al. [87] showed that the administration of both viable and heat-inactivated L. acidophilus L-92 also resulted in a significant improvement in AD symptoms in adults and children. In both studies, the improvement of patients treated with the L-92 strain was mainly associated with the suppression of Th2 dominant inflammation. In addition, no serious side effects were observed, showing that, at this early-stage, L-92 can be used as a food supplement to reduce the dose of steroidal anti-inflammatory ointments needed for atopic treatment.
In general, several other studies also show biological activity, demonstrating the potential of L. acidophilus L-92 to be applied as postbiotics [133,134,135,136]. However, a detailed study of the metabolic profile of this strain is needed to identify the molecules that may be associated with the beneficial effects observed by the authors [87,89,131]. These findings may open new avenues for the development of topical ointments for the treatment of mild and moderate cases, because, in addition to efficacy, postbiotics tend to present fewer side effects to the skin, thus being able to replace or reduce traditional treatments.
6.4. Wound Healing
The skin is the largest organ in the body and to perform its regulatory and barrier functions it needs to be intact. Skin breakdowns due to injury, disease, or operation are defined as wounds [137]. Although this organ can restore its integrity spontaneously, wound care is extremely important, mainly for the protection of the open site, preventing infection, dryness, and relieving pain [138]. Classically, this process of wound healing is divided into four distinct phases: hemostasis, inflammation, proliferation, and tissue remodeling. Several factors can slow or impair the healing process, among them microbial proliferation in the wound [139]. Microbial growth and maintenance of these microorganisms in wounds are usually associated with biofilm production. In advanced stages, there is a risk that the immune system will be unable to contain this infection. This process can result in a severe clinical picture of marked inflation or purulence [140].
Therefore, research is needed to develop new therapeutic agents that can assist in wound treatment, decreasing the infection rate, and accelerating the healing process. Postbiotics are valuable compounds as they have several biological properties (e.g., immunomodulatory, anti-inflammatory, antimicrobial, and angiogenic) that can favor wound healing. Recently, Golkar et al. [81] conducted a study in rats to evaluate the healing action of three new ice creams containing postbiotics derived from L. fermentum ATCC 9338, L. reuteri ATCC 23272 and B. subtilis sp. natto ATCC 15245. Although the three formulations showed a significant improvement in the healing process compared to the control, ice cream containing postbiotics derived from B. subtilis sp. natto showed more promising results. In general, this group presented a higher content of hydroxyproline, a basic component found in collagen. Thus, the increase in hydroxyproline is used as a parameter to assess the amount of collagen that has been produced, indicating greater progression in the healing process. Furthermore, histopathological analyses also revealed that the cream containing the postbiotic B. subtilis sp. natto showed less degree of inflammation while a mild fibrosis process was observed in all groups except the untreated group, which resulted in moderate fibrosis [81]. These studies provide insights for the development of new postbiotic-based products to be considered as an alternative treatment in wound healing.
7. Patents in the Field of Postbiotics
A patent search was carried out in the Derwent Innovations Index Database on 29 November 2022, using the terms [(post$biotic* OR para$probiotic*) AND (skin OR cutis OR derm* OR epiderm*)], in the field Topic, and 34 patent families were retrieved related to postbiotics for skin applications.
From the total patent documents, 88% were filed by companies, 9% by universities, and 3% by an individual. This indicates a high level of technology readiness at the industrial sector. The main assignee, with five patent documents, was the company Eligo Bioscience, founded in 2014, by scientists from the Rockefeller University and the Massachussetts Institute of Technology and based in Paris, France. Their technology is related to postbiotic compositions (microbial lysates) containing bacteriocin(s) and/or endolysin(s), heterologously secreted by Lactobacillus rhamnosus. In second position is the company Chambio from Taiwan with three documents, and in third position are the companies Biowish Technologies (Cincinnati, OH, USA), Dong-A Pharmaceutical (Seoul, Republic of Korea), Eczacibasi Tuketim Urunleri Sanayi (Gebze, Turkey), and Gallinee (London, UK), each of which has two patent documents.
Regarding the timeline of patent publications, 44% of the documents were published in 2022, 32% in 2021, 15% in 2020, and the remaining 9% were published between 2015 and 2019, which indicates that the technology is recent and still in its growing phase. A description about the postbiotic compositions and their main applications, as reported in the patent documents filed by each assignee, is presented in Table 2.

Table 2.
Patent documents related to postbiotics for skin applications.
8. Conclusions
Recent advances in high-throughput sequencing technologies have provided new insight into the complex interactions between commensal microorganisms and skin pathogens. When the balance of this microbiome is altered, either by skin breakdown or by a clinical condition of immunosuppression, these commensal relationships can turn into a pathogenic process due to the marked growth of unwanted species. It has been shown that these changes are associated with some skin pathologies, such as acne, atopic dermatitis, psoriasis, alopecia, and can slow down the wound-healing process. In general, treatments for these diseases include the use of synthetic compounds, such as immunosuppressants, corticosteroids, and antibiotics. However, the application of these compounds results in several side effects, including lymph node enlargement, blisters, headache, intense itching, weight gain, and hyperpigmentation. In recent years, several studies have highlighted the positive effect of postbiotics and the therapeutic potential of these compounds through in vitro tests. On the other hand, clinical research focused on topical applications for the treatment of skin diseases is still in its infancy and little data is available. However, the results are promising and have shown equal or superior efficacy to standard treatments, and, most importantly, no side effects. Despite scientific evidence of the beneficial effects of postbiotics in clinical trials, the mechanisms of action are not fully understood. In addition, research has neglected the effect of the postbiotic application on the skin microbiome after treatment. This information is extremely relevant and should be addressed in future randomized placebo-controlled clinical trials. Finally, the patent analysis revealed that there is a great interest in probiotics from industries in the technologies, as 88% of the patents were filed by companies, 9% by universities, and 3% by an individual.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation9030264/s1. Table S1: Skin microbial groups identified by culture-independent methods; Table S2: Patent documents related to postbiotics for skin applications.
Author Contributions
Conceptualization A.d.S.V., G.V.d.M.P. and C.R.S.; writing—original draft preparation, A.d.S.V., G.V.d.M.P., A.C.d.O., D.P.d.C.N., L.W.H., S.G.K. and V.T.S.; writing—review and editing, A.d.S.V., G.V.d.M.P., S.G.K., V.T.S. and C.R.S.; visualization, A.d.S.V. and L.W.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financed by the Coordination for the Improvement of Higher Education Personnel (CAPES) and the National Council for Scientific and Technological Development (CNPq).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- O’Brien, M.T.; O’Sullivan, O.; Claesson, M.J.; Cotter, P.D. The Athlete Gut Microbiome and Its Relevance to Health and Performance: A Review. Sports Med. 2022, 52, 119–128. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between Microbiota and Immunity in Health and Disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Hooks, K.B.; O’Malley, M.A. Contrasting Strategies: Human Eukaryotic versus Bacterial Microbiome Research. J. Eukaryot. Microbiol. 2020, 67, 279–295. [Google Scholar] [CrossRef]
- Boxberger, M.; Cenizo, V.; Cassir, N.; la Scola, B. Challenges in Exploring and Manipulating the Human Skin Microbiome. Microbiome 2021, 9, 125. [Google Scholar] [CrossRef]
- Bay, L.; Barnes, C.J.; Fritz, B.G.; Thorsen, J.; Restrup, M.E.M.; Rasmussen, L.; Sørensen, J.K.; Hesselvig, A.B.; Odgaard, A.; Hansen, A.J.; et al. Universal Dermal Microbiome in Human Skin. mBio 2020, 11, e02945-19. [Google Scholar] [CrossRef]
- Gueniche, A.; Perin, O.; Bouslimani, A.; Landemaine, L.; Misra, N.; Cupferman, S.; Aguilar, L.; Clavaud, C.; Chopra, T.; Khodr, A. Advances in Microbiome-Derived Solutions and Methodologies Are Founding a New Era in Skin Health and Care. Pathogens 2022, 11, 121. [Google Scholar] [CrossRef]
- Duarte, M.; Oliveira, A.L.; Oliveira, C.; Pintado, M.; Amaro, A.; Madureira, A.R. Current Postbiotics in the Cosmetic Market—An Update and Development Opportunities. Appl. Microbiol. Biotechnol. 2022, 106, 5879–5891. [Google Scholar] [CrossRef]
- Lee, M.-J.; Zang, Z.-L.; Choi, E.-Y.; Shin, H.-K.; Ji, G.-E. Cytoskeleton Reorganization and Cytokine Production of Macrophages by Bifidobacterial Cells and Cell-Free Extracts. J. Microbiol. Biotechnol. 2002, 12, 398–405. [Google Scholar]
- Salminen, S.; Collado, M.C.; Endo, A.; Hill, C.; Lebeer, S.; Quigley, E.M.M.; Sanders, M.E.; Shamir, R.; Swann, J.R.; Szajewska, H.; et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) Consensus Statement on the Definition and Scope of Postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 649–667. [Google Scholar] [CrossRef]
- Vinderola, G.; Sanders, M.E.; Salminen, S. The Concept of Postbiotics. Foods 2022, 11, 1077. [Google Scholar] [CrossRef]
- Ogai, K.; Nana, B.C.; Lloyd, Y.M.; Arios, J.P.; Jiyarom, B.; Awanakam, H.; Esemu, L.F.; Hori, A.; Matsuoka, A.; Nainu, F.; et al. Skin Microbiome Profile of Healthy Cameroonians and Japanese. Sci. Rep. 2022, 12, 1364. [Google Scholar] [CrossRef]
- Lee, H.-J.; Kim, M. Skin Barrier Function and the Microbiome. Int. J. Mol. Sci. 2022, 23, 13071. [Google Scholar] [CrossRef]
- Howard, B.; Bascom, C.C.; Hu, P.; Binder, R.L.; Fadayel, G.; Huggins, T.G.; Jarrold, B.B.; Osborne, R.; Rocchetta, H.L.; Swift, D.; et al. Aging-Associated Changes in the Adult Human Skin Microbiome and the Host Factors That Affect Skin Microbiome Composition. J. Investig. Dermatol. 2022, 142, 1934–1946.e21. [Google Scholar] [CrossRef]
- Pistone, D.; Meroni, G.; Panelli, S.; D’auria, E.; Acunzo, M.; Pasala, A.R.; Zuccotti, G.V.; Bandi, C.; Drago, L. A Journey on the Skin Microbiome: Pitfalls and Opportunities. Int. J. Mol. Sci. 2021, 22, 9846. [Google Scholar] [CrossRef]
- Pillsbury, D.M.; Shelley, W.B. Dermatology. Annu. Rev. Med. 1954, 5, 363–388. [Google Scholar] [CrossRef]
- Fredricks, D.N. Microbial Ecology of Human Skin in Health and Disease. J. Investig. Dermatol. Symp. Proc. 2001, 6, 167–169. [Google Scholar] [CrossRef]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The Human Skin Microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Ederveen, T.H.A.; Smits, J.P.H.; Boekhorst, J.; Schalkwijk, J.; van den Bogaard, E.H.; Zeeuwen, P.L.J.M. Skin Microbiota in Health and Disease: From Sequencing to Biology. J. Dermatol. 2020, 47, 1110–1118. [Google Scholar] [CrossRef]
- Nelson, K.E.; Weinstock, G.M.; Highlander, S.K.; Worley, K.C.; Creasy, H.H.; Wortman, J.R.; Rusch, D.B.; Mitreva, M.; Sodergren, E.; Chinwalla, A.T.; et al. A Catalog of Reference Genomes from the Human Microbiome. Science 2010, 328, 994–999. [Google Scholar] [CrossRef]
- Meslier, V.; Quinquis, B.; da Silva, K.; Plaza Oñate, F.; Pons, N.; Roume, H.; Podar, M.; Almeida, M. Benchmarking Second and Third-Generation Sequencing Platforms for Microbial Metagenomics. Sci. Data 2022, 9, 694. [Google Scholar] [CrossRef]
- Ferretti, P.; Farina, S.; Cristofolini, M.; Girolomoni, G.; Tett, A.; Segata, N. Experimental Metagenomics and Ribosomal Profiling of the Human Skin Microbiome. Exp. Dermatol. 2017, 26, 211–219. [Google Scholar] [CrossRef]
- Hammoudi, N.; Cassagne, C.; Million, M.; Ranque, S.; Kabore, O.; Drancourt, M.; Zingue, D.; Bouam, A. Investigation of Skin Microbiota Reveals Mycobacterium Ulcerans-Aspergillus Sp. Trans-Kingdom Communication. Sci. Rep. 2021, 11, 3777. [Google Scholar] [CrossRef]
- Oh, J.; Byrd, A.L.; Park, M.; Kong, H.H.; Segre, J.A. Temporal Stability of the Human Skin Microbiome. Cell 2016, 165, 854–866. [Google Scholar] [CrossRef]
- Dekio, I.; Hayashi, H.; Sakamoto, M.; Kitahara, M.; Nishikawa, T.; Suematsu, M.; Benno, Y. Detection of Potentially Novel Bacterial Components of the Human Skin Microbiota Using Culture-Independent Molecular Profiling. J. Med. Microbiol. 2005, 54, 1231–1238. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; Green, E.D.; et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef]
- Capone, K.A.; Dowd, S.E.; Stamatas, G.N.; Nikolovski, J. Diversity of the Human Skin Microbiome Early in Life. J. Investig. Dermatol. 2011, 131, 2026–2032. [Google Scholar] [CrossRef]
- Chng, K.R.; Tay, A.S.L.; Li, C.; Ng, A.H.Q.; Wang, J.; Suri, B.K.; Matta, S.A.; McGovern, N.; Janela, B.; Wong, X.F.C.C.; et al. Whole Metagenome Profiling Reveals Skin Microbiome-Dependent Susceptibility to Atopic Dermatitis Flare. Nat. Microbiol. 2016, 1, 16106. [Google Scholar] [CrossRef]
- Fyhrquist, N.; Muirhead, G.; Prast-Nielsen, S.; Jeanmougin, M.; Olah, P.; Skoog, T.; Jules-Clement, G.; Feld, M.; Barrientos-Somarribas, M.; Sinkko, H.; et al. Microbe-Host Interplay in Atopic Dermatitis and Psoriasis. Nat. Commun. 2019, 10, 4703. [Google Scholar] [CrossRef]
- Alkema, W.; Boekhorst, J.; Eijlander, R.T.; Schnittger, S.; de Gruyter, F.; Lukovac, S.; Schilling, K.; Kortman, G.A.M. Charting Host-Microbe Co-Metabolism in Skin Aging and Application to Metagenomics Data. PLoS ONE 2021, 16, e0258960. [Google Scholar] [CrossRef]
- Ahle, C.M.; Stødkilde, K.; Poehlein, A.; Bömeke, M.; Streit, W.R.; Wenck, H.; Reuter, J.H.; Hüpeden, J.; Brüggemann, H. Interference and Co-Existence of Staphylococci and Cutibacterium acnes within the Healthy Human Skin Microbiome. Commun. Biol. 2022, 5, 923. [Google Scholar] [CrossRef]
- Saheb Kashaf, S.; Proctor, D.M.; Deming, C.; Saary, P.; Hölzer, M.; Mullikin, J.; Thomas, J.; Young, A.; Bouffard, G.; Barnabas, B.; et al. Integrating Cultivation and Metagenomics for a Multi-Kingdom View of Skin Microbiome Diversity and Functions. Nat. Microbiol. 2022, 7, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Barnard, E.; Shi, B.; Kang, D.; Craft, N.; Li, H. The Balance of Metagenomic Elements Shapes the Skin Microbiome in Acne and Health. Sci. Rep. 2016, 6, 39491. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Byrd, A.L.; Deming, C.; Conlan, S.; Kong, H.H.; Segre, J.A.; Barnabas, B.; Blakesley, R.; Bouffard, G.; Brooks, S.; et al. Biogeography and Individuality Shape Function in the Human Skin Metagenome. Nature 2014, 514, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Dréno, B.; Araviiskaia, E.; Berardesca, E.; Gontijo, G.; Sanchez Viera, M.; Xiang, L.F.; Martin, R.; Bieber, T. Microbiome in Healthy Skin, Update for Dermatologists. J. Eur. Acad. Dermatol. Venereol. 2016, 30, 2038–2047. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, J.J.; Myeong, N.R.; Kim, T.; Kim, D.A.; An, S.; Kim, H.; Park, T.; Jang, S.I.; Yeon, J.H.; et al. Segregation of Age-Related Skin Microbiome Characteristics by Functionality. Sci. Rep. 2019, 9, 16748. [Google Scholar] [CrossRef]
- Roth, R.R.; James, W.D. Microbial Ecology of the Skin. Annu. Rev. Microbiol. 1988, 42, 441–464. [Google Scholar] [CrossRef]
- Paulino, L.C.; Tseng, C.H.; Strober, B.E.; Blaser, M.J. Molecular Analysis of Fungal Microbiota in Samples from Healthy Human Skin and Psoriatic Lesions. J. Clin. Microbiol. 2006, 44, 2933–2941. [Google Scholar] [CrossRef]
- Findley, K.; Grice, E.A. The Skin Microbiome: A Focus on Pathogens and Their with Skin Disease. PLoS Pathog. 2014, 10, e1004436. [Google Scholar] [CrossRef]
- Murillo, N.; Aubert, J.; Raoult, D. Microbiota of Demodex Mites from Rosacea Patients and Controls. Microb. Pathog. 2014, 71–72, 37–40. [Google Scholar] [CrossRef]
- Hannigan, G.D.; Meisel, J.S.; Tyldsley, A.S.; Zheng, Q.; Hodkinson, B.P.; Sanmiguel, A.J.; Minot, S.; Bushman, F.D.; Grice, E.A. The Human Skin Double-Stranded DNA Virome: Topographical and Temporal Diversity, Genetic Enrichment, and Dynamic Associations with the Host Microbiome. mBio 2015, 6, e01578-15. [Google Scholar] [CrossRef]
- Foulongne, V.; Sauvage, V.; Hebert, C.; Dereure, O.; Cheval, J.; Gouilh, M.A.; Pariente, K.; Segondy, M.; Burguière, A.; Manuguerra, J.C.; et al. Human Skin Microbiota: High Diversity of DNA Viruses Identified on the Human Skin by High Throughput Sequencing. PLoS ONE 2012, 7, e38499. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chan, H.H.; Ni, M.Y.; Lam, W.W.; Chan, W.M.M.; Pang, H. Bacteriophage of the Skin Microbiome in Patients with Psoriasis and Healthy Family Controls. J. Investig. Dermatol. 2020, 140, 182–190.e5. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Bushman, F.D. The Human Virome: Assembly, Composition and Host Interactions. Nat. Rev. Microbiol. 2021, 19, 514–527. [Google Scholar] [CrossRef]
- Park, S.Y.; Kim, H.S.; Lee, S.H.; Kim, S. Characterization and Analysis of the Skin Microbiota in Acne: Impact of Systemic Antibiotics. J. Clin. Med. 2020, 9, 168. [Google Scholar] [CrossRef]
- Roux, P.F.; Oddos, T.; Stamatas, G. Deciphering the Role of Skin Surface Microbiome in Skin Health: An Integrative Multiomics Approach Reveals Three Distinct Metabolite—Microbe Clusters. J. Investig. Dermatol. 2022, 142, 469–479.e5. [Google Scholar] [CrossRef]
- Tett, A.; Pasolli, E.; Farina, S.; Truong, D.T.; Asnicar, F.; Zolfo, M.; Beghini, F.; Armanini, F.; Jousson, O.; de Sanctis, V.; et al. Unexplored Diversity and Strain-Level Structure of the Skin Microbiome Associated with Psoriasis. NPJ Biofilms Microbiomes 2017, 3, 14. [Google Scholar] [CrossRef]
- Alekseyenko, A.V.; Perez-Perez, G.I.; de Souza, A.; Strober, B.; Gao, Z.; Bihan, M.; Li, K.; Methé, B.A.; Blaser, M.J. Community Differentiation of the Cutaneous Microbiota in Psoriasis. Microbiome 2013, 1, 31. [Google Scholar] [CrossRef]
- Woo, Y.R.; Lee, S.H.; Cho, S.H.; Lee, J.D.; Kim, H.S. Characterization and Analysis of the Skin Microbiota in Rosacea: Impact of Systemic Antibiotics. J. Clin. Med. 2020, 9, 185. [Google Scholar] [CrossRef]
- Vallejo-Cordoba, B.; Castro-López, C.; García, H.S.; González-Córdova, A.F.; Hernández-Mendoza, A. Postbiotics and Paraprobiotics: A Review of Current Evidence and Emerging Trends. In Advances in Food and Nutrition Research; Academic Press Inc.: Cambridge, MA, USA, 2020; Volume 94, pp. 1–34. ISBN 9780128202180. [Google Scholar]
- Cuevas-González, P.F.; Liceaga, A.M.; Aguilar-Toalá, J.E. Postbiotics and Paraprobiotics: From Concepts to Applications. Food Res. Int. 2020, 136, 109502. [Google Scholar] [CrossRef]
- Martyniak, A.; Medyńska-Przęczek, A.; Wędrychowicz, A.; Skoczeń, S.; Tomasik, P.J. Prebiotics, Probiotics, Synbiotics, Paraprobiotics and Postbiotic Compounds in IBD. Biomolecules 2021, 11, 1903. [Google Scholar] [CrossRef]
- Keshari, S.; Wang, Y.; Herr, D.R.; Wang, S.M.; Yang, W.C.; Chuang, T.H.; Chen, C.L.; Huang, C.M. Skin Cutibacterium acnes Mediates Fermentation to Suppress the Calcium Phosphate-induced Itching: A Butyric Acid Derivative with Potential for Uremic Pruritus. J. Clin. Med. 2020, 9, 312. [Google Scholar] [CrossRef]
- Pereira, G.V.d.M.; de Oliveira Coelho, B.; Magalhães Júnior, A.I.; Thomaz-Soccol, V.; Soccol, C.R. How to Select a Probiotic? A Review and Update of Methods and Criteria. Biotechnol. Adv. 2018, 36, 2060–2076. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.; Kim, J.H.; Baek, J.; Kim, W. Improvement of Cutaneous Wound Healing via Topical Application of Heat-Killed Lactococcus chungangensis Cau 1447 on Diabetic Mice. Nutrients 2021, 13, 2666. [Google Scholar] [CrossRef] [PubMed]
- Ácsová, A.; Hojerová, J.; Martiniaková, S. Efficacy of Postbiotics against Free Radicals and UV Radiation. Chem. Pap. 2022, 76, 2357–2364. [Google Scholar] [CrossRef]
- Kumar, D.; Kumar, S.; Kumar, A. Extraction and Characterization of Secondary Metabolites Produced by Bacteria Isolated from Industrial Wastewater. J. Water Process Eng. 2021, 40, 108. [Google Scholar] [CrossRef]
- Tantratian, S.; Pradeamchai, M. Select a Protective Agent for Encapsulation of Lactobacillus plantarum. LWT 2020, 123, 109075. [Google Scholar] [CrossRef]
- Willey, J.M.; Sherwood, L.M.; Woolverton, C.J. Prescott, Harley, and Klein’s Microbology, 7th ed.; Willey, J.M., Sherwood, L.M., Woolverton, C.J., Eds.; The McGraw-Hill Companies: New York, NY, USA, 2008; ISBN 978-0-07-299291-5. [Google Scholar]
- Tu, N.H.K.; Thien, P.V.M. Detection of N-Acetyl-D-Glucosamine in Hyaluronan by Thin Layer Chromatography. IFMBE Proc. 2013, 40 IFMBE, 174–177. [Google Scholar] [CrossRef]
- Ibáñez, A.B.; Bauer, S. Analytical Method for the Determination of Organic Acids in Dilute Acid Pretreated Biomass Hydrolysate by Liquid Chromatography-Time-of-Flight Mass Spectrometry. Biotechnol. Biofuels 2014, 7, 145. [Google Scholar] [CrossRef]
- Messi, P.; Bondi, M.; Sabia, C.; Battini, R.; Manicardi, G. Detection and Preliminary Characterization of a Bacteriocin (Plantaricin 35d) Produced by a Lactobacillus plantarum Strain. Artic. Int. J. Food Microbiol. 2001, 64, 193–198. [Google Scholar] [CrossRef]
- Choeisoongnern, T.; Sivamaruthi, B.S.; Sirilun, S.; Peerajan, S.; Choiset, Y.; Rabesona, H.; Haertlé, T.; Chaiyasut, C. Screening and Identification of Bacteriocin-like Inhibitory Substances Producing Lactic Acid Bacteria from Fermented Products. Food Sci. Technol. 2019, 40, 571–579. [Google Scholar] [CrossRef]
- Cicenia, A.; Scirocco, A.; Carabotti, M.; Pallotta, L.; Marignani, M.; Severi, C. Postbiotic Activities of Lactobacilli-Derived Factors. J. Clin. Gastroenterol. 2014, 48, S18–S22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, W.; Zhang, J.; Li, Z.; Ma, H.; Zhao, Z. Facile Method for Specifically Sensing Sphingomyelinase in Cells and Human Urine Based on a Ratiometric Fluorescent Nanoliposome Probe. Anal. Chem. 2021, 93, 11775–11784. [Google Scholar] [CrossRef] [PubMed]
- MacFarlane, B. Common Cosmetic Ingredients: Chemistry, Actions, Safety and Products. In Cosmetic Formulation: Principles and Practice; Benson, H.A.E., Roberts, M.S., Leite-Silva, V.R., Walters, K.A., Eds.; CRC Press: Boca Raton, FL, USA, 2019; pp. 105–118. ISBN 9781482235395. [Google Scholar]
- Goyal, N.; Jerold, F. Biocosmetics: Technological Advances and Future Outlook. Environ. Sci. Pollut. Res. 2021, 1–22. [Google Scholar] [CrossRef]
- Lourith, N.; Kanlayavattanakul, M. Natural Surfactants Used in Cosmetics: Glycolipids. Int. J. Cosmet. Sci. 2009, 31, 255–261. [Google Scholar] [CrossRef]
- Nowak, K.; Jabłońska, E.; Ratajczak-Wrona, W. Controversy around Parabens: Alternative Strategies for Preservative Use in Cosmetics and Personal Care Products. Environ. Res. 2021, 198, 110488. [Google Scholar] [CrossRef]
- Lane, M.E. Skin Penetration Enhancers. Int. J. Pharm. 2013, 447, 12–21. [Google Scholar] [CrossRef]
- Mavranezouli, I.; Daly, C.H.; Welton, N.J.; Deshpande, S.; Berg, L.; Bromham, N.; Arnold, S.; Phillippo, D.M.; Wilcock, J.; Xu, J.; et al. A Systematic Review and Network Meta-Analysis of Topical Pharmacological, Oral Pharmacological, Physical and Combined Treatments for Acne Vulgaris. Br. J. Dermatol. 2022, 187, 639–649. [Google Scholar] [CrossRef]
- Bakshi, H.; Nagpal, M.; Singh, M.; Dhingra, G.A.; Aggarwal, G. Treatment of Psoriasis: A Comprehensive Review of Entire Therapies. Curr. Drug Saf. 2020, 15, 82–104. [Google Scholar] [CrossRef]
- Calabrese, G.; Licata, G.; Gambardella, A.; De Rosa, A.; Alfano, R.; Argenziano, G. Topical and Conventional Systemic Treatments in Atopic Dermatitis: Have They Gone Out of Fashion? Dermatol. Pract. Concept. 2022, 12, e2022155. [Google Scholar] [CrossRef]
- Otlewska, A.; Baran, W.; Batycka-Baran, A. Adverse Events Related to Topical Drug Treatments for Acne Vulgaris. Expert Opin. Drug Saf. 2020, 19, 513–521. [Google Scholar] [CrossRef]
- Frantz, T.; Wright, E.G.; Balogh, E.A.; Cline, A.; Adler-Neal, A.L.; Feldman, S.R. Topical and Oral Therapies for Childhood Atopic Dermatitis and Plaque Psoriasis. Children 2019, 6, 125. [Google Scholar] [CrossRef] [PubMed]
- Schmid, B.; Künstner, A.; Fähnrich, A.; Bersuch, E.; Schmid-Grendelmeier, P.; Busch, H.; Glatz, M.; Bosshard, P.P. Dysbiosis of Skin Microbiota with Increased Fungal Diversity Is Associated with Severity of Disease in Atopic Dermatitis. J. Eur. Acad. Dermatol. Venereol. 2022, 36, 1811–1819. [Google Scholar] [CrossRef] [PubMed]
- Moosbrugger-Martinz, V.; Hackl, H.; Gruber, R.; Pilecky, M.; Knabl, L.; Orth-Höller, D.; Dubrac, S. Initial Evidence of Distinguishable Bacterial and Fungal Dysbiosis in the Skin of Patients with Atopic Dermatitis or Netherton Syndrome. J. Investig. Dermatol. 2021, 141, 114–123. [Google Scholar] [CrossRef]
- Dréno, B.; Dagnelie, M.A.; Khammari, A.; Corvec, S. The Skin Microbiome: A New Actor in Inflammatory Acne. Am. J. Clin. Dermatol. 2020, 21, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; You, Z.X.; Lin, Y.X.; Liu, H.Y.; Su, J. Skin Microbiome Differences Relate to the Grade of Acne Vulgaris. J. Dermatol. 2019, 46, 787–790. [Google Scholar] [CrossRef] [PubMed]
- Stehlikova, Z.; Kostovcik, M.; Kostovcikova, K.; Kverka, M.; Juzlova, K.; Rob, F.; Hercogova, J.; Bohac, P.; Pinto, Y.; Uzan, A.; et al. Dysbiosis of Skin Microbiota in Psoriatic Patients: Co-Occurrence of Fungal and Bacterial Communities. Front. Microbiol. 2019, 10, 438. [Google Scholar] [CrossRef] [PubMed]
- Visser, M.J.E.; Kell, D.B.; Pretorius, E. Bacterial Dysbiosis and Translocation in Psoriasis Vulgaris. Front. Cell Infect. Microbiol. 2019, 9, 7. [Google Scholar] [CrossRef] [PubMed]
- Golkar, N.; Ashoori, Y.; Heidari, R.; Omidifar, N.; Abootalebi, S.N.; Mohkam, M.; Gholami, A. A Novel Effective Formulation of Bioactive Compounds for Wound Healing: Preparation, in Vivo Characterization, and Comparison of Various Postbiotics Cold Creams in a Rat Model. Evid.-Based Complement. Altern. Med. 2021, 2021, 8577116. [Google Scholar] [CrossRef]
- Majeed, M.; Nagabhushanam, K.; Arumugam, S.; Ali, F. Method of Producing Partially Purified Extracellular Metabolite Products from Bacillus Coagulans and Biological Applications Thereof. U.S. Patent 9,596,861, 21 March 2017. [Google Scholar]
- Majeed, M.; Majeed, S.; Nagabhushanam, K.; Mundkur, L.; Rajalakshmi, H.R.; Shah, K.; Beede, K. Novel Topical Application of a Postbiotic, Lactosporin®, in Mild to Moderate Acne: A Randomized, Comparative Clinical Study to Evaluate Its Efficacy, Tolerability and Safety. Cosmetics 2020, 7, 70. [Google Scholar] [CrossRef]
- Łętocha, A.; Miastkowska, M.; Sikora, E. Preparation and Characteristics of Alginate Microparticles for Food, Pharmaceutical and Cosmetic Applications. Polymers 2022, 14, 3834. [Google Scholar] [CrossRef]
- Ashoori, Y.; Mohkam, M.; Heidari, R.; Abootalebi, S.N.; Mousavi, S.M.; Hashemi, S.A.; Golkar, N.; Gholami, A. Development and in Vivo Characterization of Probiotic Lysate-Treated Chitosan Nanogel as a Novel Biocompatible Formulation for Wound Healing. Biomed Res. Int. 2020, 2020, 8868618. [Google Scholar] [CrossRef] [PubMed]
- Catic, T.; Pehlivanovic, B.; Pljakic, N.; Balicevac, A. The Moisturizing Efficacy of a Proprietary Dermo-Cosmetic Product (CLS02021) versus Placebo in a 4-Week Application Period. Med. Arch. 2022, 76, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Kambara, T.; Murata, N.; Komori-Yamaguchi, J.; Matsukura, S.; Takahashi, Y.; Ikezawa, Z.; Aihara, M. Effects of Oral Administration of Lactobacillus acidophilus L-92 on the Symptoms and Serum Cytokines of Atopic Dermatitis in Japanese Adults: A Double-Blind, Randomized, Clinical Trial. Int. Arch. Allergy Immunol. 2014, 165, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, F.; Trink, A.; Pinto, D. Efficacy of Postbiotics in a PRP-Like Cosmetic Product for the Treatment of Alopecia Area Celsi: A Randomized Double-Blinded Parallel-Group Study. Dermatol. Ther. 2020, 10, 483–493. [Google Scholar] [CrossRef]
- Hoang, B.X.; Shaw, G.; Pham, P.; Levine, S.A. Lactobacillus rhamnosus Cell Lysate in the Management of Resistant Childhood Atopic Eczema. Inflamm. Allergy-Drug Targets 2010, 9, 192–196. [Google Scholar] [CrossRef]
- Darwin, E.; Hirt, P.; Fertig, R.; Doliner, B.; Delcanto, G.; Jimenez, J. Alopecia Areata: Review of Epidemiology, Clinical Features, Pathogenesis, and New Treatment Options. Int. J. Trichology 2018, 10, 51–60. [Google Scholar] [CrossRef]
- Segal-Engelchin, D.; Shvarts, S. Does Severity of Hair Loss Matter? Factors Associated with Mental Health Outcomes in Women Irradiated for Tinea Capitis in Childhood. Int. J. Environ. Res. Public Health 2020, 17, 7388. [Google Scholar] [CrossRef]
- Chiang, K.S.; Mesinkovska, N.A.; Piliang, M.P.; Bergfeld, W.F. Clinical Efficacy of Diphenylcyclopropenone in Alopecia Areata: Retrospective Data Analysis of 50 Patients. J. Investig. Dermatol. Symp. Proc. 2015, 17, 50–55. [Google Scholar] [CrossRef]
- Gupta, A.K.; Carviel, J.; Abramovits, W. Treating Alopecia Areata: Current Practices versus New Directions. Am. J. Clin. Dermatol. 2017, 18, 67–75. [Google Scholar] [CrossRef]
- Jahn-Bassler, K.; Bauer, W.M.; Karlhofer, F.; Vossen, M.G.; Stingl, G. Sequential High- and Low-Dose Systemic Corticosteroid Therapy for Severe Childhood Alopecia Areata. JDDG—J. Ger. Soc. Dermatol. 2017, 15, 42–47. [Google Scholar] [CrossRef]
- Darwin, E.; Arora, H.; Hirt, P.A.; Wikramanayake, T.C.; Jimenez, J.J. A Review of Monochromatic Light Devices for the Treatment of Alopecia Areata. Lasers Med. Sci. 2018, 33, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Damsky, W.; King, B.A. JAK Inhibitors in Dermatology: The Promise of a New Drug Class. J. Am. Acad. Dermatol. 2017, 76, 736–744. [Google Scholar] [CrossRef] [PubMed]
- Garg, S.; Manchanda, S. Platelet-Rich Plasma-an “Elixir” for Treatment of Alopecia: Personal Experience on 117 Patients with Review of Literature. Stem. Cell Investig. 2017, 4, 64. [Google Scholar] [CrossRef]
- Rinaldi, F.; Marzani, B.; Pinto, D.; Sorbellini, E. Randomized Controlled Trial on a PRP-like Cosmetic, Biomimetic Peptides Based, for the Treatment of Alopecia Areata. J. Dermatol. Treat. 2019, 30, 588–593. [Google Scholar] [CrossRef] [PubMed]
- Kimura-Ueki, M.; Oda, Y.; Oki, J.; Komi-Kuramochi, A.; Honda, E.; Asada, M.; Suzuki, M.; Imamura, T. Hair Cycle Resting Phase Is Regulated by Cyclic Epithelial FGF18 Signaling. J. Investig. Dermatol. 2012, 132, 1338–1345. [Google Scholar] [CrossRef] [PubMed]
- Kawano, M.; Komi-Kuramochi, A.; Asada, M.; Suzuki, M.; Oki, J.; Jiang, J.; Imamura, T. Comprehensive Analysis of FGF and FGFR Expression in Skin: FGF18 Is Highly Expressed in Hair Follicles and Capable of Inducing Anagen from Telogen Stage Hair Follicles. J. Investig. Dermatol. 2005, 124, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Marx, R.E. Platelet-Rich Plasma: Evidence to Support Its Use. J. Oral Maxillofac. Surg. 2004, 62, 489–496. [Google Scholar] [CrossRef]
- Giordano, S.; Romeo, M.; di Summa, P.; Salval, A.; Lankinen, P. A Meta-Analysis on Evidence of Platelet-Rich Plasma for Androgenetic Alopecia. Int. J. Trichology 2018, 10, 1–10. [Google Scholar] [CrossRef]
- Dohan Ehrenfest, D.M.; Rasmusson, L.; Albrektsson, T. Classification of Platelet Concentrates: From Pure Platelet-Rich Plasma (P-PRP) to Leucocyte- and Platelet-Rich Fibrin (L-PRF). Trends Biotechnol. 2009, 27, 158–167. [Google Scholar] [CrossRef]
- Marzani, B.; Pinto, D.; Minervini, F.; Calasso, M.; di Cagno, R.; Giuliani, G.; Gobbetti, M.; de Angelis, M. The Antimicrobial Peptide Pheromone Plantaricin A Increases Antioxidant Defenses of Human Keratinocytes and Modulates the Expression of Filaggrin, Involucrin, β-Defensin 2 and Tumor Necrosis Factor-α Genes. Exp. Dermatol. 2012, 21, 665–671. [Google Scholar] [CrossRef]
- Pinto, D.; Marzani, B.; Minervini, F.; Calasso, M.; Giuliani, G.; Gobbetti, M.; de Angelis, M. Plantaricin A Synthesized by Lactobacillus plantarum Induces in Vitro Proliferation and Migration of Human Keratinocytes and Increases the Expression of TGF-Β1, FGF7, VEGF-A and IL-8 Genes. Peptides 2011, 32, 1815–1824. [Google Scholar] [CrossRef] [PubMed]
- di Cagno, R.; Filannino, P.; Cantatore, V.; Gobbetti, M. Novel Solid-State Fermentation of Bee-Collected Pollen Emulating the Natural Fermentation Process of Bee Bread. Food Microbiol. 2019, 82, 218–230. [Google Scholar] [CrossRef]
- Williams, H.C.; Dellavalle, R.P.; Garner, S. Acne Vulgaris. Lancet 2012, 379, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Bagatin, E.; Timpano, D.L.; Guadanhim, L.R.d.S.; Nogueira, V.M.A.; Terzian, L.R.; Steiner, D.; Florez, M. Acne Vulgaris: Prevalence and Clinical Forms in Adolescents from São Paulo, Brazil. An. Bras. Dermatol. 2014, 89, 428–435. [Google Scholar] [CrossRef]
- Tan, J.K.L.; Bhate, K. A Global Perspective on the Epidemiology of Acne. Br. J. Dermatol. 2015, 172, 3–12. [Google Scholar] [CrossRef]
- Toyoda, M.; Morohashi, M. Pathogenesis of Acne. Med. Electron. Microsc. 2001, 34, 29–40. [Google Scholar] [CrossRef]
- Chilicka, K.; Rusztowicz, M.; Rogowska, A.M.; Szyguła, R.; Asanova, B.; Nowicka, D. Efficacy of Hydrogen Purification and Cosmetic Acids in the Treatment of Acne Vulgaris: A Preliminary Report. J. Clin. Med. 2022, 11, 6269. [Google Scholar] [CrossRef]
- Chilicka, K.; Rogowska, A.M.; Szyguła, R.; Rusztowicz, M.; Nowicka, D. Efficacy of Oxybrasion in the Treatment of Acne Vulgaris: A Preliminary Report. J. Clin. Med. 2022, 11, 3824. [Google Scholar] [CrossRef]
- See, J.A.; Goh, C.L.; Hayashi, N.; Suh, D.H.; Casintahan, F.A. Optimizing the Use of Topical Retinoids in Asian Acne Patients. J. Dermatol. 2018, 45, 522–528. [Google Scholar] [CrossRef]
- Fluhr, J.W.; Degitz, K. Antibiotika, Azelainsäure Und Benzoylperoxid in Der Topische Aknetherapie. JDDG—J. Ger. Soc. Dermatol. 2010, 8, S24–S30. [Google Scholar] [CrossRef] [PubMed]
- Nast, A.; Dréno, B.; Bettoli, V.; Degitz, K.; Erdmann, R.; Finlay, A.Y.; Ganceviciene, R.; Haedersdal, M.; Layton, A.; López-Estebaranz, J.L.; et al. European Evidence-Based (S3) Guidelines for the Treatment of Acne. J. Eur. Acad. Dermatol. Venereol. 2012, 26, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Strauss, J.S.; Krowchuk, D.P.; Leyden, J.J.; Lucky, A.W.; Shalita, A.R.; Siegfried, E.C.; Thiboutot, D.M.; van Voorhees, A.S.; Beutner, K.A.; Sieck, C.K.; et al. Guidelines of Care for Acne Vulgaris Management. J. Am. Acad. Dermatol. 2007, 56, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Simonart, T. Newer Approaches to the Treatment of Acne Vulgaris. Am. J. Clin. Dermatol. 2012, 13, 357–364. [Google Scholar] [CrossRef]
- Majeed, M.; Majeed, S.; Nagabhushanam, K.; Lawrence, L.; Arumugam, S.; Mundkur, L. Skin Protective Activity of Lactosporin-the Extracellular Metabolite from Bacillus coagulans Mtcc 5856. Cosmetics 2020, 7, 76. [Google Scholar] [CrossRef]
- Lambrechts, I.A.; de Canha, M.N.; Lall, N. Exploiting Medicinal Plants as Possible Treatments for Acne Vulgaris. In Medicinal Plants for Holistic Health and Well-Being; Elsevier: Amsterdam, The Netherlands, 2017; pp. 117–143. ISBN 9780128124758. [Google Scholar]
- Chung, H.J.; Lee, H.; Kim, M.; Lee, J.W.; Saeed, M.; Lee, H.; Jung, S.H.; Shim, J.J.; Lee, J.L.; Heo, K.; et al. Development and Metabolic Profiling of a Postbiotic Complex Exhibiting Antibacterial Activity against Skin Microorganisms and Anti-Inflammatory Effect on Human Keratinocytes. Food Sci. Biotechnol. 2022, 31, 1325–1334. [Google Scholar] [CrossRef]
- Kim, J.; Kim, B.E.; Leung, D.Y.M. Pathophysiology of Atopic Dermatitis: Clinical Implications. Allergy Asthma Proc. 2019, 40, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.E.; Leung, D.Y.M. Epidermal Barrier in Atopic Dermatitis. Allergy Asthma Immunol. Res. 2012, 4, 12–16. [Google Scholar] [CrossRef]
- Silverberg, J.I. Public Health Burden and Epidemiology of Atopic Dermatitis. Dermatol. Clin. 2017, 35, 283–289. [Google Scholar] [CrossRef]
- Guttman-Yassky, E.; Suárez-Fariñas, M.; Chiricozzi, A.; Nograles, K.E.; Shemer, A.; Fuentes-Duculan, J.; Cardinale, I.; Lin, P.; Bergman, R.; Bowcock, A.M.; et al. Broad Defects in Epidermal Cornification in Atopic Dermatitis Identified through Genomic Analysis. J. Allergy Clin. Immunol. 2009, 124, 1235–1244.e58. [Google Scholar] [CrossRef]
- Blicharz, L.; Rudnicka, L.; Czuwara, J.; Waśkiel-Burnat, A.; Goldust, M.; Olszewska, M.; Samochocki, Z. The Influence of Microbiome Dysbiosis and Bacterial Biofilms on Epidermal Barrier Function in Atopic Dermatitis—An Update. Int. J. Mol. Sci. 2021, 22, 8403. [Google Scholar] [CrossRef]
- Brunner, P.M.; Leung, D.Y.M.; Guttman-Yassky, E. Immunologic, Microbial, and Epithelial Interactions in Atopic Dermatitis. Ann. Allergy Asthma Immunol. 2018, 120, 34–41. [Google Scholar] [CrossRef]
- Langan, S.M.; Irvine, A.D.; Weidinger, S. Atopic Dermatitis. Lancet 2020, 396, 345–360. [Google Scholar] [CrossRef]
- Liang, Y.; Chang, C.; Lu, Q. The Genetics and Epigenetics of Atopic Dermatitis—Filaggrin and Other Polymorphisms. Clin. Rev. Allergy Immunol. 2016, 51, 315–328. [Google Scholar] [CrossRef]
- Renert-Yuval, Y.; Guttman-Yassky, E. New Treatments for Atopic Dermatitis Targeting beyond IL-4/IL-13 Cytokines. Ann. Allergy Asthma Immunol. 2020, 124, 28–35. [Google Scholar] [CrossRef]
- Wollenberg, A.; Oranje, A.; Deleuran, M.; Simon, D.; Szalai, Z.; Kunz, B.; Svensson, A.; Barbarot, S.; von Kobyletzki, L.; Taieb, A.; et al. ETFAD/EADV Eczema Task Force 2015 Position Paper on Diagnosis and Treatment of Atopic Dermatitis in Adult and Paediatric Patients. J. Eur. Acad. Dermatol. Venereol. 2016, 30, 729–747. [Google Scholar] [CrossRef] [PubMed]
- Torii, S.; Torii, A.; Itoh, K.; Urisu, A.; Terada, A.; Fujisawa, T.; Yamada, K.; Suzuki, H.; Ishida, Y.; Nakamura, F.; et al. Effects of Oral Administration of Lactobacillus acidophilus L-92 on the Symptoms and Serum Markers of Atopic Dermatitis in Children. Int. Arch. Allergy Immunol. 2011, 154, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Ishida, Y.; Nakamura, F.; Kanzato, H.; Sawada, D.; Yamamoto, N.; Kagata, H.; Oh-Ida, M.; Takeuchi, H.; Fujiwara, S. Effect of Milk Fermented with Lactobacillus acidophilus Strain L-92 on Symptoms of Japanese Cedar Pollen Allergy: A Randomized Placebo-Controlled Trial. Biosci. Biotechnol. Biochem. 2005, 69, 1652–1660. [Google Scholar] [CrossRef] [PubMed]
- Kuwana, R.; Yamamoto, N. Increases in GroES and GroEL from Lactobacillus acidophilus L-92 in Response to a Decrease in Medium PH, and Changes in Cytokine Release from Splenocytes: Transcriptome and Proteome Analyses. J. Biosci. Bioeng. 2012, 114, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.M.; Miyamoto, Y.; Yamada, Y.; Yamashita, H.; Tanaka, H.; Ezaki, T.; Nagai, H.; Inagaki, N. Orally Supplemented Lactobacillus acidophilus Strain L-92 Inhibits Passive and Active Cutaneous Anaphylaxis as Well as 2,4-Dinitroflurobenzene and Mite Fecal Antigen Induced Atopic Dermatitis-like Skin Lesions in Mice. Microbiol. Immunol. 2010, 54, 523–533. [Google Scholar] [CrossRef]
- Kanzato, H.; Fujiwara, S.; Ise, W.; Kaminogawa, S.; Sato, R.; Hachimura, S. Lactobacillus acidophilus Strain L-92 Induces Apoptosis of Antigen-Stimulated T Cells by Modulating Dendritic Cell Function. Immunobiology 2008, 213, 399–408. [Google Scholar] [CrossRef]
- Shah, M.M.; Saio, M.; Yamashita, H.; Tanaka, H.; Takami, T.; Ezaki, T.; Inagaki, N. Lactobacillus acidophilus Strain L-92 Induces CD4+CD25+Foxp3+ Regulatory T Cells and Suppresses Allergic Contact Dermatitis. Biol. Pharm. Bull. 2012, 35, 612–616. [Google Scholar] [CrossRef] [PubMed]
- Tsiouris, C.G.; Tsiouri, M.G. Human Microflora, Probiotics and Wound Healing. Wound Med. 2017, 19, 33–38. [Google Scholar] [CrossRef]
- Yu, R.; Zhang, H.; Guo, B. Conductive Biomaterials as Bioactive Wound Dressing for Wound Healing and Skin Tissue Engineering. Nanomicro Lett. 2022, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Holmes, C.J.; Plichta, J.K.; Gamelli, R.L.; Radek, K.A. Dynamic Role of Host Stress Responses in Modulating the Cutaneous Microbiome: Implications for Wound Healing and Infection. Adv. Wound Care 2015, 4, 24–37. [Google Scholar] [CrossRef]
- Percival, S.L.; McCarty, S.M.; Lipsky, B. Biofilms and Wounds: An Overview of the Evidence. Adv. Wound Care 2015, 4, 373–381. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).