Casting Light on the Micro-Organisms in Digestate: Diversity and Untapped Potential
Abstract
:1. Introduction
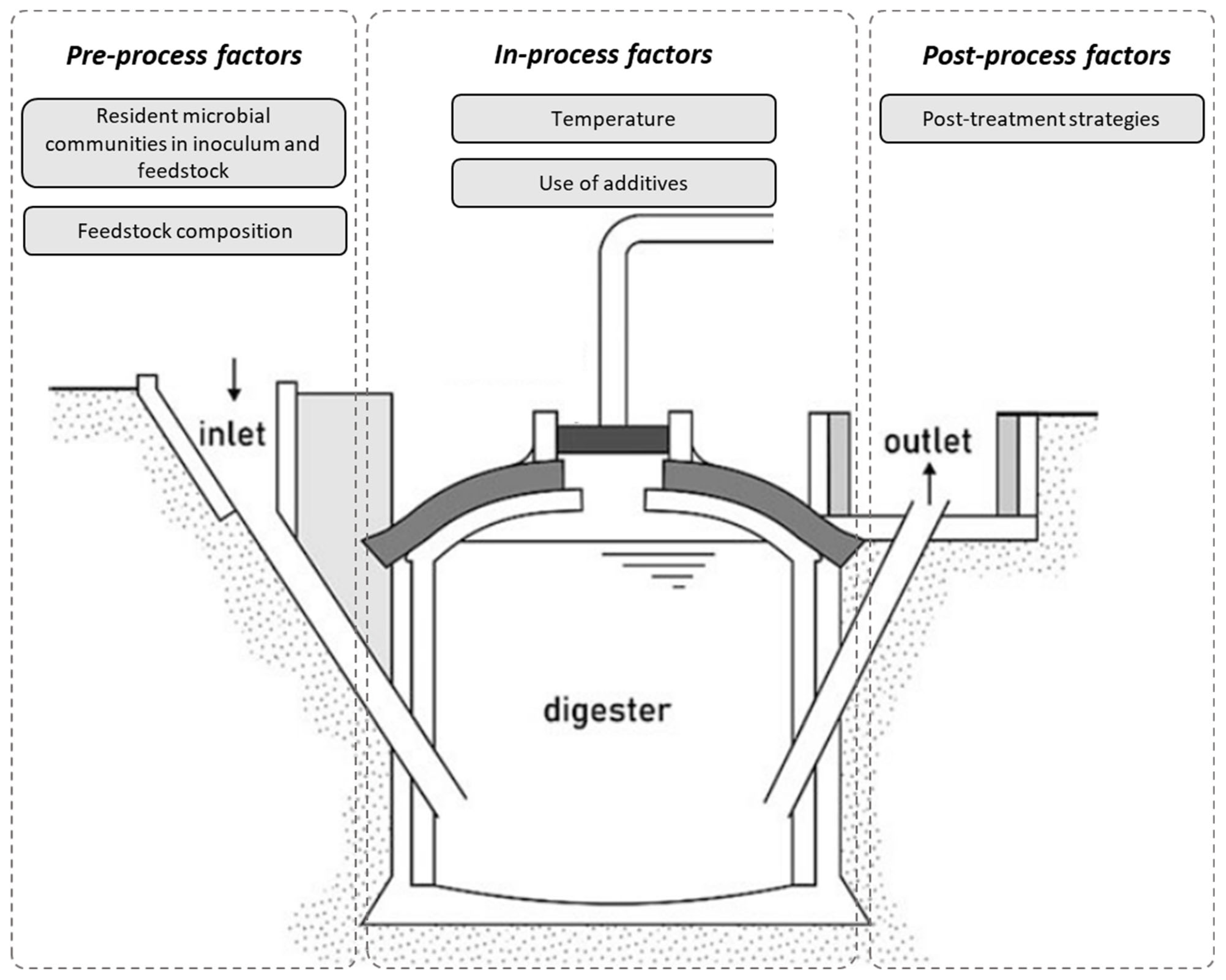
2. Factors Shaping Microbial Composition of Digestate
2.1. Resident Microbial Communities in Inoculum and Feedstock
2.2. Feedstock Composition
2.3. Temperature
2.4. Use of Additives
2.5. Post-Treatment Strategies
3. Digestate Microbial Communities and Associated Functions
| Feedstock | Treatment | Digester | Duration | Temperature | Bacterial Taxa | Archaeal Taxa | Fungal Taxa | References |
|---|---|---|---|---|---|---|---|---|
| Pig manure and plant silage | Industrial mesophilic biogas plant | 10 days | Mesophilic (37 °C) | Streptococcus, Clostridium, Syntrophaceticus, Candidatus cloacimonas, Chitinispirillum, Lactobacillus, Treponema, Bacteroides, Ruminiclostridium, Nitrosomonas, Sedimentibacter, Prevotella, Paenibacillus, Bacillus, Arcobacter, Herbinix, Pseudomonas, Candidatus riflebacteria | Methanobacterium, Methanosaeta, Methanosarcina, Methanospirillum, Methanoculleus, Methanobrevibacter, Methanothermobacter | [104] | ||
| Pig manure and plant silage digestate | H2 | Batch—bio power to methane reactor | 4 weeks | Mesophilic (37 °C) | Streptococcus, Clostridium, Syntrophaceticus, Candidatus cloacimonas, Lactobacillus, Treponema, Bacteroides, Ruminiclostridium, Nitrosomonas, Sedimentibacter, Prevotella | Methanobacterium, Methanosaeta, Methanosarcina, Methanospirillum, Methanoculleus, Methanobrevibacter | [104] | |
| Pig manure and plant silage digestate + α-Cellulose | H2 | Batch—bio power to methane reactor | 4 weeks | Mesophilic (37 °C) | Streptococcus, Clostridium, Syntrophaceticus, Candidatus cloacimonas, Lactobacillus, Treponema, Bacteroides, Ruminiclostridium, Nitrosomonas, Sedimentibacter, Prevotella, Paenibacillus, Herbinix | Methanobacterium, Methanosaeta, Methanosarcina, Methanospirillum, Methanoculleus, Methanobrevibacter | [104] | |
| Pig manure and plant silage digestate | N2 | Batch—bio power to methane reactor | 4 weeks | Mesophilic (37 °C) | Streptococcus, Clostridium, Syntrophaceticus, Lactobacillus, Treponema, Bacteroides, Ruminiclostridium, Nitrosomonas, Prevotella, Pseudomonas | Methanobacterium, Methanosaeta, Methanosarcina, Methanospirillum, Methanoculleus, Methanobrevibacter | [104] | |
| Pig manure and plant silage digestate + α-Cellulose | N2 | Batch—bio power to methane reactor | 4 weeks | Mesophilic (37 °C) | Streptococcus, Clostridium, Syntrophaceticus, Chitinispirillum, Lactobacillus, Treponema, Bacteroides, Ruminiclostridium, Nitrosomonas, Sedimentibacter, Prevotella, Pseudomonas | Methanobacterium, Methanosaeta, Methanosarcina, Methanospirillum, Methanoculleus, Methanobrevibacter | [104] | |
| Pig manure and plant silage digestate | H2 and CO2 | Batch—bio power to methane reactor | 12 weeks | Mesophilic (37 °C) | Streptococcus, Clostridium, Syntrophaceticus, Ruminiclostridium | Methanobacterium, Methanoculleus, Methanosarcina, Methanobrevibacter | [104] | |
| Pig manure and plant silage digestate | N2 | Batch—bio power to methane reactor | 12 weeks | Mesophilic (37 °C) | Streptococcus, Clostridium, Syntrophaceticus, Candidatus cloacimonas, Ruminiclostridium, Herbinix | Methanobacterium, Methanosaeta, Methanosarcina, Methanobrevibacter | [104] | |
| Dairy cow manure containing wheat straw and woodchip | Commercially operating biogas plant | Unknown | Peptostreptococcaceae, Family XI, Ruminococcaceae, Bacteroidales UCG-001, Rikenellaceae, Porphyromonadaceae, Helicobacteraceae, Clostridiaceae 1, Carnobacteriaceae, Lachnospiraceae, Syntrophomonadaceae, Erysipelotrichaceae | Methanobacteriaceae, Methanosarcinaceae | [105] | |||
| Food waste from the dairy industry | Pasteurization pre-digestion | Commercial biogas plant | 70 days (HRT) | Mesophilic | Clostridium, Cloacimonas, Bacteroides, Acetivibrio, Rikenella, Eubacterium, Bacillus, Proteiniphilum, Symbiobacterium, Petrotoga, Sporobacter, Syntrophomonas, Cytophaga, Acidaminococcus, Bellilinea | Methanobacterium, Methanobrevibacter, Methanoculleus | Hanseniaspora, Cryptococcus, Guehomyces | [21] |
| Food waste and pig slurry | Commercial biogas plant | 90 days (HRT) | Mesophilic (40 °C) | Clostridium, Cloacimonas, Bacteroides, Acetivibrio, Rikenella, Pseudoalteromonas, Eubacterium, Bacillus, Proteiniphilum, Symbiobacterium, Syntrophomonas, Bellilinea, Verrucomicrobium | Methanobacterium, Methanobrevibacter, Methanosarcina, Methanoculleus, Methanothermobacter | Acaulospora, Kazachstania, Penicillium, Saccharomyces | [21] | |
| Farm and food industry wastes | Commercial biogas plant | 54 days (HRT) | Mesophilic (38 °C) | Clostridium, Cloacimonas, Bacteroides, Acetivibrio, Rikenella, Pseudoalteromonas, Eubacterium, Bacillus, Proteiniphilum, Symbiobacterium, Natronoanaerobium, Cytophaga, Syntrophomonas, Bellilinea, Verrucomicrobium | Methanobacterium, Methanobrevibacter, Methanosaeta, Methanospirillum, Methanosarcina, Methanoculleus | Acaulospora, Scedosporium, Cyllamyces | [21] | |
| Food waste and municipal wastes | Commercial biogas plant | 60 days (HRT) | Mesophilic (37–42 °C) | Clostridium, Cloacimonas, Bacteroides, Acetivibrio, Pseudoalteromonas, Eubacterium, Bacillus, Proteiniphilum, Symbiobacterium, Natronoanaerobium, Cytophaga, Syntrophomonas, Bellilinea, Verrucomicrobium, Acidaminococcus | Methanobacterium, Methanosarcina, Methanobrevibacter, Methanoculleus, Methanothermobacter | Acaulospora, Penicillium, Paramicro-sporidium, Mucor | [21] | |
| Green waste (leaves, grass, prunings and trimmings, branches and stumps) and food waste (dog food) mixture with digestate sludge inoculum | C/N ratio of 17 | Batch anaerobic digester | 14–15 days | Thermophilic (55 °C) | Thermotogaceae, Halanaerobiaceae, Lachnospiraceae, Caldicoprobacteraceae, Tissierellaceae, Clostridiaceae, Ruminococcaceae, Porphyromonadaceae, Anaerobaculaceae | Methanosarcinaceae | [43] | |
| Green waste (leaves, grass, prunings and trimmings, branches and stumps) and food waste (dog food) mixture with digestate sludge inoculum | C/N ratio of 20 | Batch anaerobic digester | 14–15 days | Thermophilic (55 °C) | Thermotogaceae, Halanaerobiaceae, Lachnospiraceae, Caldicoprobacteraceae, Tissierellaceae, Clostridiaceae, Ruminococcaceae, Porphyromonadaceae, Anaerobaculaceae | Methanosarcinaceae | [43] | |
| Green waste (leaves, grass, prunings and trimmings, branches and stumps) and food waste (dog food) mixture with digestate sludge inoculum | C/N ratio of 23 | Batch anaerobic digester | 14–15 days | Thermophilic (55 °C) | Thermotogaceae, Halanaerobiaceae, Lachnospiraceae, Clostridiaceae, Ruminococcaceae, Tissierellaceae, Syntrophomonadaceae | Methanosarcinaceae | [43] | |
| Green waste (leaves, grass, prunings and trimmings, branches and stumps) and food waste (dog food) mixture with digestate sludge inoculum | C/N ratio of 27 | Batch anaerobic digester | 14–15 days | Thermophilic (55 °C) | Thermotogaceae, Halanaerobiaceae, Lachnospiraceae, Clostridiaceae, Ruminococcaceae, Caldicoprobacteracea, Tissierellaceae, Porphyromonadaceae, Anaerobaculaceae, Syntrophomonadaceae | Methanosarcinaceae | [43] | |
| Green waste (leaves, grass, prunings and trimmings, branches and stumps) and food waste (dog food) mixture with digestate sludge inoculum | C/N ratio of 34 | Batch anaerobic digester | 14–15 days | Thermophilic (55 °C) | Thermotogaceae, Lachnospiraceae, Clostridiaceae, Ruminococcaceae, Halanaerobiaceae, Porphyromonadaceae, Anaerobaculaceae | Methanosarcinaceae | [43] | |
| Unknown | Unknown | Biogas plant | Unknown | Unknown | Psychrobacter, Mycobacterium, Acinetobacter, Microbacterium | [23] | ||
| Corn stover | NaOH pretreatment | Continuously stirred tank reactor | 1–60 days | Mesophilic (35 °C) | Bacteroidetes, Firmicutes, WS6, Spirochaetae, Verrucomicrobia, Synergistetes, Proteobacteria, Cloacimonetes, Saccharibacteria, Acidobacteria, Chloroflexi | Methanosarcina, Methanobacterium | [106] | |
| Livestock effluent and agricultural waste | Unknown | Unknown | Unknown | Unknown | Bacillales, Bacteroidales, Clostridiales, Acholeplasmatales | Methanomicrobiales | [107] | |
| Mixture of sewage sludge (53% total solids [TS]) and municipal solid waste (47% TS) | Industrial-scale plant | 19 days (HRT) | Mesophilic (37 °C) | Holophagae, Actinobacteria, Anaerolineae, Bacteroidetes_vadinHA17, Bacteroidia, Sphingobacteriia, Cloacimonetes, Fibrobacteria, Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria, Clostridia, Synergistia | Methanobacterium, Methanobrevibacter, Methanosaeta, Methanomassiliicoccus | [108] | ||
| 50% digestate and 50% mixture of sewage sludge (53% total solids [TS]) and municipal solid waste (47% TS) | Industrial-scale plant | 10 days (HRT) | Mesophilic (37 °C) | Holophagae, Anaerolineae, Bacteroidetes_vadinHA17, Bacteroidia, Sphingobacteriia, Fibrobacteria, Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria, Clostridia, Synergistia | Methanobacterium, Methanobrevibacter, Methanosaeta, Methanomassiliicoccus, Methanospirillum | [108] | ||
| Mixture of sewage sludge (53% total solids [TS]) and municipal solid waste (47% TS) | Thermal hydrolysis processing | Industrial-scale plant | 23 days (HRT) | Mesophilic (40–42 °C) | Anaerolineae, Bacteroidetes_vadinHA17, Bacteroidia, Sphingobacteriia, W27, Clostridia, Thermotogae, LD1-PB3, Synergistia | Methanobacterium, Methanosaeta, Methanomassiliicoccus | [108] | |
| 50% digestate and 50% mixture of sewage sludge (53% total solids [TS]) and municipal solid waste (47% TS) | Thermal hydrolysis processing | Industrial-scale plant | 20 days (HRT) | Mesophilic (39 °C) | Anaerolineae, Bacteroidetes_vadinHA17, Bacteroidia, Sphingobacteriia, W27, W5, Clostridia, Thermotogae, LD1-PB3, Synergistia | Methanobacterium, Methanosaeta, Methanomassiliicoccus | [108] | |
| Organic household waste 37%, manure 31%, slaughter residues 19%, other organic food waste 13% and iron chloride 0.03% | Biogas plant | Unknown | Pseudomonas, Sporosarcina, Leuconostoc, Romboutsia | [109] | ||||
| Organic household waste 37%, manure 31%, slaughter residues 19%, other organic food waste 13% and iron chloride 0.03% | Nitrification | Biogas plant | Unknown | Lactobacillus, Ralstonia, Pseudomonas, Mycobacterium, Chujaibacter, Romboutsia | [109] | |||
| Sewage sludge produced by wastewater treatment plants from urban sites | Organic waste treatment plants | 34–38 days | Mesophilic (32–42 °C) | Acidobacteria, Actinobacteria, Atribacteria, Bacteroidetes, Chlorobi, Chloroflexi, Deferribacteres, Firmicutes, Lentisphaerae, Nitrospirae, Planctomycetes, Proteobacteria, Spirochaetae, Verrucomicrobia | [37] | |||
| Animal manure or slurry from agricultural sites and a mix of food industry waste, harvest residues, livestock effluents or sewage sludge from central sites | Organic waste treatment plants | 30–90 days | Mesophilic (32–42 °C) | Actinobacteria, Atribacteria, Bacteroidetes, Cloacimonetes, Firmicutes, Planctomycetes, Proteobacteria, Spirochaetae, Synergistetes, Tenericutes | [37] | |||
| Mixed sludge | Anaerobic digestion plant | Unknown | Mesophilic (36 °C) | Clostridia, Bacillus, Bacteroidia, Sphingobacteriia, Cytophagia, Flavobacteriia, Alphaproteobacteria, Gammaproteobacteria, Betaproteobacteria | [110] | |||
| Cattle manure digestate inoculum, pig manure (85%) and food waste (15%), volatile solids basis | Anaerobic digestion reactor (semi-continous) | 41 days (HRT) | Mesophilic (39 °C) | Acholeplasma, Treponema, Sphaerochaeta, Succinivibrio, RFN20, ML1228J-1, Sedimentibacter, Syntrophomonas, Coprococcus, Clostridium, Alkaliphilus, Caldicoprobacter, Leuconostoc, Lactobacillus, Corynebacterium | Methanosarcina, Methanoculleus, Methanosphaera, Methanobrevibacter, Methanobacterium | [111] | ||
| Cattle manure digestate inoculum, pig manure (62.5%) and food waste (37.5%), volatile solids basis | Anaerobic digestion reactor (semi-continous) | 41 days (HRT) | Mesophilic (39 °C) | Acholeplasma, Treponema, Sphaerochaeta, Succinivibrio, RFN20, ML1228J-1, Sedimentibacter, Syntrophomonas, Coprococcus, Clostridium, Alkaliphilus, Caldicoprobacter, Leuconostoc, Lactobacillus, Corynebacterium | Methanosarcina, Methanoculleus, Methanosphaera, Methanobrevibacter, Methanobacterium | [111] | ||
| Cattle manure digestate inoculum, pig manure (40%) and food waste (60%), volatile solids basis | Anaerobic digestion reactor (semi-continous) | 41 days (HRT) | Mesophilic (39 °C) | Acholeplasma, Treponema, Sphaerochaeta, Succinivibrio, RFN20, ML1228J-1, Sedimentibacter, Syntrophomonas, Coprococcus, Clostridium, Alkaliphilus, Caldicoprobacter, Leuconostoc, Lactobacillus, Corynebacterium | Methanosarcina, Methanoculleus, Methanosphaera, Methanobrevibacter, Methanobacterium | [111] | ||
| Cattle manure digestate inoculum, pig manure (85%) and food waste (15%), volatile solids basis | Anaerobic digestion reactor (semi-continous) | 29 days (HRT) | Mesophilic (39 °C) | Acholeplasma, Treponema, Sphaerochaeta, RFN20, ML1228J-1, Sedimentibacter, Syntrophomonas, Coprococcus, Clostridium, Alkaliphilus, Caldicoprobacter, Leuconostoc, Lactobacillus, Propionicimonas, Corynebacterium | Methanomassiliicoccus, Methanosarcina, Methanospirillum, Methanogenium, Methanoculleus, Methanosphaera, Methanobrevibacter, Methanobacterium | [111] | ||
| Cattle manure digestate inoculum, pig manure (62.5%) and food waste (37.5%), volatile solids basis | Anaerobic digestion reactor (semi-continous) | 29 days (HRT) | Mesophilic (39 °C) | Acholeplasma, Treponema, Sphaerochaeta, RFN20, ML1228J-1, Sedimentibacter, Syntrophomonas, Coprococcus, Clostridium, Alkaliphilus, Caldicoprobacter, Leuconostoc, Lactobacillus, Propionicimonas, Corynebacterium | Methanomassiliicoccus, Methanosarcina, Methanogenium, Methanoculleus, Methanosphaera, Methanobrevibacter | [111] | ||
| Cattle manure digestate inoculum, pig manure (40%) and food waste (60%), volatile solids basis | Anaerobic digestion reactor (semi-continous) | 29 days (HRT) | Mesophilic (39 °C) | Acholeplasma, Treponema, Sphaerochaeta, RFN20, ML1228J-1, Sedimentibacter, Syntrophomonas, Coprococcus, Clostridium, Alkaliphilus, Caldicoprobacter, Leuconostoc, Lactobacillus, Propionicimonas, Corynebacterium | Methanomassiliicoccus, Methanosarcina, Methanosaeta, Methanoculleus, Methanosphaera, Methanobrevibacter | [111] | ||
| Cattle manure digestate inoculum, pig manure (85%) and food waste (15%), volatile solids basis | Anaerobic digestion reactor (semi-continous) | 21 days (HRT) | Mesophilic (39 °C) | Acholeplasma, Treponema, Sphaerochaeta, RFN20, ML1228J-1, Sedimentibacter, Syntrophomonas, Coprococcus, Clostridium, Alkaliphilus, Caldicoprobacter, Leuconostoc, Lactobacillus, Propionicimonas, Corynebacterium | Methanomassiliicoccus, Methanosarcina, Methanospirillum, Methanogenium, Methanoculleus, Methanosphaera, Methanobrevibacter, Methanobacterium | [111] | ||
| Cattle manure digestate inoculum, pig manure (62.5%) and food waste (37.5%), volatile solids basis | Anaerobic digestion reactor (semi-continous) | 21 days (HRT) | Mesophilic (39 °C) | Acholeplasma, Treponema, Sphaerochaeta, RFN20, ML1228J-1, Sedimentibacter, Syntrophomonas, Coprococcus, Clostridium, Alkaliphilus, Caldicoprobacter, Leuconostoc, Lactobacillus, Corynebacterium | Methanomassiliicoccus, Methanosarcina, Methanogenium, Methanoculleus, Methanosphaera, Methanobrevibacter | [111] | ||
| Cattle manure digestate inoculum, pig manure (40%) and food waste (60%), volatile solids basis | Anaerobic digestion reactor (semi-continous) | 21 days (HRT) | Mesophilic (39 °C) | Acholeplasma, Treponema, Sphaerochaeta, RFN20, ML1228J-1, Sedimentibacter, Syntrophomonas, Coprococcus, Clostridium, Alkaliphilus, Caldicoprobacter, Leuconostoc, Lactobacillus, Corynebacterium | Methanomassiliicoccus, Methanosarcina, Methanosaeta, Methanoculleus, Methanosphaera, Methanobrevibacter | [111] | ||
| Cow manure, wood straw and wood chip | Biogas plant | Unknown | Unknown | Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Plantomycetes, Proteobacteria, OP9, Synergistetes | Crenarchaeota, Euryarchaeota | [112] | ||
| Mixture of dairy cattle manure (70%), energy crops (maize silage up to 30%) and waste from agro-food industries | Biogas plant | Mesophilic | Acidimicrobiia, Actinobacteria, Alphaproteobacteria, Bacilli, Bacteroidia, Clostridia, Deltaproteobacteria, Gammaproteobacteria, Gemmatimonadetes, Mollicutes, Phycisphaerae, Saccharomonadia, Verrucomicrobiae | Methanosarcina | Ascomycota, Basidiomycota | [113] | ||
| Anaerobically digested sludge inoculum, food waste and waste activated sludge (TS: 5%) | Continuous stirred tank reactors | 30 days | Mesophilic (35 °C) | Mycobacterium, Thermovirga, Mesotoga, Erysipelatoclostridium, Eubacterium, Aminobacterium, Bacillus, Clostridium | Methanobrevibacter, Methanofollis | [58] | ||
| Anaerobically digested sludge inoculum, food waste and waste activated sludge (TS: 5%) | Wheat straw pellet biochar | Continuous stirred tank reactors | 30 days | Mesophilic (35 °C) | Mycobacterium, Thermovirga, Mesotoga, Erysipelatoclostridium, Eubacterium, Bacteroides, Aminobacterium, Clostridium | Methanosarcina, Methanomicrobiales, Methanofollis, Methanoculleus, Methanolinea | [58] | |
| Anaerobically digested sludge inoculum, food waste and waste activated sludge (TS: 7.5%) | Continuous stirred tank reactors | 30 days | Mesophilic (35 °C) | Thermovirga, Mesotoga, Erysipelatoclostridium, Syntrophomonas, Bacteroides, Aminobacterium | Methanocorpusculum, Methanobrevibacter, Methanosarcina | [58] | ||
| Anaerobically digested sludge inoculum, food waste and waste activated sludge (TS: 7.5%) | Wheat straw pellet biochar | Continuous stirred tank reactors | 30 days | Mesophilic (35 °C) | Thermovirga, Mesotoga, Erysipelatoclostridium, Eubacterium, Aminobacterium, Bacillus, Clostridium | Methanobrevibacter, Methanosarcina | [58] |
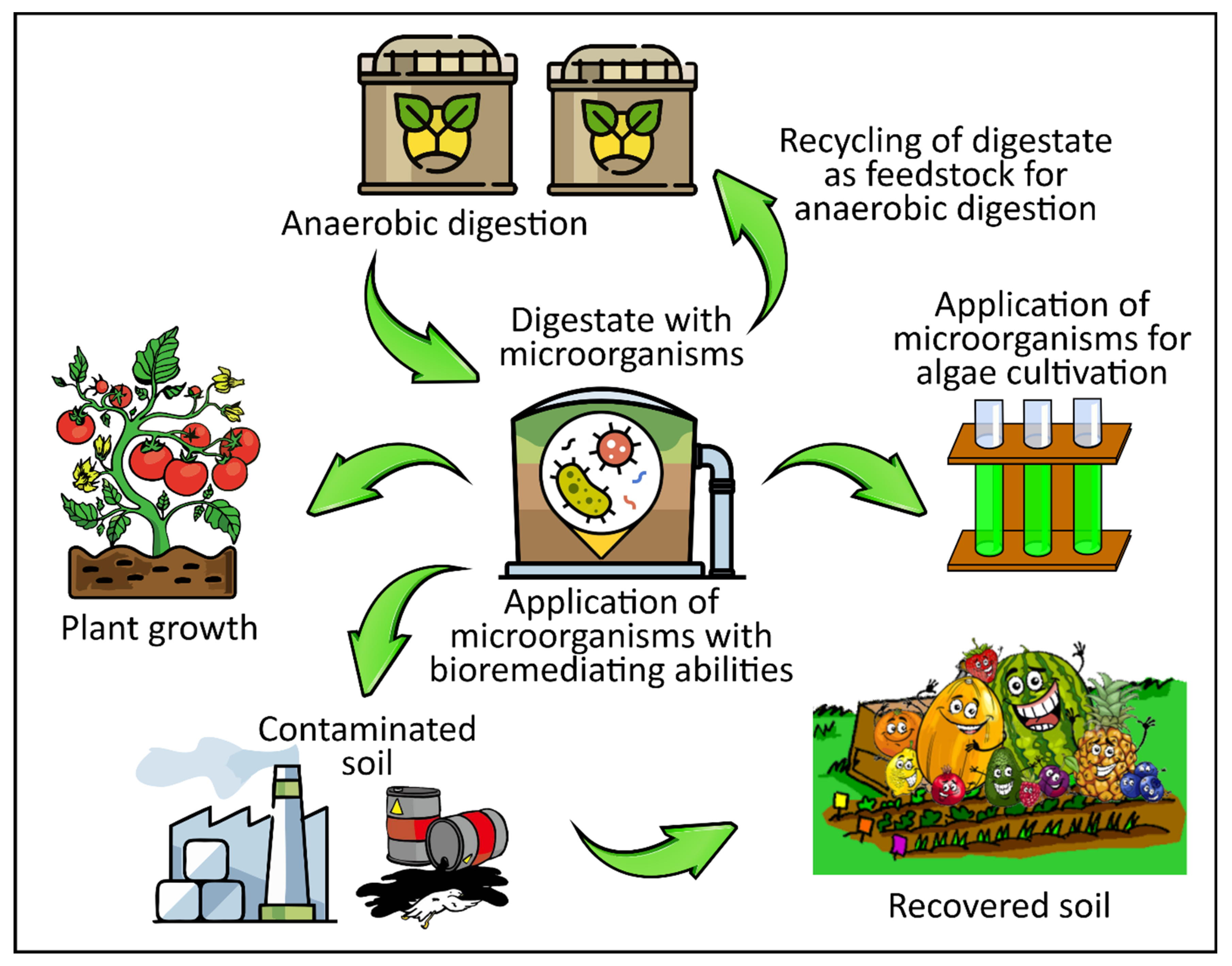
4. Applications of Digestate in Relation to Microbial Composition
4.1. Digestate Use as a Biofertilizer
4.2. Digestate Use in Aquaculture
4.3. Digestate Use in Biofuel Production—Digestate Recirculation
4.4. Digestate Use for Bioremediation
5. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhatt, A.H.; Tao, L. Economic Perspectives of Biogas Production via Anaerobic Digestion. Bioengineering 2020, 7, 74. [Google Scholar] [CrossRef]
- Roopnarain, A.; Adeleke, R. Current Status, Hurdles and Future Prospects of Biogas Digestion Technology in Africa. Renew. Sustain. Energy Rev. 2017, 67, 1162–1179. [Google Scholar] [CrossRef]
- Edwards, J.; Othman, M.; Burn, S. A Review of Policy Drivers and Barriers for the Use of Anaerobic Digestion in Europe, the United States and Australia. Renew. Sustain. Energy Rev. 2015, 52, 815–828. [Google Scholar] [CrossRef]
- Rasapoor, M.; Young, B.; Brar, R.; Sarmah, A.; Zhuang, W.-Q.; Baroutian, S. Recognizing the Challenges of Anaerobic Digestion: Critical Steps toward Improving Biogas Generation. Fuel 2020, 261, 116497. [Google Scholar] [CrossRef]
- Harirchi, S.; Wainaina, S.; Sar, T.; Nojoumi, S.A.; Parchami, M.; Parchami, M.; Varjani, S.; Khanal, S.K.; Wong, J.; Awasthi, M.K.; et al. Microbiological Insights into Anaerobic Digestion for Biogas, Hydrogen or Volatile Fatty Acids (VFAs): A Review. Bioengineered 2022, 13, 6521–6557. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, R.; Baitha, R. Biogas and Methane Yield Enhancement Using Graphene Oxide Nanoparticles and Ca(OH) 2 Pre-Treatment in Anaerobic Digestion. Int. J. Ambient Energy 2021, 42, 618–625. [Google Scholar] [CrossRef]
- Sandhu, S.; Kaushal, R. Anaerobic Co-Digestion of Food Wastes, Algae, Pond Sludge and Cow Dung for Biogas Yield Enhancement as a Potent Approach to Reduce Carbon Footprints. Aust. J. Mech. Eng. 2021, 1–20. [Google Scholar] [CrossRef]
- Alburquerque, J.A.; de la Fuente, C.; Ferrer-Costa, A.; Carrasco, L.; Cegarra, J.; Abad, M.; Bernal, M.P. Assessment of the Fertiliser Potential of Digestates from Farm and Agroindustrial Residues. Biomass Bioenergy 2012, 40, 181–189. [Google Scholar] [CrossRef]
- Wang, W.; Lee, D.-J. Valorization of Anaerobic Digestion Digestate: A Prospect Review. Bioresour. Technol. 2021, 323, 124626. [Google Scholar] [CrossRef]
- Peng, W.; Lü, F.; Hao, L.; Zhang, H.; Shao, L.; He, P. Digestate Management for High-Solid Anaerobic Digestion of Organic Wastes: A Review. Bioresour. Technol. 2020, 297, 122485. [Google Scholar] [CrossRef]
- Cesaro, A. The Valorization of the Anaerobic Digestate from the Organic Fractions of Municipal Solid Waste: Challenges and Perspectives. J. Environ. Manag. 2021, 280, 111742. [Google Scholar] [CrossRef]
- Karaeva, J.V.; Timofeeva, S.S.; Bashkirov, V.N.; Bulygina, K.S. Thermochemical Processing of Digestate from Biogas Plant for Recycling Dairy Manure and Biomass. Biomass Conv. Bioref. 2023, 13, 685–695. [Google Scholar] [CrossRef]
- Satpathy, P.; Steinigeweg, S.; Cypionka, H.; Engelen, B. Different Substrates and Starter Inocula Govern Microbial Community Structures in Biogas Reactors. Environ. Technol. 2016, 37, 1441–1450. [Google Scholar] [CrossRef]
- Nelson, M.C.; Morrison, M.; Yu, Z. A Meta-Analysis of the Microbial Diversity Observed in Anaerobic Digesters. Bioresour. Technol. 2011, 102, 3730–3739. [Google Scholar] [CrossRef]
- Niu, Q.; Kobayashi, T.; Takemura, Y.; Kubota, K.; Li, Y.-Y. Evaluation of Functional Microbial Community’s Difference in Full-Scale and Lab-Scale Anaerobic Digesters Feeding with Different Organic Solid Waste: Effects of Substrate and Operation Factors. Bioresour. Technol. 2015, 193, 110–118. [Google Scholar] [CrossRef]
- Coelho, J.J.; Prieto, M.L.; Hennessy, A.; Casey, I.; Woodcock, T.; Kennedy, N. Determination of Microbial Numbers in Anaerobically Digested Biofertilisers. Environ. Technol. 2021, 42, 753–763. [Google Scholar] [CrossRef]
- Sahlström, L. A Review of Survival of Pathogenic Bacteria in Organic Waste Used in Biogas Plants. Bioresour. Technol. 2003, 87, 161–166. [Google Scholar] [CrossRef]
- Seruga, P.; Krzywonos, M.; Paluszak, Z.; Urbanowska, A.; Pawlak-Kruczek, H.; Niedźwiecki, Ł.; Pińkowska, H. Pathogen Reduction Potential in Anaerobic Digestion of Organic Fraction of Municipal Solid Waste and Food Waste. Molecules 2020, 25, 275. [Google Scholar] [CrossRef]
- Smith, S.R.; Lang, N.L.; Cheung, K.H.M.; Spanoudaki, K. Factors Controlling Pathogen Destruction during Anaerobic Digestion of Biowastes. Waste Manag. 2005, 25, 417–425. [Google Scholar] [CrossRef]
- Jiang, Y.; Dennehy, C.; Lawlor, P.G.; Hu, Z.; Zhan, X.; Gardiner, G.E. Inactivation of Enteric Indicator Bacteria and System Stability during Dry Co-Digestion of Food Waste and Pig Manure. Sci. Total Environ. 2018, 612, 293–302. [Google Scholar] [CrossRef]
- Coelho, J.J.; Hennessy, A.; Casey, I.; Bragança, C.R.S.; Woodcock, T.; Kennedy, N. Biofertilisation with Anaerobic Digestates: A Field Study of Effects on Soil Microbial Abundance and Diversity. Appl. Soil Ecol. 2020, 147, 103403. [Google Scholar] [CrossRef]
- Qi, G.; Pan, Z.; Andriamanohiarisoamanana, F.J.; Yamashiro, T.; Iwasaki, M.; Kawamoto, K.; Umetsu, K. Isolation and Characterization of Plant Growth Promoting Bacteria (PGPB) from Anaerobic Digestate and Their Effect on Common Wheat (Triticum Aestivum) Seedling Growth. Int. J. Environ. Agric. Res. 2017, 3, 46–52. [Google Scholar] [CrossRef]
- Gielnik, A.; Pechaud, Y.; Huguenot, D.; Cébron, A.; Esposito, G.; van Hullebusch, E.D. Functional Potential of Sewage Sludge Digestate Microbes to Degrade Aliphatic Hydrocarbons during Bioremediation of a Petroleum Hydrocarbons Contaminated Soil. J. Environ. Manag. 2021, 280, 111648. [Google Scholar] [CrossRef] [PubMed]
- Bankston, E.M.; Higgins, B.T. Anaerobic Microbial Communities Can Influence Algal Growth and Nutrient Removal from Anaerobic Digestate. Bioresour. Technol. 2020, 297, 122445. [Google Scholar] [CrossRef] [PubMed]
- Baştabak, B.; Koçar, G. A Review of the Biogas Digestate in Agricultural Framework. J. Mater. Cycles Waste Manag. 2020, 22, 1318–1327. [Google Scholar] [CrossRef]
- Roopnarain, A.; Ndaba, B.; Rama, H.; Obi, L.; Bello-Akinosho, M.; Akindolire, M. Liquid Gold: Harnessing the Potential of Digestate to Enhance Smallholder Farmer Food Security and Livelihood. In Food Security for African Smallholder Farmers; Mupambwa, H.A., Nciizah, A.D., Nyambo, P., Muchara, B., Gabriel, N.N., Eds.; Sustainability Sciences in Asia and Africa; Springer Nature: Singapore, 2022; pp. 313–341. ISBN 9789811667701. [Google Scholar]
- Li, Y.; Xu, H.; Hua, D.; Zhao, B.; Mu, H.; Jin, F.; Meng, G.; Fang, X. Two-Phase Anaerobic Digestion of Lignocellulosic Hydrolysate: Focusing on the Acidification with Different Inoculum to Substrate Ratios and Inoculum Sources. Sci. Total Environ. 2020, 699, 134226. [Google Scholar] [CrossRef]
- Rico, C.; Montes, J.A.; Rico, J.L. Evaluation of Different Types of Anaerobic Seed Sludge for the High Rate Anaerobic Digestion of Pig Slurry in UASB Reactors. Bioresour. Technol. 2017, 238, 147–156. [Google Scholar] [CrossRef]
- Han, S.; Liu, Y.; Zhang, S.; Luo, G. Reactor Performances and Microbial Communities of Biogas Reactors: Effects of Inoculum Sources. Appl. Microbiol. Biotechnol. 2016, 100, 987–995. [Google Scholar] [CrossRef]
- Duan, N.; Kougias, P.G.; Campanaro, S.; Treu, L.; Angelidaki, I. Evolution of the Microbial Community Structure in Biogas Reactors Inoculated with Seeds from Different Origin. Sci. Total Environ. 2021, 773, 144981. [Google Scholar] [CrossRef]
- De Jonge, N.; Davidsson, Å.; la Cour Jansen, J.; Nielsen, J.L. Characterisation of Microbial Communities for Improved Management of Anaerobic Digestion of Food Waste. Waste Manag. 2020, 117, 124–135. [Google Scholar] [CrossRef]
- Liu, T.; Sun, L.; Müller, B.; Schnürer, A. Importance of Inoculum Source and Initial Community Structure for Biogas Production from Agricultural Substrates. Bioresour. Technol. 2017, 245, 768–777. [Google Scholar] [CrossRef]
- Tonanzi, B.; Crognale, S.; Gianico, A.; Della Sala, S.; Miana, P.; Zaccone, M.C.; Rossetti, S. Microbial Community Successional Changes in a Full-Scale Mesophilic Anaerobic Digester from the Start-Up to the Steady-State Conditions. Microorganisms 2021, 9, 2581. [Google Scholar] [CrossRef]
- Kirkegaard, R.H.; McIlroy, S.J.; Kristensen, J.M.; Nierychlo, M.; Karst, S.M.; Dueholm, M.S.; Albertsen, M.; Nielsen, P.H. The Impact of Immigration on Microbial Community Composition in Full-Scale Anaerobic Digesters. Sci. Rep. 2017, 7, 9343. [Google Scholar] [CrossRef]
- Jiang, F.; Wang, S.; Zhang, Y.; Ma, S.; Huang, Y.; Fan, H.; Li, Q.; Wang, H.; Wang, A.; Liu, H.; et al. Variation of Metagenome From Feedstock to Digestate in Full-Scale Biogas Plants. Front. Microbiol. 2021, 12, 660225. [Google Scholar] [CrossRef]
- Pampillón-González, L.; Ortiz-Cornejo, N.L.; Luna-Guido, M.; Dendooven, L.; Navarro-Noya, Y.E. Archaeal and Bacterial Community Structure in an Anaerobic Digestion Reactor (Lagoon Type) Used for Biogas Production at a Pig Farm. Microb. Physiol. 2017, 27, 306–317. [Google Scholar] [CrossRef]
- Aigle, A.; Bourgeois, E.; Marjolet, L.; Houot, S.; Patureau, D.; Doelsch, E.; Cournoyer, B.; Galia, W. Relative Weight of Organic Waste Origin on Compost and Digestate 16S RRNA Gene Bacterial Profilings and Related Functional Inferences. Front. Microbiol. 2021, 12, 667043. [Google Scholar] [CrossRef]
- Wang, P.; Yu, Z.; Zhao, J.; Zhang, H. Do Microbial Communities in an Anaerobic Bioreactor Change with Continuous Feeding Sludge into a Full-Scale Anaerobic Digestion System? Bioresour. Technol. 2018, 249, 89–98. [Google Scholar] [CrossRef]
- Li, J.; Rui, J.; Yao, M.; Zhang, S.; Yan, X.; Wang, Y.; Yan, Z.; Li, X. Substrate Type and Free Ammonia Determine Bacterial Community Structure in Full-Scale Mesophilic Anaerobic Digesters Treating Cattle or Swine Manure. Front. Microbiol. 2015, 6, 1337. [Google Scholar] [CrossRef]
- Tapadia-Maheshwari, S.; Pore, S.; Engineer, A.; Shetty, D.; Dagar, S.S.; Dhakephalkar, P.K. Illustration of the Microbial Community Selected by Optimized Process and Nutritional Parameters Resulting in Enhanced Biomethanation of Rice Straw without Thermo-Chemical Pretreatment. Bioresour. Technol. 2019, 289, 121639. [Google Scholar] [CrossRef]
- Li, C.; He, P.; Hao, L.; Lü, F.; Shao, L.; Zhang, H. Diverse Acetate-Oxidizing Syntrophs Contributing to Biogas Production from Food Waste in Full-Scale Anaerobic Digesters in China. Renew. Energy 2022, 193, 240–250. [Google Scholar] [CrossRef]
- Calusinska, M.; Goux, X.; Fossépré, M.; Muller, E.E.L.; Wilmes, P.; Delfosse, P. A Year of Monitoring 20 Mesophilic Full-Scale Bioreactors Reveals the Existence of Stable but Different Core Microbiomes in Bio-Waste and Wastewater Anaerobic Digestion Systems. Biotechnol. Biofuels 2018, 11, 196. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Bayo, J.D.; Simmons, C.W.; VanderGheynst, J.S. Characterization of Digestate Microbial Community Structure Following Thermophilic Anaerobic Digestion with Varying Levels of Green and Food Wastes. J. Ind. Microbiol. Biotechnol. 2020, 47, 1031–1044. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, P.; Yang, X.; Lin, P.; Wang, Y.; Cheng, M.; Ren, L. Process Performance and Microbial Communities in Anaerobic Co-Digestion of Sewage Sludge and Food Waste with a Lower Range of Carbon/Nitrogen Ratio. Bioenerg. Res. 2022, 15, 1664–1674. [Google Scholar] [CrossRef]
- Esercizio, N.; Lanzilli, M.; Vastano, M.; Landi, S.; Xu, Z.; Gallo, C.; Nuzzo, G.; Manzo, E.; Fontana, A.; d’Ippolito, G. Fermentation of Biodegradable Organic Waste by the Family Thermotogaceae. Resources 2021, 10, 34. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, L.; Guo, B.; Liu, Y. Mesophiles Outperform Thermophiles in the Anaerobic Digestion of Blackwater with Kitchen Residuals: Insights into Process Limitations. Waste Manag. 2020, 105, 279–288. [Google Scholar] [CrossRef]
- Tanuwidjaja, I.; Vogel, C.; Pronk, G.J.; Schöler, A.; Kublik, S.; Vestergaard, G.; Kögel-Knabner, I.; Mrkonjic Fuka, M.; Schloter, M.; Schulz, S. Microbial Key Players Involved in P Turnover Differ in Artificial Soil Mixtures Depending on Clay Mineral Composition. Microb. Ecol. 2021, 81, 897–907. [Google Scholar] [CrossRef]
- Rajagopal, R.; Massé, D.I.; Singh, G. A Critical Review on Inhibition of Anaerobic Digestion Process by Excess Ammonia. Bioresour. Technol. 2013, 143, 632–641. [Google Scholar] [CrossRef]
- Vrieze, J.D.; Saunders, A.M.; He, Y.; Fang, J.; Nielsen, P.H.; Verstraete, W.; Boon, N. Ammonia and Temperature Determine Potential Clustering in the Anaerobic Digestion Microbiome. Water Res. 2015, 75, 312–323. [Google Scholar] [CrossRef]
- Alsouleman, K.; Linke, B.; Klang, J.; Klocke, M.; Krakat, N.; Theuerl, S. Reorganisation of a Mesophilic Biogas Microbiome as Response to a Stepwise Increase of Ammonium Nitrogen Induced by Poultry Manure Supply. Bioresour. Technol. 2016, 208, 200–204. [Google Scholar] [CrossRef]
- Wang, H.; Fotidis, I.A.; Angelidaki, I. Ammonia Effect on Hydrogenotrophic Methanogens and Syntrophic Acetate-Oxidizing Bacteria. FEMS Microbiol. Ecol. 2015, 91, fiv130. [Google Scholar] [CrossRef]
- Ruiz-Sánchez, J.; Campanaro, S.; Guivernau, M.; Fernández, B.; Prenafeta-Boldú, F.X. Effect of Ammonia on the Active Microbiome and Metagenome from Stable Full-Scale Digesters. Bioresour. Technol. 2018, 250, 513–522. [Google Scholar] [CrossRef]
- Monlau, F.; Sambusiti, C.; Barakat, A.; Quéméneur, M.; Trably, E.; Steyer, J.-P.; Carrère, H. Do Furanic and Phenolic Compounds of Lignocellulosic and Algae Biomass Hydrolyzate Inhibit Anaerobic Mixed Cultures? A Comprehensive Review. Biotechnol. Adv. 2014, 32, 934–951. [Google Scholar] [CrossRef]
- Kim, J.R.; Karthikeyan, K.G. Effects of Severe Pretreatment Conditions and Lignocellulose-Derived Furan Byproducts on Anaerobic Digestion of Dairy Manure. Bioresour. Technol. 2021, 340, 125632. [Google Scholar] [CrossRef]
- Levén, L.; Nyberg, K.; Schnürer, A. Conversion of Phenols during Anaerobic Digestion of Organic Solid Waste—A Review of Important Microorganisms and Impact of Temperature. J. Environ. Manag. 2012, 95, S99–S103. [Google Scholar] [CrossRef]
- Chapleur, O.; Madigou, C.; Civade, R.; Rodolphe, Y.; Mazéas, L.; Bouchez, T. Increasing Concentrations of Phenol Progressively Affect Anaerobic Digestion of Cellulose and Associated Microbial Communities. Biodegradation 2016, 27, 15–27. [Google Scholar] [CrossRef]
- Poirier, S.; Bize, A.; Bureau, C.; Bouchez, T.; Chapleur, O. Community Shifts within Anaerobic Digestion Microbiota Facing Phenol Inhibition: Towards Early Warning Microbial Indicators? Water Res. 2016, 100, 296–305. [Google Scholar] [CrossRef]
- Johnravindar, D.; Kaur, G.; Liang, J.; Lou, L.; Zhao, J.; Manu, M.K.; Kumar, R.; Varjani, S.; Wong, J.W.C. Impact of Total Solids Content on Biochar Amended Co-Digestion of Food Waste and Sludge: Microbial Community Dynamics, Methane Production and Digestate Quality Assessment. Bioresour. Technol. 2022, 361, 127682. [Google Scholar] [CrossRef]
- Jiang, J.; Wu, P.; Sun, Y.; Guo, Y.; Song, B.; Huang, Y.; Xing, T.; Li, L. Comparison of Microbial Communities during Anaerobic Digestion of Kitchen Waste: Effect of Substrate Sources and Temperatures. Bioresour. Technol. 2020, 317, 124016. [Google Scholar] [CrossRef]
- Akindolire, M.A.; Rama, H.; Roopnarain, A. Psychrophilic Anaerobic Digestion: A Critical Evaluation of Microorganisms and Enzymes to Drive the Process. Renew. Sustain. Energy Rev. 2022, 161, 112394. [Google Scholar] [CrossRef]
- Chen, C.; Sun, M.; Liu, Z.; Zhang, J.; Xiao, K.; Zhang, X.; Song, G.; Chang, J.; Liu, G.; Wang, H.; et al. Robustness of Granular Activated Carbon-Synergized Anaerobic Membrane Bioreactor for Pilot-Scale Application over a Wide Seasonal Temperature Change. Water Res. 2021, 189, 116552. [Google Scholar] [CrossRef]
- Guo, B.; Zhang, Y.; Zhang, L.; Zhou, Y.; Liu, Y. RNA-Based Spatial Community Analysis Revealed Intra-Reactor Variation and Expanded Collection of Direct Interspecies Electron Transfer Microorganisms in Anaerobic Digestion. Bioresour. Technol. 2020, 298, 122534. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Ni, J.; Ohtsu, A.; Isozumi, N.; Hu, Y.; Du, R.; Chen, Y.; Qin, Y.; Kubota, K.; Li, Y.-Y. Important Effects of Temperature on Treating Real Municipal Wastewater by a Submerged Anaerobic Membrane Bioreactor: Removal Efficiency, Biogas, and Microbial Community. Bioresour. Technol. 2021, 336, 125306. [Google Scholar] [CrossRef] [PubMed]
- Ribera-Pi, J.; Campitelli, A.; Badia-Fabregat, M.; Jubany, I.; Martínez-Lladó, X.; McAdam, E.; Jefferson, B.; Soares, A. Hydrolysis and Methanogenesis in UASB-AnMBR Treating Municipal Wastewater Under Psychrophilic Conditions: Importance of Reactor Configuration and Inoculum. Front. Bioeng. Biotechnol. 2020, 8, 567695. [Google Scholar] [CrossRef] [PubMed]
- Ao, T.; Xie, Z.; Zhou, P.; Liu, X.; Wan, L.; Li, D. Comparison of Microbial Community Structures between Mesophilic and Thermophilic Anaerobic Digestion of Vegetable Waste. Bioprocess. Biosyst. Eng. 2021, 44, 1201–1214. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Loh, K.-C.; Sarvanantharajah, S.; Tong, Y.W.; Wang, C.-H.; Dai, Y. Mesophilic and Thermophilic Anaerobic Digestion of Soybean Curd Residue for Methane Production: Characterizing Bacterial and Methanogen Communities and Their Correlations with Organic Loading Rate and Operating Temperature. Bioresour. Technol. 2019, 288, 121597. [Google Scholar] [CrossRef]
- Wang, P.; Wang, X.; Chen, X.; Ren, L. Effects of Bentonite on Antibiotic Resistance Genes in Biogas Slurry and Residue from Thermophilic and Mesophilic Anaerobic Digestion of Food Waste. Bioresour. Technol. 2021, 336, 125322. [Google Scholar] [CrossRef]
- Shin, J.; Rhee, C.; Shin, J.; Jang, H.M.; Shin, S.G.; Kim, Y.M. Determining the Composition of Bacterial Community and Relative Abundance of Specific Antibiotics Resistance Genes via Thermophilic Anaerobic Digestion of Sewage Sludge. Bioresour. Technol. 2020, 311, 123510. [Google Scholar] [CrossRef]
- Wen, Q.; Yang, S.; Chen, Z. Mesophilic and Thermophilic Anaerobic Digestion of Swine Manure with Sulfamethoxazole and Norfloxacin: Dynamics of Microbial Communities and Evolution of Resistance Genes. Front. Environ. Sci. Eng. 2021, 15, 94. [Google Scholar] [CrossRef]
- Xu, R.; Zhang, Y.; Xiong, W.; Sun, W.; Fan, Q.; Yang, Z. Metagenomic Approach Reveals the Fate of Antibiotic Resistance Genes in a Temperature-Raising Anaerobic Digester Treating Municipal Sewage Sludge. J. Clean. Prod. 2020, 277, 123504. [Google Scholar] [CrossRef]
- Romero-Güiza, M.S.; Vila, J.; Mata-Alvarez, J.; Chimenos, J.M.; Astals, S. The Role of Additives on Anaerobic Digestion: A Review. Renew. Sustain. Energy Rev. 2016, 58, 1486–1499. [Google Scholar] [CrossRef]
- Yao, Y.; Zhou, J.; An, L.; Kafle, G.K.; Chen, S.; Qiu, L. Role of Soil in Improving Process Performance and Methane Yield of Anaerobic Digestion with Corn Straw as Substrate. Energy 2018, 151, 998–1006. [Google Scholar] [CrossRef]
- Tabatabaei, M.; Aghbashlo, M.; Valijanian, E.; Panahi, H.K.S.; Nizami, A.-S.; Ghanavati, H.; Sulaiman, A.; Mirmohamadsadeghi, S.; Karimi, K. A Comprehensive Review on Recent Biological Innovations to Improve Biogas Production, Part 1: Upstream Strategies. Renew. Energy 2020, 146, 1204–1220. [Google Scholar] [CrossRef]
- Shen, Y.; Forrester, S.; Koval, J.; Urgun-Demirtas, M. Yearlong Semi-Continuous Operation of Thermophilic Two-Stage Anaerobic Digesters Amended with Biochar for Enhanced Biomethane Production. J. Clean. Prod. 2017, 167, 863–874. [Google Scholar] [CrossRef]
- Yan, M.; Treu, L.; Campanaro, S.; Tian, H.; Zhu, X.; Khoshnevisan, B.; Tsapekos, P.; Angelidaki, I.; Fotidis, I.A. Effect of Ammonia on Anaerobic Digestion of Municipal Solid Waste: Inhibitory Performance, Bioaugmentation and Microbiome Functional Reconstruction. Chem. Eng. J. 2020, 401, 126159. [Google Scholar] [CrossRef]
- Li, Y.; Li, L.; Sun, Y.; Yuan, Z. Bioaugmentation Strategy for Enhancing Anaerobic Digestion of High C/N Ratio Feedstock with Methanogenic Enrichment Culture. Bioresour. Technol. 2018, 261, 188–195. [Google Scholar] [CrossRef]
- Xu, X.; Sun, Y.; Sun, Y.; Li, Y. Bioaugmentation Improves Batch Psychrophilic Anaerobic Co-Digestion of Cattle Manure and Corn Straw. Bioresour. Technol. 2022, 343, 126118. [Google Scholar] [CrossRef]
- Linsong, H.; Lianhua, L.; Ying, L.; Changrui, W.; Yongming, S. Bioaugmentation with Methanogenic Culture to Improve Methane Production from Chicken Manure in Batch Anaerobic Digestion. Chemosphere 2022, 303, 135127. [Google Scholar] [CrossRef]
- Gurmessa, B.; Milanovic, V.; Pedretti, E.F.; Corti, G.; Ashworth, A.J.; Aquilanti, L.; Ferrocino, I.; Corvaglia, M.R.; Cocco, S. Post-Digestate Composting Shifts Microbial Composition and Degrades Antimicrobial Resistance Genes. Bioresour. Technol. 2021, 340, 125662. [Google Scholar] [CrossRef]
- Tamang, J.P. Biochemical and Modern Identification Techniques | Microfloras of Fermented Foods. In Encyclopedia of Food Microbiology; Elsevier: Amsterdam, The Netherlands; Academic Press: Cambridge, MA, USA, 2014; pp. 250–258. [Google Scholar]
- Hashemi, B.; Sarker, S.; Lamb, J.J.; Lien, K.M. Yield Improvements in Anaerobic Digestion of Lignocellulosic Feedstocks. J. Clean. Prod. 2021, 288, 125447. [Google Scholar] [CrossRef]
- Fell, J.W.; Guého-Kellermann, E. Chapter 143—Guehomyces Fell & Scorzetti (2004). In The Yeasts, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: London, UK, 2011; pp. 1773–1775. ISBN 978-0-444-52149-1. [Google Scholar]
- Hess, M.; Paul, S.S.; Puniya, A.K.; van der Giezen, M.; Shaw, C.; Edwards, J.E.; Fliegerová, K. Anaerobic Fungi: Past, Present, and Future. Front. Microbiol. 2020, 11, 584893. [Google Scholar] [CrossRef]
- Qin, S.; Wainaina, S.; Awasthi, S.K.; Mahboubi, A.; Liu, T.; Liu, H.; Zhou, Y.; Liu, H.; Zhang, Z.; Taherzadeh, M.J.; et al. Fungal Dynamics during Anaerobic Digestion of Sewage Sludge Combined with Food Waste at High Organic Loading Rates in Immersed Membrane Bioreactors. Bioresour. Technol. 2021, 335, 125296. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.M.-H.; Boss, P.K.; Walker, M.E.; Sumby, K.M.; Jiranek, V. Influence of Kazachstania Spp. on the Chemical and Sensory Profile of Red Wines. Int. J. Food Microbiol. 2022, 362, 109496. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, K.J.G.; Fernandes, J.A.L.; Magurno, F.; Leandro, L.B.A.; Goto, B.T.; Theodoro, R.C. Phylogenetic Review of Acaulospora (Diversisporales, Glomeromycota) and the Homoplasic Nature of Its Ornamentations. JoF 2022, 8, 892. [Google Scholar] [CrossRef] [PubMed]
- Kazda, M.; Langer, S.; Bengelsdorf, F.R. Fungi Open New Possibilities for Anaerobic Fermentation of Organic Residues. Energy Sustain. Soc. 2014, 4, 6. [Google Scholar] [CrossRef]
- Langer, S.G.; Gabris, C.; Einfalt, D.; Wemheuer, B.; Kazda, M.; Bengelsdorf, F.R. Different Response of Bacteria, Archaea and Fungi to Process Parameters in Nine Full-scale Anaerobic Digesters. Microb. Biotechnol. 2019, 12, 1210–1225. [Google Scholar] [CrossRef]
- Hagen, L.H.; Brooke, C.G.; Shaw, C.A.; Norbeck, A.D.; Piao, H.; Arntzen, M.Ø.; Olson, H.M.; Copeland, A.; Isern, N.; Shukla, A.; et al. Proteome Specialization of Anaerobic Fungi during Ruminal Degradation of Recalcitrant Plant Fiber. ISME J. 2021, 15, 421–434. [Google Scholar] [CrossRef]
- Zanellati, A.; Spina, F.; Poli, A.; Rollé, L.; Varese, G.C.; Dinuccio, E. Fungal Pretreatment of Non-Sterile Maize Silage and Solid Digestate with a Cephalotrichum Stemonitis Strain Selected from Agricultural Biogas Plants to Enhance Anaerobic Digestion. Biomass Bioenergy 2021, 144, 105934. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Z.; Li, S.; He, Q.; Peng, X.; Du, X.; Feng, K.; Wang, S.; Deng, Y. Fungal Dynamics and Potential Functions during Anaerobic Digestion of Food Waste. Environ. Res. 2022, 212, 113298. [Google Scholar] [CrossRef]
- Cheng, Y.F.; Edwards, J.E.; Allison, G.G.; Zhu, W.-Y.; Theodorou, M.K. Diversity and Activity of Enriched Ruminal Cultures of Anaerobic Fungi and Methanogens Grown Together on Lignocellulose in Consecutive Batch Culture. Bioresour. Technol. 2009, 100, 4821–4828. [Google Scholar] [CrossRef]
- Cheng, Y.; Shi, Q.; Sun, R.; Liang, D.; Li, Y.; Li, Y.; Jin, W.; Zhu, W. The Biotechnological Potential of Anaerobic Fungi on Fiber Degradation and Methane Production. World J. Microbiol. Biotechnol. 2018, 34, 155. [Google Scholar] [CrossRef]
- Jin, W.; Cheng, Y.-F.; Mao, S.-Y.; Zhu, W.-Y. Isolation of Natural Cultures of Anaerobic Fungi and Indigenously Associated Methanogens from Herbivores and Their Bioconversion of Lignocellulosic Materials to Methane. Bioresour. Technol. 2011, 102, 7925–7931. [Google Scholar] [CrossRef]
- Li, Y.; Meng, Z.; Xu, Y.; Shi, Q.; Ma, Y.; Aung, M.; Cheng, Y.; Zhu, W. Interactions between Anaerobic Fungi and Methanogens in the Rumen and Their Biotechnological Potential in Biogas Production from Lignocellulosic Materials. Microorganisms 2021, 9, 190. [Google Scholar] [CrossRef]
- Panahi, H.K.S.; Dehhaghi, M.; Guillemin, G.J.; Gupta, V.K.; Lam, S.S.; Aghbashlo, M.; Tabatabaei, M. A Comprehensive Review on Anaerobic Fungi Applications in Biofuels Production. Sci. Total Environ. 2022, 829, 154521. [Google Scholar] [CrossRef]
- Wang, X.; He, X.; Liang, J. Succession of Microbial Community during the Co-Composting of Food Waste Digestate and Garden Waste. IJERPH 2022, 19, 9945. [Google Scholar] [CrossRef]
- Meng, X.; Yan, J.; Zuo, B.; Wang, Y.; Yuan, X.; Cui, Z. Full-Scale of Composting Process of Biogas Residues from Corn Stover Anaerobic Digestion: Physical-Chemical, Biology Parameters and Maturity Indexes during Whole Process. Bioresour. Technol. 2020, 302, 122742. [Google Scholar] [CrossRef]
- Su, H.; Liu, L.; Wang, S.; Wang, Q.; Jiang, Y.; Hou, X.; Tan, T. Semi-Continuous Anaerobic Digestion for Biogas Production: Influence of Ammonium Acetate Supplement and Structure of the Microbial Community. Biotechnol. Biofuels 2015, 8, 13. [Google Scholar] [CrossRef]
- Yan, M.; Wang, B.; Xu, X.; der Meister, T.; Tabγač, H.; Hwang, F.; Liu, Z. Extrusion of Dissolved Oxygen by Exopolysaccharide From Leuconostoc Mesenteroides and Its Implications in Relief of the Oxygen Stress. Front. Microbiol. 2018, 9, 2467. [Google Scholar] [CrossRef]
- Moestedt, J.; Müller, B.; Nagavara Nagaraj, Y.; Schnürer, A. Acetate and Lactate Production During Two-Stage Anaerobic Digestion of Food Waste Driven by Lactobacillus and Aeriscardovia. Front. Energy Res. 2020, 8, 105. [Google Scholar] [CrossRef]
- Cai, G.; Zhao, L.; Wang, T.; Lv, N.; Li, J.; Ning, J.; Pan, X.; Zhu, G. Variation of Volatile Fatty Acid Oxidation and Methane Production during the Bioaugmentation of Anaerobic Digestion System: Microbial Community Analysis Revealing the Influence of Microbial Interactions on Metabolic Pathways. Sci. Total Environ. 2021, 754, 142425. [Google Scholar] [CrossRef]
- Wang, S.; Li, D.; Zhang, K.; Ma, Y.; Liu, F.; Li, Z.; Gao, X.; Gao, W.; Du, L. Effects of Initial Volatile Fatty Acid Concentrations on Process Characteristics, Microbial Communities, and Metabolic Pathways on Solid-State Anaerobic Digestion. Bioresour. Technol. 2023, 369, 128461. [Google Scholar] [CrossRef]
- Ács, N.; Szuhaj, M.; Wirth, R.; Bagi, Z.; Maróti, G.; Rákhely, G.; Kovács, K. Microbial Community Rearrangements in Power-to-Biomethane Reactors Employing Mesophilic Biogas Digestate. Front. Energy Res. 2019, 7, 132. [Google Scholar] [CrossRef]
- Akari, M.; Uchida, Y. Survival Rates of Microbial Communities from Livestock Waste to Soils: A Comparison between Compost and Digestate. Appl. Environ. Soil Sci. 2021, 2021, 6645203. [Google Scholar] [CrossRef]
- Li, Y.; Liu, C.; Wachemo, A.C.; Li, X. Effects of Liquid Fraction of Digestate Recirculation on System Performance and Microbial Community Structure during Serial Anaerobic Digestion of Completely Stirred Tank Reactors for Corn Stover. Energy 2018, 160, 309–317. [Google Scholar] [CrossRef]
- Mang, S.M.; Trotta, V.; Scopa, A.; Camele, I. Metagenomic Analysis of Bacterial Community Structure and Dynamics of a Digestate and a More Stabilized Digestate-Derived Compost from Agricultural Waste. Processes 2022, 10, 379. [Google Scholar] [CrossRef]
- Westerholm, M.; Castillo, M.D.P.; Andersson, A.C.; Nilsen, P.J.; Schnürer, A. Effects of Thermal Hydrolytic Pre-Treatment on Biogas Process Efficiency and Microbial Community Structure in Industrial- and Laboratory-Scale Digesters. Waste Manag. 2019, 95, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Södergren, J.; Larsson, C.U.; Wadsö, L.; Bergstrand, K.-J.; Asp, H.; Hultberg, M.; Schelin, J. Food Waste to New Food: Risk Assessment and Microbial Community Analysis of Anaerobic Digestate as a Nutrient Source in Hydroponic Production of Vegetables. J. Clean. Prod. 2022, 333, 130239. [Google Scholar] [CrossRef]
- Díaz, A.I.; Oulego, P.; Collado, S.; Laca, A.; González, J.M.; Díaz, M. Impact of Anaerobic Digestion and Centrifugation/Decanting Processes in Bacterial Communities Fractions. J. Biosci. Bioeng. 2018, 126, 742–749. [Google Scholar] [CrossRef]
- Dennehy, C.; Lawlor, P.G.; McCabe, M.S.; Cormican, P.; Sheahan, J.; Jiang, Y.; Zhan, X.; Gardiner, G.E. Anaerobic Co-Digestion of Pig Manure and Food Waste; Effects on Digestate Biosafety, Dewaterability, and Microbial Community Dynamics. Waste Manag. 2018, 71, 532–541. [Google Scholar] [CrossRef]
- Madegwa, Y.M.; Uchida, Y. Liming Improves the Stability of Soil Microbial Community Structures against the Application of Digestate Made from Dairy Wastes. J. Environ. Manag. 2021, 297, 113356. [Google Scholar] [CrossRef]
- Garbini, G.L.; Grenni, P.; Rauseo, J.; Patrolecco, L.; Pescatore, T.; Spataro, F.; Barra Caracciolo, A. Insights into Structure and Functioning of a Soil Microbial Community Amended with Cattle Manure Digestate and Sulfamethoxazole. J. Soils Sediments 2022, 22, 2158–2173. [Google Scholar] [CrossRef]
- Franke-Whittle, I.H.; Confalonieri, A.; Insam, H.; Schlegelmilch, M.; Körner, I. Changes in the Microbial Communities during Co-Composting of Digestates. Waste Manag. 2014, 34, 632–641. [Google Scholar] [CrossRef]
- Lim, J.W.; Park, T.; Tong, Y.W.; Yu, Z. Chapter One—The Microbiome Driving Anaerobic Digestion and Microbial Analysis. In Advances in Bioenergy; Li, Y., Khanal, S.K., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; Volume 5, pp. 1–61. [Google Scholar]
- Roopnarain, A.; Rama, H.; Ndaba, B.; Bello-Akinosho, M.; Bamuza-Pemu, E.; Adeleke, R. Unravelling the Anaerobic Digestion ‘Black Box’: Biotechnological Approaches for Process Optimization. Renew. Sustain. Energy Rev. 2021, 152, 111717. [Google Scholar] [CrossRef]
- Risberg, K.; Cederlund, H.; Pell, M.; Arthurson, V.; Schnürer, A. Comparative Characterization of Digestate versus Pig Slurry and Cow Manure—Chemical Composition and Effects on Soil Microbial Activity. Waste Manag. 2017, 61, 529–538. [Google Scholar] [CrossRef]
- Mohammadi, K.; Sohrabi, Y. Bacterial Biofertilizers for Sustainable Crop Production: A Review. ARPN J. Agric. Biol. Sci. 2012, 7, 307–316. [Google Scholar]
- Guo Gao, T.; Yuan Xu, Y.; Jiang, F.; Zhen Li, B.; Shui Yang, J.; Tao Wang, E.; Li Yuan, H. Nodulation Characterization and Proteomic Profiling of Bradyrhizobium Liaoningense CCBAU05525 in Response to Water-Soluble Humic Materials. Sci. Rep. 2015, 5, 10836. [Google Scholar] [CrossRef]
- Caracciolo, A.B.; Bustamante, M.A.; Nogues, I.; Lenola, M.D.; Luprano, M.L.; Grenni, P. Changes in Microbial Community Structure and Functioning of a Semiarid Soil Due to the Use of Anaerobic Digestate Derived Composts and Rosemary Plants. Geoderma 2015, 245–246, 89–97. [Google Scholar] [CrossRef]
- Walsh, J.J.; Rousk, J.; Edwards-Jones, G.; Jones, D.L.; Williams, A.P. Fungal and Bacterial Growth Following the Application of Slurry and Anaerobic Digestate of Livestock Manure to Temperate Pasture Soils. Biol. Fertil Soils 2012, 48, 889–897. [Google Scholar] [CrossRef]
- Fernandes, F.; Silkina, A.; Gayo-Peláez, J.I.; Kapoore, R.V.; de la Broise, D.; Llewellyn, C.A. Microalgae Cultivation on Nutrient Rich Digestate: The Importance of Strain and Digestate Tailoring under PH Control. Appl. Sci. 2022, 12, 5429. [Google Scholar] [CrossRef]
- Higgins, B.T.; Gennity, I.; Fitzgerald, P.S.; Ceballos, S.J.; Fiehn, O.; VanderGheynst, J.S. Algal–Bacterial Synergy in Treatment of Winery Wastewater. NPJ Clean Water 2018, 1, 6. [Google Scholar] [CrossRef]
- Tawfik, A.; Eraky, M.; Alhajeri, N.S.; Osman, A.I.; Rooney, D.W. Cultivation of Microalgae on Liquid Anaerobic Digestate for Depollution, Biofuels and Cosmetics: A Review. Environ. Chem. Lett. 2022, 20, 3631–3656. [Google Scholar] [CrossRef]
- Uggetti, E.; Sialve, B.; Latrille, E.; Steyer, J.-P. Anaerobic Digestate as Substrate for Microalgae Culture: The Role of Ammonium Concentration on the Microalgae Productivity. Bioresour. Technol. 2014, 152, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Lü, F.; He, P.; Shao, L. Impact of liquid volume of recycled methanogenic effluent on anaerobic hydrolysis. Huan Jing Ke Xue 2008, 29, 2637–2642. [Google Scholar]
- Maria, F.D.; Barratta, M.; Bianconi, F.; Placidi, P.; Passeri, D. Solid Anaerobic Digestion Batch with Liquid Digestate Recirculation and Wet Anaerobic Digestion of Organic Waste: Comparison of System Performances and Identification of Microbial Guilds. Waste Manag. 2017, 59, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Yu, Z.; Li, Y. Sequential Batch Thermophilic Solid-State Anaerobic Digestion of Lignocellulosic Biomass via Recirculating Digestate as Inoculum—Part II: Microbial Diversity and Succession. Bioresour. Technol. 2017, 241, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Bello-Akinosho, M.; Adeleke, R.; Swanevelder, D.; Thantsha, M. Draft Genome Sequence of Pseudomonas Sp. Strain 10-1B, a Polycyclic Aromatic Hydrocarbon Degrader in Contaminated Soil. Genome Announc. 2015, 3, e00325-15. [Google Scholar] [CrossRef]
- Abtahi, H.; Parhamfar, M.; Saeedi, R.; Villaseñor, J.; Sartaj, M.; Kumar, V.; Coulon, F.; Parhamfar, M.; Didehdar, M.; Seifi, H.; et al. Effect of Competition between Petroleum-Degrading Bacteria and Indigenous Compost Microorganisms on the Efficiency of Petroleum Sludge Bioremediation: Field Application of Mineral-Based Culture in the Composting Process. J. Environ. Manag. 2020, 258, 110013. [Google Scholar] [CrossRef]
- Kataki, S.; Hazarika, S.; Baruah, D.C. Assessment of By-Products of Bioenergy Systems (Anaerobic Digestion and Gasification) as Potential Crop Nutrient. Waste Manag. 2017, 59, 102–117. [Google Scholar] [CrossRef]
- Wang, Y.; Pandey, P.K.; Kuppu, S.; Pereira, R.; Aly, S.; Zhang, R. Degradation of Antibiotic Resistance Genes and Mobile Gene Elements in Dairy Manure Anerobic Digestion. PLoS ONE 2021, 16, e0254836. [Google Scholar] [CrossRef]
- Sun, W.; Qian, X.; Gu, J.; Wang, X.-J.; Duan, M.-L. Mechanism and Effect of Temperature on Variations in Antibiotic Resistance Genes during Anaerobic Digestion of Dairy Manure. Sci. Rep. 2016, 6, 30237. [Google Scholar] [CrossRef]
- Flores-Orozco, D.; Patidar, R.; Levin, D.B.; Sparling, R.; Kumar, A.; Çiçek, N. Effect of Mesophilic Anaerobic Digestion on the Resistome Profile of Dairy Manure. Bioresour. Technol. 2020, 315, 123889. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roopnarain, A.; Akindolire, M.A.; Rama, H.; Ndaba, B. Casting Light on the Micro-Organisms in Digestate: Diversity and Untapped Potential. Fermentation 2023, 9, 160. https://doi.org/10.3390/fermentation9020160
Roopnarain A, Akindolire MA, Rama H, Ndaba B. Casting Light on the Micro-Organisms in Digestate: Diversity and Untapped Potential. Fermentation. 2023; 9(2):160. https://doi.org/10.3390/fermentation9020160
Chicago/Turabian StyleRoopnarain, Ashira, Muyiwa Ajoke Akindolire, Haripriya Rama, and Busiswa Ndaba. 2023. "Casting Light on the Micro-Organisms in Digestate: Diversity and Untapped Potential" Fermentation 9, no. 2: 160. https://doi.org/10.3390/fermentation9020160
APA StyleRoopnarain, A., Akindolire, M. A., Rama, H., & Ndaba, B. (2023). Casting Light on the Micro-Organisms in Digestate: Diversity and Untapped Potential. Fermentation, 9(2), 160. https://doi.org/10.3390/fermentation9020160






