Microbial Diversity of Traditional Livno Cheese from Bosnia and Herzegovina
Abstract
1. Introduction
2. Materials and Methods
2.1. Traditional Livno Cheese Production and Sampling
2.2. Culture-Dependent Microbial Analysis
2.3. Culture-Independent Microbial Analysis
2.3.1. Extraction and Purification of Total DNA from Cheese
2.3.2. PCR-DGGE Analysis and Band Identification by Sequencing
2.3.3. ARISA
2.4. Statistical Analysis
3. Results
3.1. Enumeration of Bacteria Using Culture-Dependent Methods
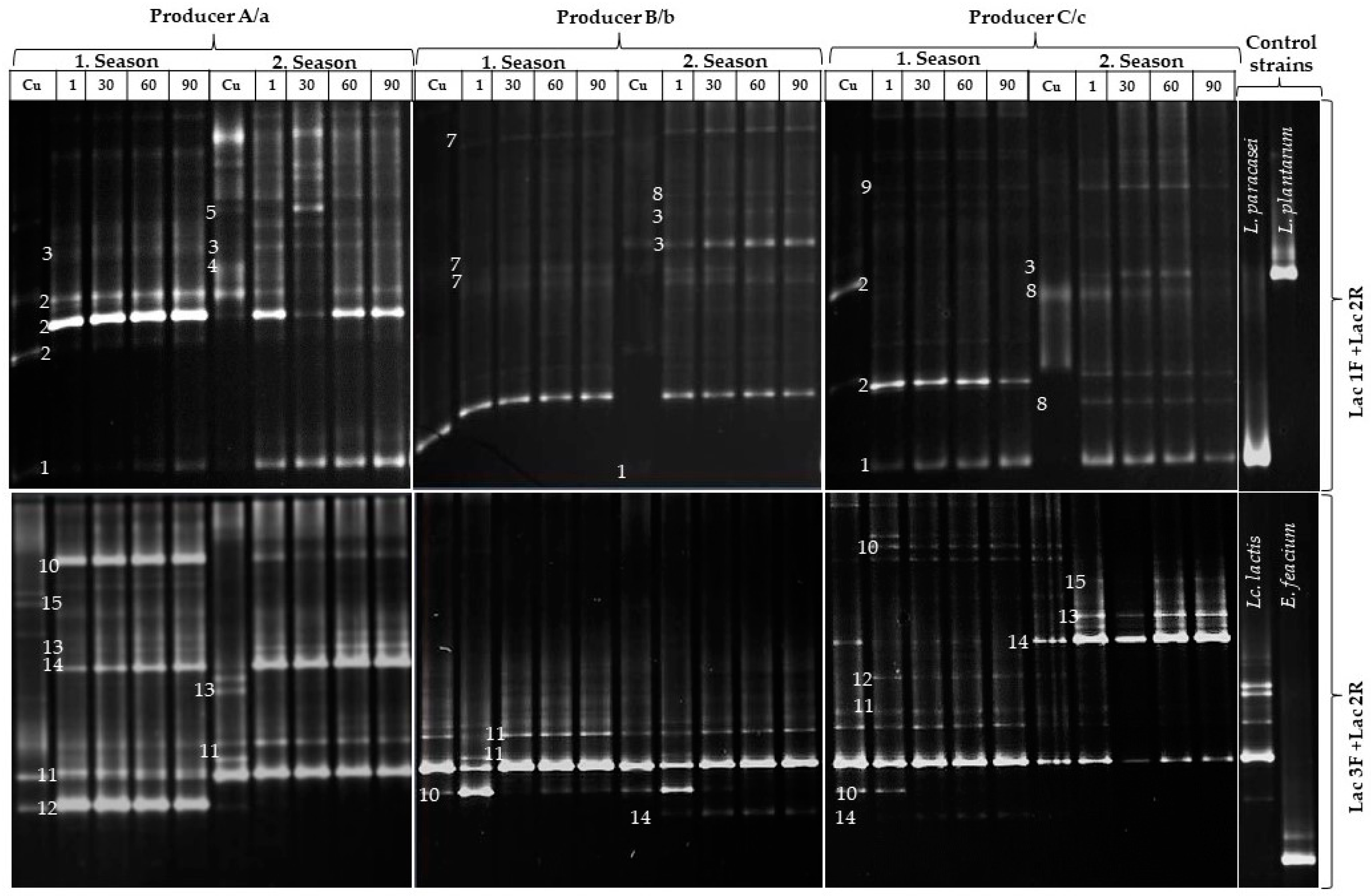
3.2. Microbial Diversity of Traditional Livno Cheese by PCR-DGGE
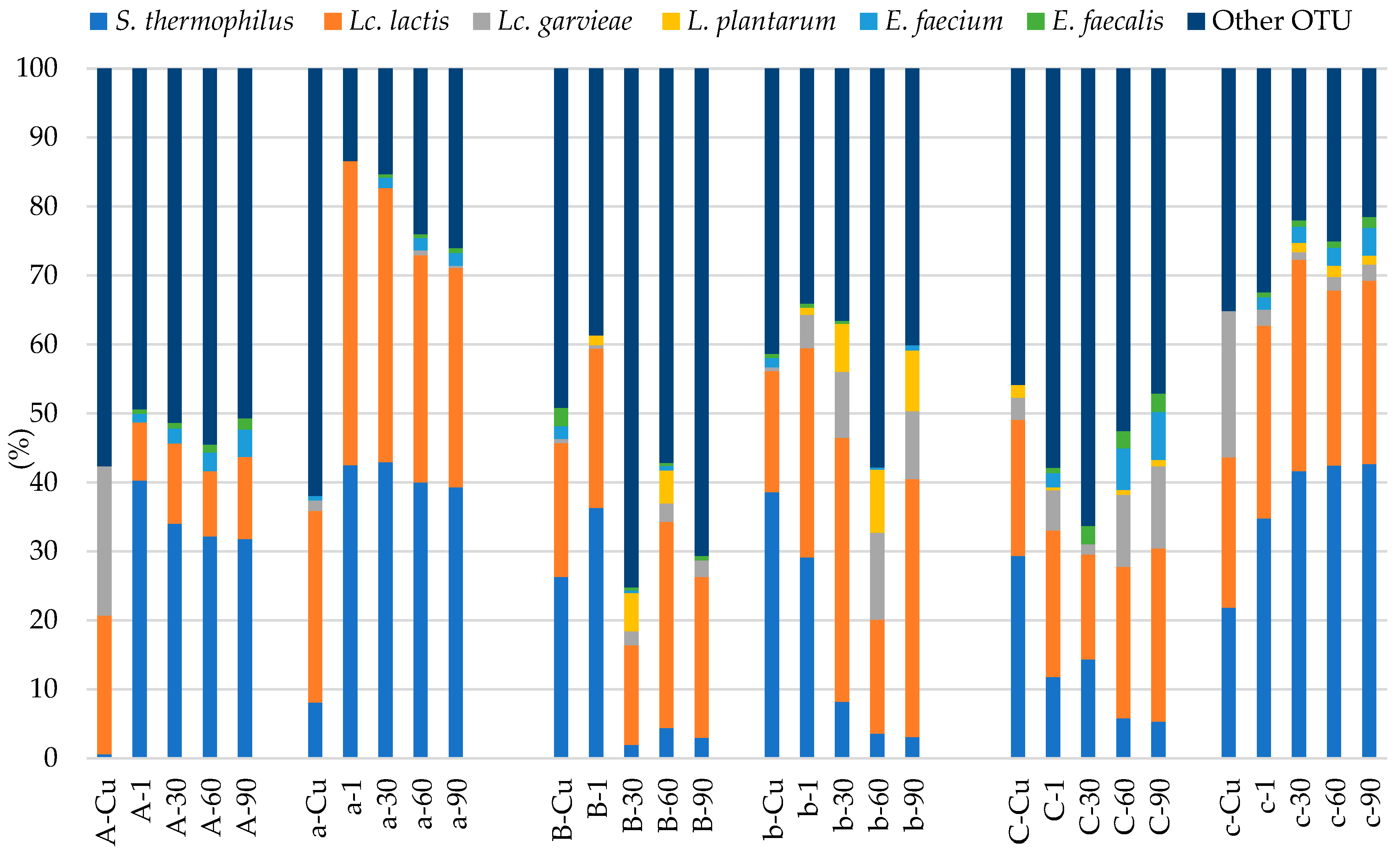
3.3. Microbial Diversity of Livno Cheese Using ARISA
4. Discussion
4.1. Enumeration of Bacteria on Agar in Livno Cheese
4.2. DGGE
4.3. ARISA
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Montel, M.C.; Buchin, S.; Mallet, A.; Delbes-Paus, C.; Vuitton, D.A.; Desmasures, N.; Berthier, F. Traditional cheeses: Rich and diverse microbiota with associated benefits. Int. J. Food Microbiol. 2014, 177, 136–154. [Google Scholar] [CrossRef] [PubMed]
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. The complex microbiota of raw milk. FEMS Microbiol. Rev. 2013, 37, 664–698. [Google Scholar] [CrossRef] [PubMed]
- Carraro, L.; Maifreni, M.; Bartolomeoli, I.; Martino, M.E.; Novelli, E.; Frigo, F.; Marino, M.; Cardazzo, B. Comparison of culture-dependent and-independent methods for bacterial community monitoring during Montasio cheese manufacturing. Res. Microbiol. 2011, 162, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Mayo, B.; Rodríguez, J.; Vázquez, L.; Flórez, A.B. Microbial interactions within the cheese ecosystem and their application to improve quality and safety. Foods 2021, 10, 602. [Google Scholar] [CrossRef]
- González, L.; Cuadrillero, A.F.; Castro, J.M.; Bernardo, A.; Tornadijo, M.E. Selection of lactic acid bacteria isolated from San Simón da Costa Cheese (PDO) in order to develop an autochthonous starter culture. Adv. Microbiol. 2015, 5, 748–759. [Google Scholar] [CrossRef][Green Version]
- Coelho, M.C.; Malcata, F.X.; Silva, C.C. Lactic acid bacteria in raw-milk cheeses: From starter cultures to probiotic functions. Foods 2022, 11, 2276. [Google Scholar] [CrossRef]
- Vásquez, A.; Molin, G.; Pettersson, B.; Antonsson, M.; Ahrné, S. DNA-based classification and sequence heterogeneities in the 16S rRNA genes of Lactobacillus casei/paracasei and related species. Syst. Appl. Microbiol. 2005, 28, 430–441. [Google Scholar] [CrossRef]
- Van Geel-Schuttená, G.H.; Flesch, F.; ten Brink, B.; Smith, M.R.; Dijkhuizen, L. Screening and characterization of Lactobacillus strains producing large amounts of exopolysaccharides. Appl. Microbiol. Biotechnol. 1998, 50, 697–703. [Google Scholar] [CrossRef]
- Hati, S.; Mandal, S.; Prajapati, J.B. Novel starters for value added fermented dairy products. Curr. Res. Nutr. Food Sci. J. 2013, 1, 83–91. [Google Scholar] [CrossRef]
- Cogan, T.M.; Barbosa, M.; Beuvier, E.; Bianchi-Salvadori, B.; Cocconcelli, P.S.; Fernandes, I.; Gomez, J.; Gomez, R.; Kalantzopoulos, G.; Ledda, A.; et al. Characterization of the lactic acid bacteria in artisanal dairy products. J. Dairy Res. 1997, 64, 409–421. [Google Scholar] [CrossRef]
- Chambers, D.H.; Esteve, E.; Retiveau, A. Effect of milk pasteurization on flavor properties of seven commercially available French cheese types. J. Sens. Stud. 2010, 25, 494–511. [Google Scholar] [CrossRef]
- Arias-Roth, E.; Bachmann, H.P.; Frölich-Wyder, M.T.; Schmidt, R.S.; Wechsler, D.; Beuvier, E.; Delbès, C. Raw milk cheeses. In Encyclopedia of Dairy Sciences, 3rd ed.; McSweeney, P.L.H., McNamara, J.P., Eds.; Elsevier: Philadelphia, PA, USA, 2022; pp. 299–308. [Google Scholar]
- Zepeda Mendoza, M.L.; Sicheritz-Ponten, T.; Gilbert, M.T.P. Environmental genes and genomes: Understanding the differences and challenges in the approaches and software for their analyses. Brief. Bioinform. 2015, 16, 745–758. [Google Scholar] [CrossRef] [PubMed]
- Gatti, M.; Bernini, V.; Lazzi, C.; Neviani, E. Fluorescence microscopy for studying the viability of micro-organisms in natural whey starters. Lett. Appl. Microbiol. 2006, 42, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Rowan, N.J. Viable but non-culturable forms of food and waterborne bacteria: Quo Vadis? Trends Food Sci. Technol. 2004, 15, 462–467. [Google Scholar] [CrossRef]
- Feligini, M.; Panelli, S.; Buffoni, J.N.; Bonacina, C.; Andrighetto, C.; Lombardi, A. Identification of microbiota present on the surface of Taleggio cheese using PCR-DGGE and RAPD-PCR. J. Food Sci. 2012, 77, 609–615. [Google Scholar] [CrossRef]
- Ndoye, B.; Rasolofo, E.A.; LaPointe, G.; Roy, D. A review of the molecular approaches to investigate the diversity and activity of cheese microbiota. Dairy Sci. Technol. 2011, 91, 495–524. [Google Scholar] [CrossRef]
- Ercolini, D.; Moschetti, G.; Blaiotta, G.; Coppola, S. The potential of a polyphasic PCR-DGGE Approach in evaluating microbial diversity of natural whey cultures for water-buffalo mozzarella cheese production: Bias of culture-dependent and culture-independent analyses. Syst. Appl. Microbiol. 2001, 24, 610–617. [Google Scholar] [CrossRef]
- Ferrocino, I.; Rantsiou, K.; Cocolin, L. Investigating dairy microbiome: An opportunity to ensure quality, safety and typicity. Curr. Opin. Biotechnol. 2022, 73, 164–170. [Google Scholar] [CrossRef]
- Bonizzi, I.; Feligini, M.; Aleandri, R.; Enne, G. Genetic traceability of the geographical origin of typical Italian water buffalo Mozzarella cheese: A preliminary approach. J. Appl. Microbiol. 2007, 102, 667–673. [Google Scholar] [CrossRef]
- Arteau, M.; Labrie, S.; Roy, D. Terminal-restriction fragment length polymorphism and automated ribosomal intergenic spacer analysis profiling of fungal communities in Camembert cheese. Int. Dairy J. 2010, 20, 545–554. [Google Scholar] [CrossRef]
- Feligini, M.; Brambati, E.; Panelli, S.; Ghitti, M.; Sacchi, R.; Capelli, E.; Bonacina, C. One-year investigation of Clostridium spp. occurrence in raw milk and curd of Grana Padano cheese by the automated ribosomal intergenic spacer analysis. Food Control 2014, 42, 71–77. [Google Scholar] [CrossRef]
- Porcellato, D.; Johnson, M.E.; Houck, K.; Skeie, S.B.; Mills, D.A.; Kalanetra, K.M.; Steele, J.L. Potential of Lactobacillus curvatus LFC1 to produce slits in Cheddar cheese. Food Microbiol. 2015, 49, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Picozzi, C.; Clagnan, E.; Musatti, A.; Rollini, M.; Brusetti, L. Characterization of Two Zymomonas mobilis Wild Strains and Analysis of Populations Dynamics during Their Leavening of Bread-like Doughs. Foods 2022, 11, 2768. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Hudagula, H.; Minami, N.; Maeno, N.; Yoshida, K.; Onodera, S.; Takeda, Y.; Tobiyama, T.; Nakamura, T.; Hanai, J.; et al. A model study for contributing factors of the fermentation of qvevri wine. Food Control 2023, 148, 109668. [Google Scholar] [CrossRef]
- Florczyk, M.; Cydzik-Kwiatkowska, A.; Ziembinska-Buczynska, A.; Ciesielski, S. Comparison of Three DNA Extraction Kits for Assessment of Bacterial Diversity in Activated Sludge, Biofilm, and Anaerobic Digestate. Appl. Sci. 2022, 12, 9797. [Google Scholar] [CrossRef]
- Bigot, C.; Meile, J.; Remize, F.; Strub, C. Applications of metagenomics to fermented foods. In Fermented Foods, Part I: Biochemistry and Biotechnology; Montet, D., Ray, R.C., Eds.; CRC Press: Boca Raton, FL, USA, 2015; pp. 333–346. [Google Scholar]
- Hornik, B.; Czarny, J.; Staninska-Pięta, J.; Wolko, Ł.; Cyplik, P.; Piotrowska-Cyplik, A. The raw milk microbiota from semi-subsistence farms characteristics by NGS analysis method. Molecules 2021, 26, 5029. [Google Scholar] [CrossRef]
- Porcellato, D.; Grønnevik, H.; Rudi, K.; Narvhus, J.; Skeie, S.B. Rapid lactic acid bacteria identification in dairy products by high-resolution melt analysis of DGGE bands. Lett. Appl. Microbiol. 2012, 54, 344–351. [Google Scholar] [CrossRef]
- Walter, J.; Hertel, C.; Tannock, G.W.; Lis, C.M.; Munro, K.; Hammes, W.P. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 2001, 67, 2578–2585. [Google Scholar] [CrossRef]
- Endo, A.; Okada, S. Monitoring the lactic acid bacterial diversity during shochu fermentation by PCR-denaturing gradient gel electrophoresis. J. Biosci. Bioeng. 2005, 99, 216–221. [Google Scholar] [CrossRef]
- Porcellato, D.; Brighton, C.; McMahon, D.J.; Oberg, C.J.; Lefevre, M.; Broadbent, J.R.; Steele, J.L. Application of ARISA to assess the influence of salt content and cation type on microbiological diversity of Cheddar cheese. Lett. Appl. Microbiol. 2014, 59, 207–216. [Google Scholar] [CrossRef]
- Fisher, M.M.; Triplett, E.W. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 1999, 65, 4630–4636. [Google Scholar] [CrossRef] [PubMed]
- Schirone, M.; Tofalo, R.; Fasoli, G.; Perpetuini, G.; Corsetti, A.; Manetta, A.C.; Suzzi, G. High content of biogenic amines in Pecorino cheeses. Food Microbiol. 2012, 34, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Di Cagno, R.; Banks, J.; Sheehan, L.; Fox, P.F.; Brechany, E.Y.; Corsetti, A.; Gobbetti, M. Comparison of the microbiological, compositional, biochemical, volatile profile and sensory characteristics of three Italian PDO ewes’ milk cheeses. Int. Dairy J. 2003, 13, 961–972. [Google Scholar] [CrossRef]
- Callon, C.; Berdagué, J.L.; Dufour, E.; Montel, M.C. The effect of raw milk microbial flora on the sensory characteristics of Salers-type cheeses. J. Dairy Sci. 2005, 88, 3840–3850. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, M.; Corsetti, A.; Tosti, N.; Rossi, J.; Corbo, M.R.; Gobbetti, M. Characterization of non-starter lactic acid bacteria from Italian ewe cheeses based on phenotypic, genotypic, and cell wall protein analyses. Appl. Environ. Microbiol. 2001, 67, 2011–2020. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Mills, D.A. Next-generation approaches to the microbial ecology of food fermentations. BMB Rep. 2012, 45, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Fitzsimons, N.A.; Cogan, T.M.; Condon, S.; Beresford, T. Spatial and temporal distribution of non-starter lactic acid bacteria in Cheddar cheese. J. Appl. Microbiol. 2001, 90, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Beresford, T.; Williams, A. The microbiology of cheese ripening. In Cheese: Chemistry, Physics and Microbiology, Vol 1: General Aspects, 3rd ed.; Fox, P.F., McSweeney, P.L.H., Cogan, T.M., Guinee, T.P., Eds.; Academic Press: Cambridge, MA, USA, 2004; pp. 287–318. [Google Scholar]
- Gobbetti, M.; De Angelis, M.; Di Cagno, R.; Mancini, L.; Fox, P.F. Pros and cons for using non-starter lactic acid bacteria (NSLAB) as secondary/adjunct starters for cheese ripening. Trends Food Sci. Technol. 2015, 45, 167–178. [Google Scholar] [CrossRef]
- Guarcello, R.; Diviccaro, A.; Barbera, M.; Giancippoli, E.; Settanni, L.; Minervini, F.; Giancarlo, M.; Gobbetti, M. A survey of the main technology, biochemical and microbiological features influencing the concentration of biogenic amines of twenty Apulian and Sicilian (Southern Italy) cheeses. Int. Dairy J. 2015, 43, 61–69. [Google Scholar] [CrossRef][Green Version]
- Arizcun, C.; Barcina, Y.; Torre, P. Identification of lactic acid bacteria isolated from Roncal and Idiazabal cheeses. Le Lait 1997, 77, 729–736. [Google Scholar] [CrossRef]
- Florez, A.B.; Mayo, B. Microbial diversity and succession during the manufacture and ripening of traditional, Spanish, blue-veined Cabrales cheese, as determined by PCR-DGGE. Int. J. Food Microbiol. 2006, 110, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Litopoulou-Tzanetaki, E.; Tzanetakis, N. Microbiological characteristics of Greek traditional cheeses. Small Rumin. Res. 2011, 101, 17–32. [Google Scholar] [CrossRef]
- Bouton, Y.; Guyot, P.; Grappin, R. Preliminary characterization of microflora of Comté cheese. J. Appl. Microbiol. 1998, 85, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Giraffa, G. Functionality of enterococci in dairy products. Int. J. Food Microbiol. 2003, 88, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Sarantinopoulos, P.; Leroy, F.; Leontopoulou, E.; Georgalaki, M.D.; Kalantzopoulos, G.; Tsakalidou, E.; De Vuyst, L. Bacteriocin production by Enterococcus faecium FAIR-E 198 in view of its application as adjunct starter in Greek Feta cheese making. Int. J. Food Microbiol. 2002, 72, 125–136. [Google Scholar] [CrossRef]
- Mundt, J.O. Enterococci. In Bergey’s Manual of Systematic Bacteriology. Volume 2; Sneath, P.H.A., Mair, N.S., Sharpe, M.E., Holt, J.G., Eds.; Williams &Wilkins: Baltimore, MD, USA, 1986; pp. 1063–1065. [Google Scholar]
- Cosentino, S.; Pisano, M.B.; Corda, A.; Fadda, M.E.; Piras, C. Genotypic and technological characterization of enterococci isolated from artisanal Fiore Sardo cheese. J. Dairy Res. 2004, 71, 444–450. [Google Scholar] [CrossRef]
- Majhenič, A.Č.; Rogelj, I.; Perko, B. Enterococci from Tolminc cheese: Population structure, antibiotic susceptibility and incidence of virulence determinants. Int. J. Food Microbiol. 2005, 102, 239–244. [Google Scholar] [CrossRef]
- Nieto-Arribas, P.; Seseña, S.; Poveda, J.M.; Chicón, R.; Cabezas, L.; Palop, L. Enterococcus populations in artisanal Manchego cheese: Biodiversity, technological and safety aspects. Food Microbiol. 2011, 28, 891–899. [Google Scholar] [CrossRef]
- Bonetta, S.; Bonetta, S.; Carraro, E.; Rantsiou, K.; Cocolin, L. Microbiological characterisation of Robiola di Roccaverano cheese using PCR–DGGE. Food Microbiol. 2008, 25, 786–792. [Google Scholar] [CrossRef]
- Gala, E.; Landi, S.; Solieri, L.; Nocetti, M.; Pulvirenti, A.; Giudici, P. Diversity of lactic acid bacteria population in ripened Parmigiano Reggiano cheese. Int. J. Food Microbiol. 2008, 125, 347–351. [Google Scholar] [CrossRef]
- Randazzo, C.L.; Pitino, I.; Ribbera, A.; Caggia, C. Pecorino Crotonese cheese: Study of bacterial population and flavour compounds. Food Microbiol. 2010, 27, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Demirci, T.; Akin, N.; Atik, D.S.; Özkan, E.R.; Dertli, E.; Akyol, İ. Lactic acid bacteria diversity and dynamics during ripening of traditional Turkish goatskin Tulum cheese produced in Mut region assessed by culturing and PCR-DGGE. LWT 2021, 138, 110701. [Google Scholar] [CrossRef]
- Chombo-Morales, P.; Kirchmayr, M.; Gschaedler, A.; Lugo-Cervantes, E.; Villanueva-Rodríguez, S. Effects of controlling ripening conditions on the dynamics of the native microbial population of Mexican artisanal Cotija cheese assessed by PCR-DGGE. LWT-Food Sci. Technol. 2016, 65, 1153–1161. [Google Scholar] [CrossRef]
- Randazzo, C.L.; Torriani, S.; Akkermans, A.D.; de Vos, W.M.; Vaughan, E.E. Diversity, dynamics, and activity of bacterial communities during production of an artisanal Sicilian cheese as evaluated by 16S rRNA analysis. Appl. Environ. Microbiol. 2002, 68, 1882–1892. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, C.L.; Caggia, C.; Neviani, E. Application of molecular approaches to study lactic acid bacteria in artisanal cheeses. J. Microbiol. Methods 2009, 78, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Fuka, M.M.; Engel, M.; Skelin, A.; Redžepović, S.; Schloter, M. Bacterial communities associated with the production of artisanal Istrian cheese. Int. J. Food Microbiol. 2010, 142, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Havranek, J.; Mikulec, N.; Antunac, N.; Iveković, D.; Jakaša, I.; Rogelj, I.; Perko, B.; Trmčić, A.; Majhenić, A.Č.; Sarić, Z.; et al. Croatian cheeses. In An Atlas of Sheep Cheeses of the Countries of the Western Balkans; Sveučilišna Tiskara: Zagreb, Croatia, 2012; pp. 17–27. [Google Scholar]
- Pogačić, T.; D’Andrea, M.; Kagkli, D.M.; Corich, V.; Giacomini, A.; Baldan, E.; Samaržija, D. Biodiversity of microbial consortia isolated from traditional fresh sheep cheese Karakačanski skakutanac. Mljekarstvo 2011, 61, 208–219. [Google Scholar]
- Edalatian, M.R.; Najafi, M.B.H.; Mortazavi, S.A.; Alegría, Á.; Nassiri, M.R.; Bassami, M.R.; Mayo, B. Microbial diversity of the traditional Iranian cheeses Lighvan and Koozeh, as revealed by polyphasic culturing and culture-independent approaches. Dairy Sci. Technol. 2012, 92, 75–90. [Google Scholar] [CrossRef]
- Ogier, J.C.; Lafarge, V.; Girard, V.; Rault, A.; Maladen, V.; Gruss, A.; Leveau, J.-Y.; Delacroix-Buchet, A. Molecular fingerprinting of dairy microbial ecosystems by use of temporal temperature and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 2004, 70, 5628–5643. [Google Scholar] [CrossRef]
- González-Cortés, J.J.; Valle, A.; Ramírez, M.; Cantero, D. Characterization of bacterial and Archaeal communities by DGGE and next generation sequencing (NGS) of nitrification bioreactors using two different intermediate landfill leachates as ammonium substrate. Waste Biomass Valorization 2022, 13, 3753–3766. [Google Scholar] [CrossRef]
- Al-Mailem, D.M.; Kansour, M.K.; Radwan, S.S. Capabilities and limitations of DGGE for the analysis of hydrocarbonoclastic prokaryotic communities directly in environmental samples. Microbiologyopen 2017, 6, e00495. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, C.L.; Vaughan, E.E.; Caggia, C. Artisanal and experimental Pecorino Siciliano cheese: Microbial dynamics during manufacture assessed by culturing and PCR–DGGE analyses. Int. J. Food Microbiol. 2006, 109, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, P.; Alessandria, V.; Rantsiou, K.; Rolle, L.; Zeppa, G.; Cocolin, L. Microbial dynamics of Castelmagno PDO, a traditional Italian cheese, with a focus on lactic acid bacteria ecology. Int. J. Food Microbiol. 2008, 122, 302–311. [Google Scholar]
- Neilson, J.W.; Jordan, F.L.; Maier, R.M. Analysis of artifacts suggests DGGE should not be used for quantitative diversity analysis. J. Microbiol. Methods 2013, 92, 256–263. [Google Scholar] [CrossRef]
| Total Count of Aerobic Bacteria | Presumptive Mesophilic Lactococci | Presumptive Thermophilic Streptococci | Presumptive Mesophilic Lactobacilli | Presumptive Thermophilic Lactobacilli | Leuconostoc spp. | Enterococcus spp. | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Days | 1.season | 2.season | 1.season | 2.season | 1.season | 2.season | 1.season | 2.season | 1.season | 2.season | 1.season | 2.season | 1.season | 2.season | |
| Producer A | Cu | 6.1 | 6.7 | 6.0 | 6.9 | 5.3 | 6.0 | 6.5 | 6.0 | 3.9 | 4.5 | 4.1 | 4.3 | 4.9 | 5.0 |
| 1 | 8.9 | 10.2 | 8.7 | 9.7 | 8.3 | 8.7 | 7.0 | 7.3 | 6.9 | 8.4 | 7.1 | 8.3 | 6.4 | 8.0 | |
| 30 | 7.3 | 9.4 | 7.4 | 8.6 | 7.4 | 8.9 | 6.4 | 7.6 | 5.7 | 7.2 | 6.1 | 7.4 | 4.9 | 6.4 | |
| 60 | 6.8 | 8.8 | 6.4 | 8.6 | 5.9 | 8.8 | 6.2 | 7.3 | 6.1 | 5.6 | 6.5 | 7.5 | 6.3 | 7.1 | |
| 90 | 7.8 | 8.7 | 6.9 | 8.9 | 6.9 | 9.1 | 6.7 | 7.8 | 6.7 | 4.9 | 7.3 | 7.7 | 6.9 | 8.1 | |
| Producer B | Cu | 6.2 | 5.9 | 7.1 | 6.6 | 6.4 | 6.6 | 6.9 | 6.8 | 4.5 | 6.3 | 4.8 | 4.0 | 6.4 | 6.8 |
| 1 | 7.8 | 10.4 | 7.7 | 9.6 | 7.4 | 9.4 | 7.6 | 7.6 | 5.8 | 7.6 | 8.2 | 7.9 | 6.9 | 8.1 | |
| 30 | 7.9 | 9.1 | 8.6 | 8.4 | 5.7 | 8.9 | 7.8 | 8.0 | 5.8 | 6.6 | 7.8 | 7.4 | 6.8 | 7.9 | |
| 60 | 8.0 | 8.3 | 7.7 | 8.2 | 6.6 | 8.4 | 7.4 | 7.2 | 5.2 | 7.2 | 7.1 | 7.2 | 5.7 | 7.5 | |
| 90 | 7.5 | 8.2 | 7.2 | 7.5 | 7.2 | 8.4 | 7.2 | 7.2 | 5.4 | 6.8 | 7.3 | 7.3 | 5.3 | 7.3 | |
| Producer C | Cu | 6.3 | 7.0 | 6.7 | 6.9 | 6.9 | 5.9 | 5.9 | 5.5 | 4.1 | 4.4 | 5.8 | 5.0 | 5.9 | 7.1 |
| 1 | 9.8 | 8.7 | 8.6 | 9.0 | 9.0 | 9.1 | 7.1 | 7.5 | 6.9 | 6.9 | 7.4 | 7.6 | 6.4 | 8.4 | |
| 30 | 7.7 | 9.2 | 7.8 | 8.3 | 6.4 | 8.8 | 7.4 | 7.6 | 7.2 | 6.3 | 6.7 | 7.3 | 7.0 | 6.9 | |
| 60 | 9.4 | 8.8 | 6.5 | 8.1 | 6.9 | 7.9 | 6.7 | 7.3 | 6.7 | 6.4 | 6.0 | 7.4 | 6.7 | 7.4 | |
| 90 | 8.0 | 8.7 | 7.9 | 7.2 | 7.9 | 8.3 | 7.4 | 7.3 | 7.8 | 6.7 | 7.9 | 6.9 | 7.9 | 7.3 | |
| Producer 1 | Band | Identification 2 | NCBI-BLAST Accession Number Access Date: 28 October 2023 | Identity (%) |
|---|---|---|---|---|
| A, B, C, a, b, c | 1 | Lacticaseibacillus paracasei | OR267407.1 | 99 |
| A, C, a, c | 2 | Lactobacillus helveticus | MT538439.1 | 98 |
| A, B, C, a, c | 3 | Lactiplantibacillus plantarum | EF597125.1 | 98 |
| A, a | 4 | Leuconostoc citreum | ON631287.1 | 99 |
| a | 5 | Leuconostoc pseudomesenteroides | OM943117.1 | 98 |
| B, b | 6 | Lactobacillus delbrueckii subsp. bulgaricus | MT516034.1 | 99 |
| B, b | 7 | Pediococcus pentosaceus | OR518626.1 | 97 |
| A, C, a, c | 8 | Leuconostoc mesenteroides subsp. mesenteroides | OK135479.1 | 98 |
| C | 9 | Lactobacillus rhamnosus | CP101845.1 | 98 |
| A, B, C, a, b, c | 10 | Streptococcus thermophilus | KX926522.1 | 99 |
| A, B, C, a, b, c | 11 | Lactococcus lactis subsp. lactis | JN851797.1 | 99 |
| A, C, a, c | 12 | Streptococcus parauberis | MT597919.1 | 97 |
| C, c | 13 | Streptococcus gallolyticus | OP714497.1 | 97 |
| A, C, a, c | 14 | Streptococcus macedonicus | MN305794.1 | 96 |
| A, c | 15 | Lactococcus garvieae | OR502221.1 | 98 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dizdarević, T.; Sakić-Dizdarević, S.; Porcellato, D.; Sarić, Z.; Alkić-Subašić, M.; Abrahamsen, R.K.; Narvhus, J.A. Microbial Diversity of Traditional Livno Cheese from Bosnia and Herzegovina. Fermentation 2023, 9, 1006. https://doi.org/10.3390/fermentation9121006
Dizdarević T, Sakić-Dizdarević S, Porcellato D, Sarić Z, Alkić-Subašić M, Abrahamsen RK, Narvhus JA. Microbial Diversity of Traditional Livno Cheese from Bosnia and Herzegovina. Fermentation. 2023; 9(12):1006. https://doi.org/10.3390/fermentation9121006
Chicago/Turabian StyleDizdarević, Tarik, Svijetlana Sakić-Dizdarević, Davide Porcellato, Zlatan Sarić, Mersiha Alkić-Subašić, Roger K. Abrahamsen, and Judith A. Narvhus. 2023. "Microbial Diversity of Traditional Livno Cheese from Bosnia and Herzegovina" Fermentation 9, no. 12: 1006. https://doi.org/10.3390/fermentation9121006
APA StyleDizdarević, T., Sakić-Dizdarević, S., Porcellato, D., Sarić, Z., Alkić-Subašić, M., Abrahamsen, R. K., & Narvhus, J. A. (2023). Microbial Diversity of Traditional Livno Cheese from Bosnia and Herzegovina. Fermentation, 9(12), 1006. https://doi.org/10.3390/fermentation9121006






