Saccharification of Agricultural Wastes and Clarification of Orange Juice by Penicillium rolfsii CCMB 714 Pectinase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture Maintenance
2.2. Agroindustrial Waste Obtention
2.3. Solid-State Fermentation and the Crude Enzyme Extract Obtention
2.4. Enzyme Assay
2.5. Study of Fermentation Conditions
2.6. Physicochemical Characterization of Pectinases
2.7. Application of Enzyme Extract with Pectinase Activity
2.7.1. Saccharification of Agroindustrial Waste
2.7.2. Orange Juice Clarification
3. Results and Discussion
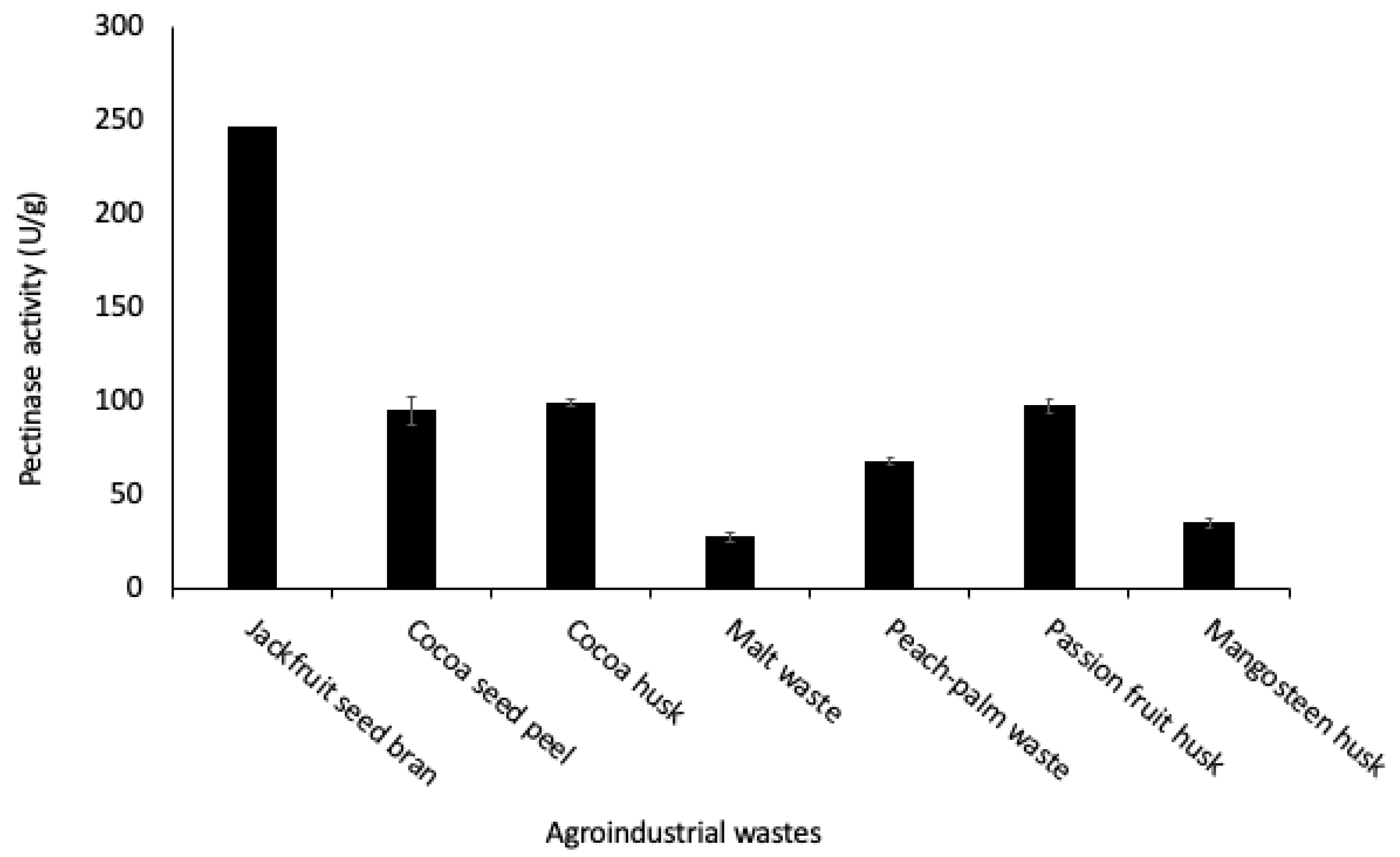
3.1. Screening of Pectinase Production Using Agro-Industrial Wastes as Substrates
3.2. Study of Fermentation Conditions
- p-value = 0.900.
- where Pec: pectinase, CSP: cocoa seed peel, YE: yeast extract and AP: ammonium phosphate.
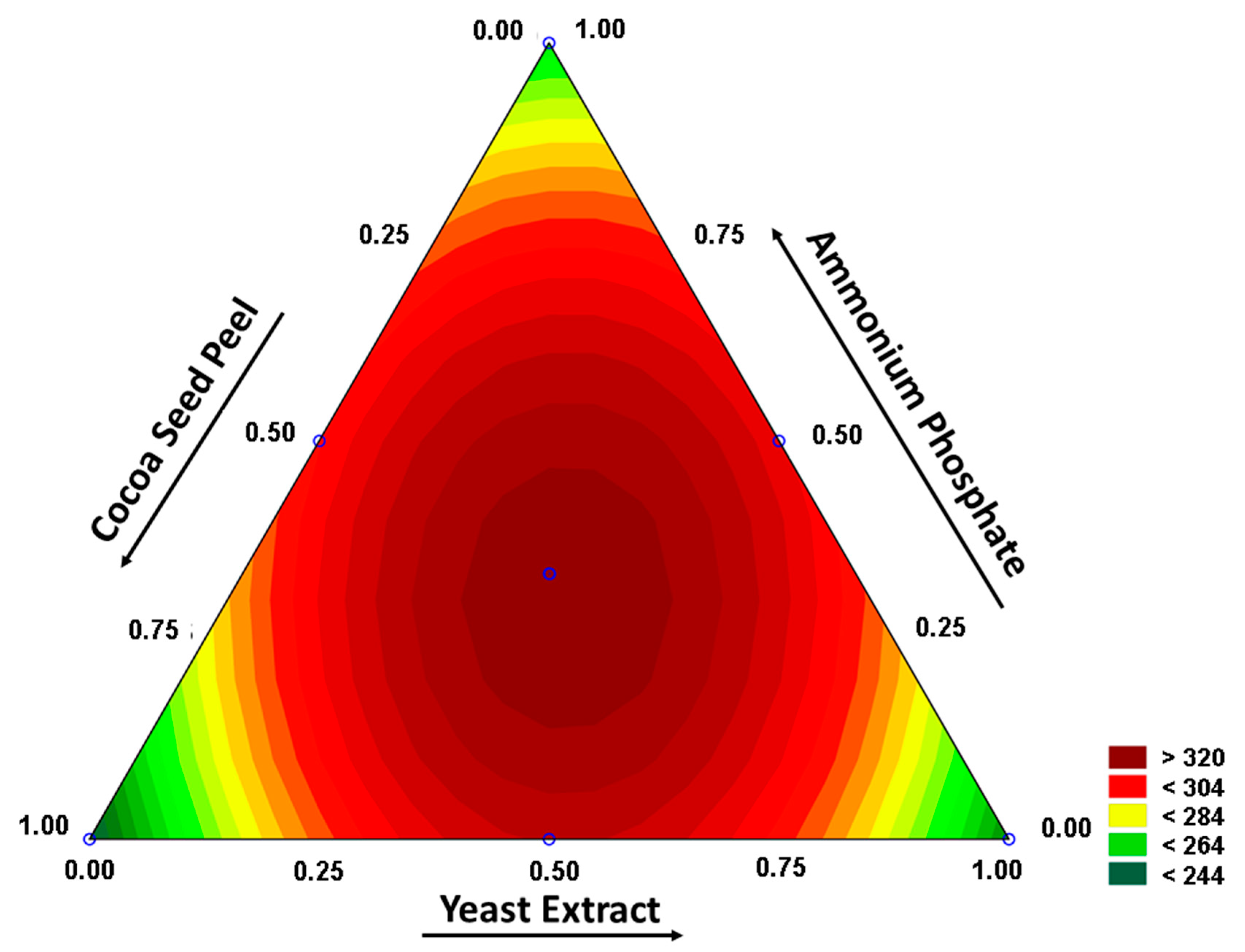
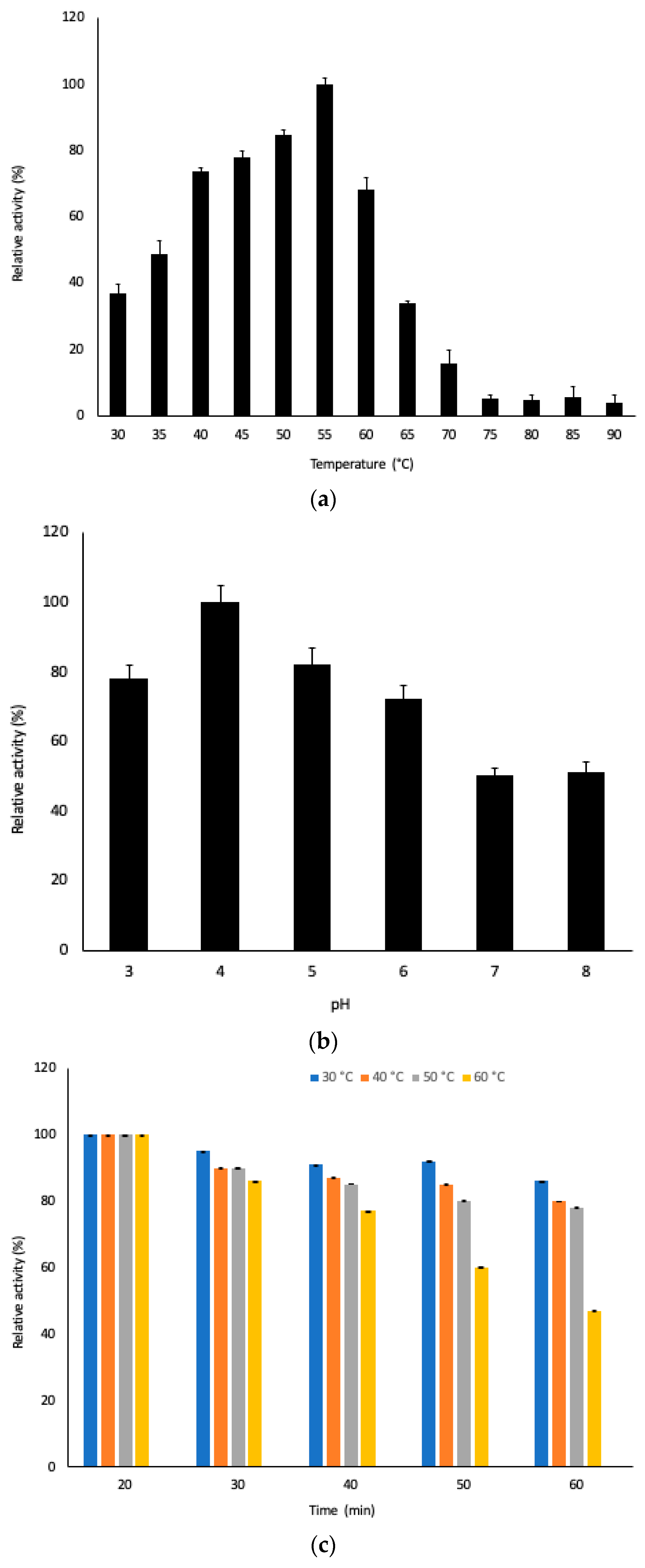
3.3. Physicochemical Characterization of Pectinase in the Extract
3.4. Application of Pectinase
3.4.1. Saccharification of Agricultural Wastes
3.4.2. Orange Juice Clarification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- John, J.; Kaimal, K.S.; Smith, M.L.; Rahman, P.K.; Chellam, P.V. Advances in upstream and downstream strategies of pectinase bioprocessing: A review. Int. J. Biol. Macromol. 2020, 162, 1086–1099. [Google Scholar] [CrossRef] [PubMed]
- Yapo, B.M. Pectic substances: From simple pectic polysaccharides to complex pectins—A new hypothetical model. Carbohydr. Polym. 2011, 2, 373–385. [Google Scholar] [CrossRef]
- Amin, F.; Bhatti, H.N.; Bilal, M. Recent advances in the production strategies of microbial pectinases—A review. Int. J. Biol. Macromol. 2019, 122, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Martín, M.C.; López, O.V.; Ciolino, A.E.; Morata, V.I.; Villar, M.A.; Ninago, M.D. Immobilization of enological pectinase in calcium alginate hydrogels: A T potential biocatalyst for winemaking. Biocatal. Agric. Biotechnol. 2019, 18, 101091. [Google Scholar] [CrossRef]
- Salim, D.; Anwar, Z.; Zafar, M.; Anjum, A.; Bhatti, K.H.; Irshad, M. Pectinolytic cocktail: Induced yield and its exploitation for lignocellulosic materials saccharification and fruit juice clarification. Food Biosci. 2018, 22, 154–164. [Google Scholar] [CrossRef]
- Wang, J.; Chio, C.; Chen, X.; Su, E.; Cao, F.; Jin, Y.; Qin, W. Efficient saccharification of agave biomass using Aspergillus niger produced low-cost enzyme cocktail with hyperactive pectinase activity. Bioresour. Technol. 2018, 272, 26–33. [Google Scholar] [CrossRef]
- Schneider, W.D.H.; Gonçalves, T.A.; Uchima, C.A.; dos Reis, L.; Fontana, R.C.; Squina, F.M.; Dillon, A.J.P.; Camassola, M. Comparison of the production of enzymes to cell wall hydrolysis using different carbon sources by Penicillium echinulatum strains and its hydrolysis potential for lignocelullosic biomass. Process. Biochem. 2018, 66, 162–170. [Google Scholar] [CrossRef]
- Terrasan, C.R.F.; Temer, B.; Duarte, M.C.T.; Carmona, E.C. Production of xylanolytic enzymes by Penicillium janczewskii. Bioresour. Technol. 2010, 101, 4139–4143. [Google Scholar] [CrossRef]
- Chávez, R.; Roa, A.; Navarrete, K.; Trebotich, J.; Espinosa, Y.; Vaca, I. Evaluation of properties of several cheese-ripening fungi for potential biotechnological applications. Mycoscience 2010, 51, 84–87. [Google Scholar] [CrossRef]
- Leitão, A.L. Potential of Penicillium Species in the Bioremediation Field. Int. J. Environ. Res. Public Health 2009, 6, 1393–1417. [Google Scholar] [CrossRef]
- Niyonzima, F.N. Production of Microbial Industrial Enzymes. Acta Sci. Microbiol. 2019, 2, 75–89. [Google Scholar] [CrossRef]
- Lee, K.C.; Arai, T.; Ibrahim, D.; Prawitwong, P.; Deng, L.; Murata, Y.; Mori, Y.; Kosugi, A. Purification and Characterization of a Xylanase from the Newly Isolated Penicillium rolfsii c3-2(1) IBRL. Bioresources 2014, 10, 1627–1643. [Google Scholar] [CrossRef]
- Lee, K.C.; Arai, T.; Ibrahim, D.; Deng, L.; Murata, Y.; Mori, Y.; Kosugi, A. Characterization of oil-palm trunk residue degradation enzymes derived from the isolated fungus, Penicillium rolfsii c3-2(1) IBRL. Environ. Technol. 2016, 37, 1550–1558. [Google Scholar] [CrossRef] [PubMed]
- Andrade, P.M.L.; Baptista, L.; Britto, J.S.; Uetenabaro, A.P.T.; da Costa, A.M. Co-production of tannase and gallic acid by a novel Penicillium rolfsii (CCMB 714). Prep. Biochem. Biotechnol. 2018, 48, 700–706. [Google Scholar] [CrossRef]
- Ahmed, A.; Khan, M.N.; Ahmad, A.; Khan, S.A.; Sohail, M. Optimization of pectinase production from Geotrichum candidum AA15 using response surface methodology. Pak. J. Bot. 2018, 51, 743–750. [Google Scholar] [CrossRef] [PubMed]
- El-Rahim, W.M.A.; Moawad, H.; Hashem, M.M.; Gebreil, G.M.; Zakaria, M. Highly efficient fungal pectinase and laccase producers among isolates from flax retting liquor. Biocatal. Agric. Biotechnol. 2020, 25, 101570. [Google Scholar] [CrossRef]
- Handa, S.; Sharma, N.; Pathania, S. Multiple Parameter Optimization for Maximization of Pectinase Production by Rhizopus sp. C4 under Solid State Fermentation. Fermentation 2016, 2, 10. [Google Scholar] [CrossRef]
- Rodrigues, M.I.; Iemma, A.F. Experimental Design and Process Optimization; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Pakalapati, H.; Tariq, M.A.; Arumugasamy, S.K. Optimization and modelling of enzymatic polymerization of ε-caprolactone to polycaprolactone using Candida Antartica Lipase B with response surface methodology and artificial neural network. Enzym. Microb. Technol. 2018, 122, 7–18. [Google Scholar] [CrossRef]
- De Menezes, L.H.S.; Pimentel, A.B.; Oliveira, P.C.; Tavares, I.M.D.C.; Ruiz, H.A.; Irfan, M.; Bilal, M.; das Chagas, T.P.; da Silva, E.G.P.; Salay, L.C.; et al. The Application of Chemometric Methods in the Production of Enzymes Through Solid State Fermentation Uses the Artificial Neural Network—A Review. BioEnergy Res. 2022, 16, 279–288. [Google Scholar] [CrossRef]
- Nunes, N.D.S.; Carneiro, L.L.; De Menezes, L.H.S.; De Carvalho, M.S.; Pimentel, A.B.; Silva, T.P.; Pacheco, C.S.V.; Tavares, I.M.D.C.; Santos, P.H.; Das Chagas, T.P.; et al. Simplex-Centroid Design and Artificial Neural Network-Genetic Algorithm for the Optimization of Exoglucanase Production by Penicillium Roqueforti ATCC 10110 Through Solid-State Fermentation Using a Blend of Agroindustrial Wastes. BioEnergy Res. 2020, 13, 1130–1143. [Google Scholar] [CrossRef]
- Bezerra, C.O.; Carneiro, L.L.; Carvalho, E.A.; Chagas, T.P.; Carvalho, L.R.; Uetanabaro, A.P.; Silva, G.P.; Silva, E.G.P.; Costa, A.M. Artificial intelligence as a combinatorial optimization strategy for cellulase production by Trichoderma stromaticum AM7 using peach-palm waste under solid-state fermentation. Bioenergy Res. 2021, 14, 1161–1170. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Moustafa, A. Big Data in Psychiatry and Neurology; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar]
- De Menezes, L.H.S.; Carneiro, L.L.; Tavares, I.M.C.; Santos, P.H.; Chagas, T.P.; Mendes, A.A.; Silva, E.G.P.; Franco, M.; Oliveira, J.R. Artificial neural network hybridized with a genetic algorithm for optimization of lipase production from Penicillium roqueforti ATCC 10110 in solid-state fermentation. Biocatal. Agric. Biotechnol. 2021, 31, 101885. [Google Scholar] [CrossRef]
- Oumer, O.J.; Abate, D. Characterization of Pectinase from Bacillus subtilis Strain Btk 27 and Its Potential Application in Removal of Mucilage from Coffee Beans. Enzym. Res. 2017, 2017, 7686904. [Google Scholar] [CrossRef]
- Ojwang, R.A.; Muge, E.K.; Mbatia, B.N.; Mwanza, B.K.; Ogoyi, D.O. Compositional, Elemental, Phytochemical and Antioxidant Characterization of Jackfruit (Artocarpus heterophyllus) Pulps and Seeds from Selected Regions in Kenya and Uganda. Eur. J. Med. Plants 2018, 23, 1–12. [Google Scholar] [CrossRef]
- Shafiq, M.; Mehmood, S.; Yasmin, A.; Khan, S.J.; Khan, N.H.; Ali, S. Evaluation of Phytochemical, Nutritional and Antioxidant Activity of Indigenously Grown Jackfruit (Artocarpus heterophyllus Lam). J. Sci. Res. 2017, 9, 135–143. [Google Scholar] [CrossRef]
- Ahmed, A.; Sohail, M. Characterization of pectinase from Geotrichum candidum AA15 and its potential application in orange juice clarification. J. King Saud Univ. Sci. 2020, 32, 955–961. [Google Scholar] [CrossRef]
- Begum, R.; Aziz, M.G.; Uddin, M.B.; Yusof, Y.A. Characterization of Jackfruit (Artocarpus heterophyllus) Waste Pectin as Influenced by Various Extraction Conditions. Agric. Agric. Sci. Procedia 2014, 2, 244–251. [Google Scholar] [CrossRef]
- Begum, R.; Aziz, M.G.; Yusof, Y.A.; Uddin, M.B. Extraction and characterization of pectin from jackfruit (Artocarpus heterophyllus Lam) waste. IOSR J. Pharm. Biol. Sci. 2017, 12, 42–49. [Google Scholar]
- Patil, R.; Joshi, G.; Haldankar, P.M.; More, M. Estimation of pectin content in jackfruit (Artocarpus heterophyllus). Asian J. Hort. 2022, 6, 536–537. [Google Scholar]
- Jahan, N.; Shahid, F.; Aman, A.; Mujahid, T.Y.; Qader, S.A.U. Utilization of agro waste pectin for the production of industrially important polygalacturonase. Heliyon 2017, 3, e00330. [Google Scholar] [CrossRef] [PubMed]
- Mahmoodi, M.; Najafpour, G.; Mohammadi, M. Bioconversion of agroindustrial wastes to pectinases enzyme via solid state fermentation in trays and rotating drum bioreactors. Biocatal. Agric. Biotechnol. 2019, 21, 101280. [Google Scholar] [CrossRef]
- Sethi, B.K.; Nanda, P.K.; Sahoo, S. Enhanced production of pectinase by Aspergillus terreus NCFT 4269.10 using banana peels as substrate. 3 Biotech 2016, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Viayaraghavan, P.; Kumar, S.J.; Arasu, M.V.; Al-Dhabi, N.A. Simultaneous production of commercial enzymes using agro industrial residues by statistical approach. J. Sci. Food Agric. 2019, 99, 2685–2696. [Google Scholar] [CrossRef]
- Marques, G.; Uesc; Silva, T.; Lessa, O.; De Brito, A.; Reis, N.; Fernandes, A.D.A.; Oliveira, J.; Franco, M. Production of Xylanase and Endoglucanase by Solid-State Fermentation of Jackfruit Residue. Ver. Mex. Ing. Quím. 2019, 18, 673–680. [Google Scholar] [CrossRef]
- Mehmood, T.; Saman, T.; Irfan, M.; Anwar, F.; Ikram, M.S.; Tabassam, Q. Pectinase Production from Schizophyllum commune Through Central Composite Design Using Citrus Waste and Its Immobilization for Industrial Exploitation. Waste Biomass-Valorization 2018, 10, 2527–2536. [Google Scholar] [CrossRef]
- Nagaoka, T.; Nakagawa, T.; Miyaji, T.; Tomizuka, N. A cold-active pectin lyase from the psychrophilic and basidiomycetous yeast Cystofilobasidium capitatum strain PPY-1. Biotechnol. Appl. Biochem. 2005, 42, 193–196. [Google Scholar] [CrossRef]
- Okonji, R.E.; Itakorode, B.O.; Ovumedia, J.O.; Adedeji, O.S. Purification and biochemical characterization of pectinase produced by Aspergillus fumigatus isolated from soil of decomposing plant materials. J. Appl. Biotechnol. 2019, 7, 1–8. [Google Scholar]
- Sudeep, K.C.; Jitendra, U.; Dev, R.J.; Binod, L.; Dhiraj, K.C.; Bhoj, R.P.; Tirtha, R.B.; Rajiv, D.; Santosh, K.; Niranjan, K.; et al. Production, Characterization, and Industrial Application of Pectinase Enzyme Isolated from Fungal Strains. Fermentation 2020, 6, 59. [Google Scholar] [CrossRef]
- Kadija, T.K.S.A.; Manam, W.H.S. Potential Applications of Pectinases in Food, Agricultural and Environmental Sectors. J. Pharm. Chem. Biol. Sci. 2018, 6, 23–34. [Google Scholar]
- Carvalho, E.A.; Nunes, L.V.; Goes, L.M.D.S.; Silva, E.G.P.D.; Franco, M.; Gross, E.; Costa, A.M.D. Peach-palm (Bactris gasipaes Kunth.) waste as substrate for xylanase production by Trichoderma stromaticum AM7. Chem. Eng. Commun. 2018, 205, 975–985. [Google Scholar] [CrossRef]
- Losonczi, A.; Csiszár, E.; Szakács, G.; Bezúr, L. Role of the EDTA Chelating Agent in Bioscouring of Cotton. Text. Res. J. 2005, 75, 411–417. [Google Scholar] [CrossRef]
- Hansen, J.H.; Petersen, S.V.; Andersen, K.K.; Enghild, J.J.; Damhus, T.; Otzen, D. Stable intermediates determine proteins’ primary unfolding sites in the presence of surfactants. Biopolymers 2009, 91, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Aizat, W.M.; Ahmad-Hashim, F.H.; Syed Jaafar, S.N. Valorization of mangosteen, “The Queen of Fruits,” and new advances in postharvest and in food and engineering applications: A review. J. Advan. Res. 2019, 20, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Canteri, M.H.G.; Scheer, A.P.; Ginies, C.; Renard, C.M.C.; WosiackiI, G. Importance of thermal treatment on the rind passion Fruit to pectin extraction. Rev. Bras. Tecnol. Agroindustrial. 2010, 4, 109–121. [Google Scholar]
- Locatelli, G.O.; Finkler, L.; Finkler, C.L. Orange and Passion Fruit Wastes Characterization, Substrate Hydrolysis and Cell Growth of Cupriavidus necator, as Proposal to Converting of Residues in High Value Added Product. Biological Sciences. An. Acad. Bras. Ciênc. 2019, 91, e20180058. [Google Scholar] [CrossRef] [PubMed]
- Bonvehí, J.S.; Benería, M.A. Composition of dietary fibre in cocoa husk. Z. Lebensm. Unters. Forsch. 1998, 207, 105–109. [Google Scholar]
- Parkhey, P.; Ram, A.K.; Diwan, B.; Eswari, J.S.; Gupta, P. Artificial neural network and response surface methodology: A comparative analysis for optimizing rice straw pretreatment and saccharification. Prep. Biochem. Biotechnol. 2020, 50, 768–780. [Google Scholar] [CrossRef]
- Andrade, P.M.L.; Baptista, L.; Bezerra, C.O.; Peralta, R.M.; Góes-Neto, A.; Uetanabaro, A.P.T.; da Costa, A.M. Immobilization and characterization of tannase from Penicillium rolfsii CCMB 714 and its efficiency in apple juice clarification. J. Food Meas. Charact. 2021, 15, 1005–1013. [Google Scholar] [CrossRef]


| Experiment | M (mL) | t (Days) | T (°C) | NS (%) | EA (U/g) | ANN Estimated (EEA) * | Residue (EA-EEA) | MSE | R2 |
|---|---|---|---|---|---|---|---|---|---|
| Training set | |||||||||
| 1 | 2.5 | 4 | 20 | 1 | 199.91 | 199.33 | 0.58 | 351.12 | 0.87 |
| 2 | 2.5 | 4 | 20 | 2 | 273.20 | 272.65 | 0.55 | ||
| 3 | 2.5 | 4 | 30 | 1 | 273.20 | 267.71 | 5.49 | ||
| 7 | 2.5 | 8 | 30 | 1 | 197.95 | 212.22 | −14.27 | ||
| 8 | 2.5 | 8 | 30 | 2 | 346.56 | 287.90 | 58.66 | ||
| 9 | 3.5 | 4 | 20 | 1 | 227.12 | 227.38 | −0.26 | ||
| 10 | 3.5 | 4 | 20 | 2 | 306.58 | 307.24 | −0.66 | ||
| 11 | 3.5 | 4 | 30 | 1 | 257.23 | 258.95 | −1.72 | ||
| 14 | 3.5 | 8 | 20 | 2 | 233.80 | 232.21 | 1.59 | ||
| 15 | 3.5 | 8 | 30 | 1 | 253.17 | 240.20 | 12.25 | ||
| 17 | 3 | 6 | 25 | 1.5 | 196.16 ** | 198.13 ** | −1.97 ** | ||
| 18 | 3 | 6 | 25 | 1.5 | |||||
| 19 | 3 | 6 | 25 | 1.5 | |||||
| 20 | 2 | 10 | 15 | 2.5 | 312.89 | 284.08 | 28.81 | ||
| 21 | 4 | 2 | 35 | 0.5 | 362.09 | 377.71 | −15.62 | ||
| 22 | 4.5 | 12 | 40 | 3 | 290.83 | 288.36 | 2.47 | ||
| Validation set | |||||||||
| 12 | 3.5 | 4 | 30 | 2 | 281.54 | 290.38 | −8.84 | 28.04 | 0.98 |
| 13 | 3.5 | 8 | 20 | 1 | 187.86 | 190.32 | −2.46 | ||
| 16 | 3.5 | 8 | 30 | 2 | 255.35 | 255.46 | −0.11 | ||
| Testing set | |||||||||
| 4 | 2.5 | 4 | 30 | 2 | 330.10 | 355.33 | −25.23 | 326.90 | 0.93 |
| 5 | 2.5 | 8 | 20 | 1 | 181.12 | 180.68 | 0.44 | ||
| 6 | 2.5 | 8 | 20 | 2 | 186.05 | 204.60 | −18.55 | ||
| Total Dataset | 299.02 | 0.90 | |||||||
| Variables | Responses | ||||
|---|---|---|---|---|---|
| Experiment | Time (Minutes) | Enzyme Extract Volume (mL) | Temperature (°C) | Absorbance Predict | Absorbance Experimental |
| 1 | −1 (80) | −1 (4) | −1 (30) | 0.187 | 0.181 |
| 2 | 1 (120) | −1 (4) | −1 (30) | 0.182 | 0.189 |
| 3 | −1 (80) | 1 (8) | −1 (30) | 0.119 | 0.123 |
| 4 | 1 (120) | 1 (8) | −1 (30) | 0.084 | 0.081 |
| 5 | −1 (80) | −1 (4) | 1 (50) | 0.197 | 0.215 |
| 6 | 1 (120) | −1 (4) | 1 (50) | 0.252 | 0.263 |
| 7 | −1 (80) | 1 (8) | 1 (50) | 0.113 | 0.121 |
| 8 | 1 (120) | 1 (8) | 1 (50) | 0.113 | 0.157 |
| 9 | −1.68 (66.4) | 0 (6) | 0 (40) | 0.136 | 0.113 |
| 10 | 1.68 (133.6) | 0 (6) | 0 (40) | 0.120 | 0.123 |
| 11 | 0 (100) | −1.68 (2.64) | 0 (40) | 0.285 | 0.275 |
| 12 | 0 (100) | 1.68 (9.36) | 0 (40) | 0.137 | 0.120 |
| 13 | 0 (100) | 0 (6) | −1.68 (23.2) | 0.131 | 0.114 |
| 14 | 0 (100) | 0 (6) | 1.68 (56.8) | 0.108 | 0.135 |
| 15 | 0 (100) | 0 (6) | 0 (40) | 0.162 | 0.117 |
| 16 | 0 (100) | 0 (6) | 0 (40) | 0.136 | 0.165 |
| 17 | 0 (100) | 0 (6) | 0 (40) | 0.136 | 0.130 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Macedo, K.M.; Azevedo, R.A.; da Silva, E.G.P.; das Chagas, T.P.; Salay, L.C.; Uetanabaro, A.P.T.; Aguiar-Oliveira, E.; da Costa, A.M. Saccharification of Agricultural Wastes and Clarification of Orange Juice by Penicillium rolfsii CCMB 714 Pectinase. Fermentation 2023, 9, 917. https://doi.org/10.3390/fermentation9100917
Macedo KM, Azevedo RA, da Silva EGP, das Chagas TP, Salay LC, Uetanabaro APT, Aguiar-Oliveira E, da Costa AM. Saccharification of Agricultural Wastes and Clarification of Orange Juice by Penicillium rolfsii CCMB 714 Pectinase. Fermentation. 2023; 9(10):917. https://doi.org/10.3390/fermentation9100917
Chicago/Turabian StyleMacedo, Kelly Menezes, Raquel Araújo Azevedo, Erik Galvão Paranhos da Silva, Thiago Pereira das Chagas, Luiz Carlos Salay, Ana Paula Trovatti Uetanabaro, Elizama Aguiar-Oliveira, and Andréa Miura da Costa. 2023. "Saccharification of Agricultural Wastes and Clarification of Orange Juice by Penicillium rolfsii CCMB 714 Pectinase" Fermentation 9, no. 10: 917. https://doi.org/10.3390/fermentation9100917
APA StyleMacedo, K. M., Azevedo, R. A., da Silva, E. G. P., das Chagas, T. P., Salay, L. C., Uetanabaro, A. P. T., Aguiar-Oliveira, E., & da Costa, A. M. (2023). Saccharification of Agricultural Wastes and Clarification of Orange Juice by Penicillium rolfsii CCMB 714 Pectinase. Fermentation, 9(10), 917. https://doi.org/10.3390/fermentation9100917






