Autoinducer-2: Its Role in Biofilm Formation and L-Threonine Production in Escherichia coli
Abstract
:1. Introduction
2. Results
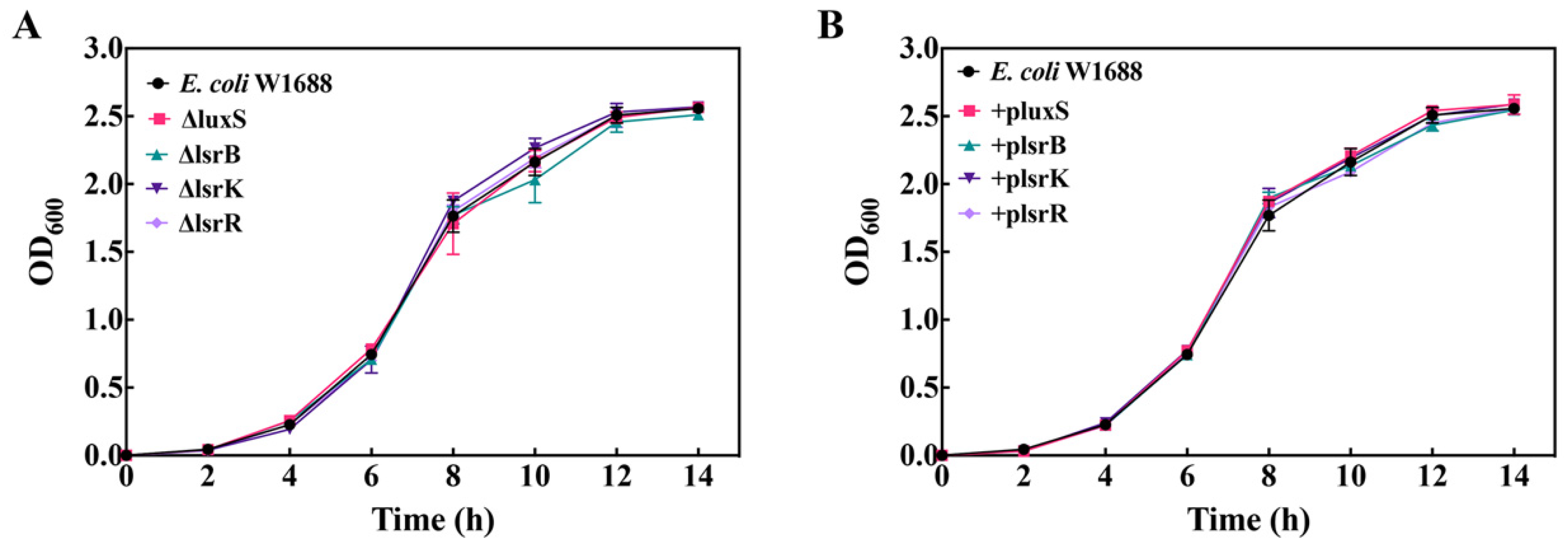
2.1. Growth of luxS, lsrB, lsrK, and lsrR Mutants
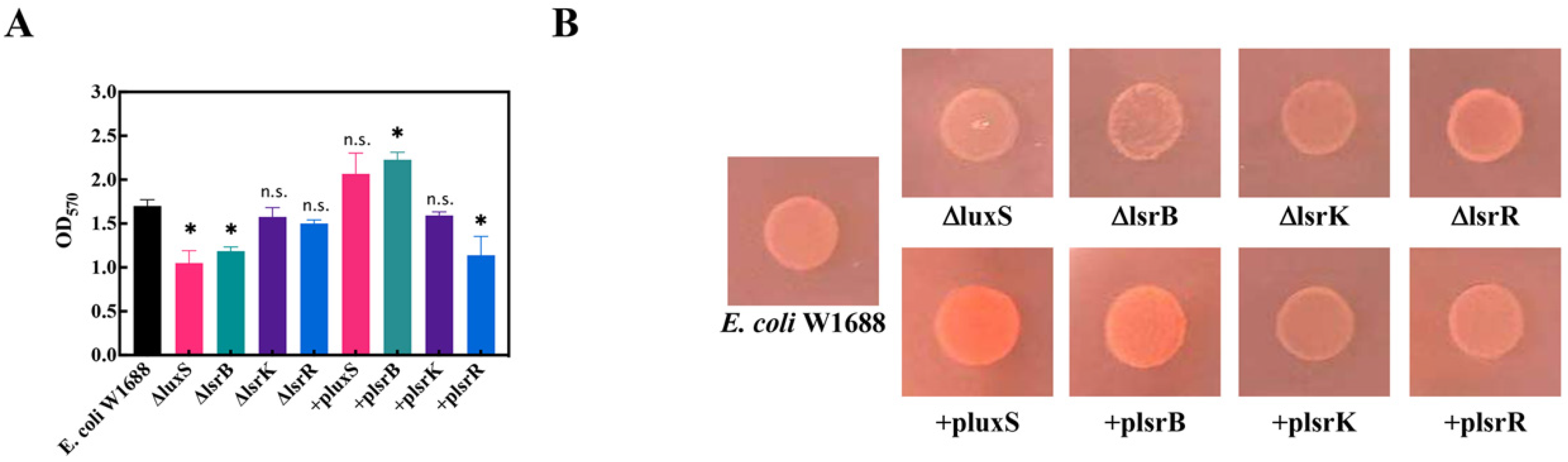
2.2. lsrB, luxS, lsrK, and lsrR affect biofilm formation in E. coli W1688
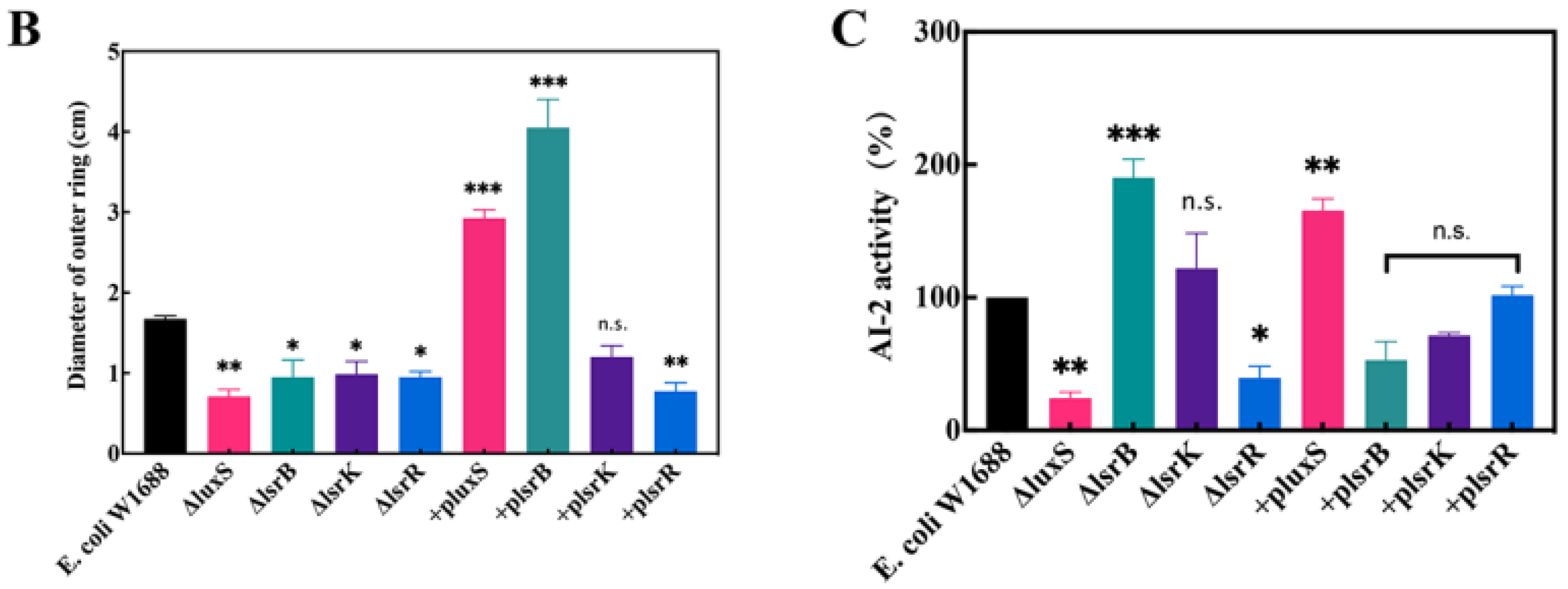
2.3. Overexpression of lsrB and luxS Affects the Motility of E. coli W1688
2.4. luxS and lsrB Are Necessary for AI-2 Production and Transport in E. coli W1688
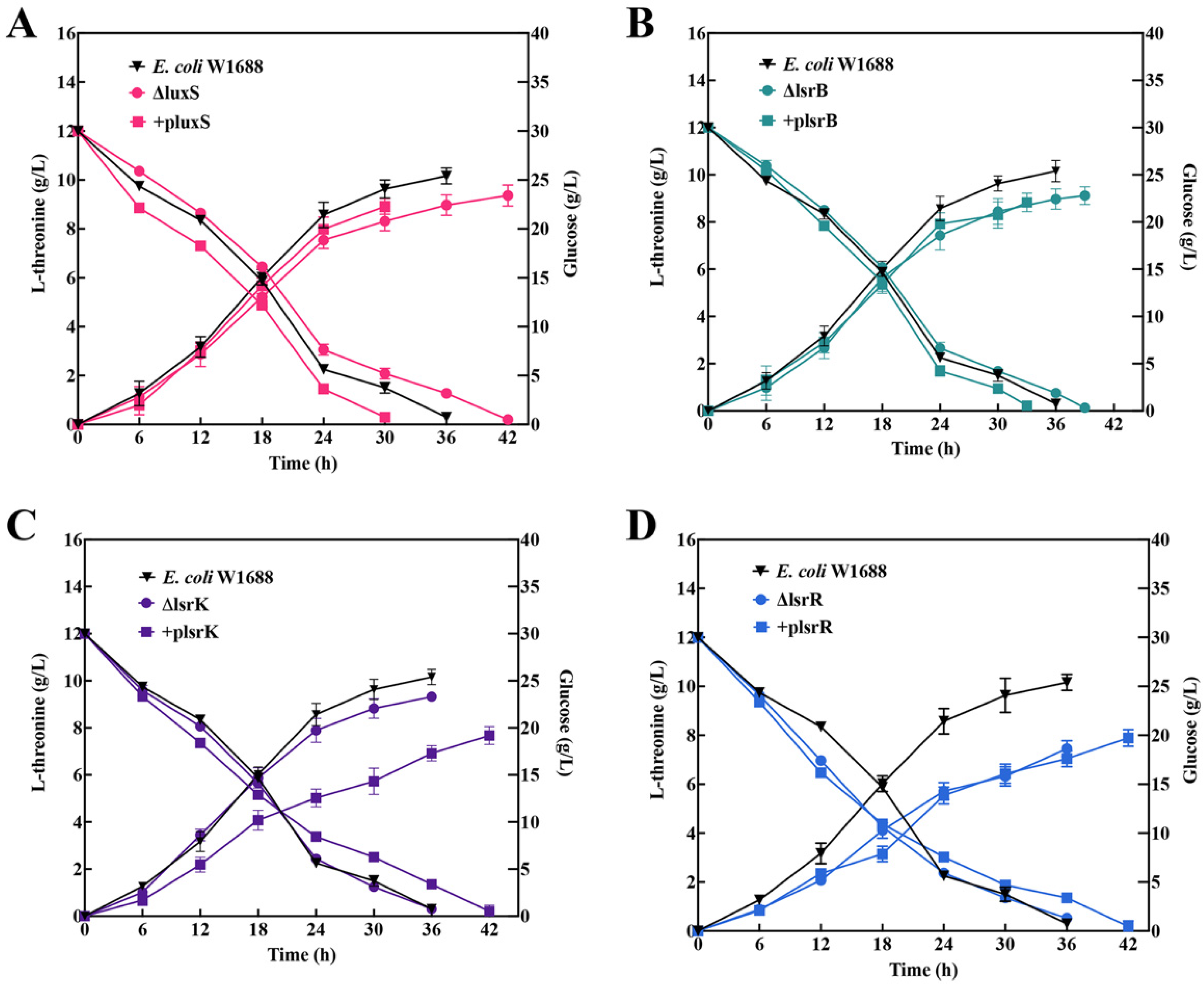
2.5. Free-Cell Fermentation of luxS, lsrB, lsrK, and lsrR Mutants
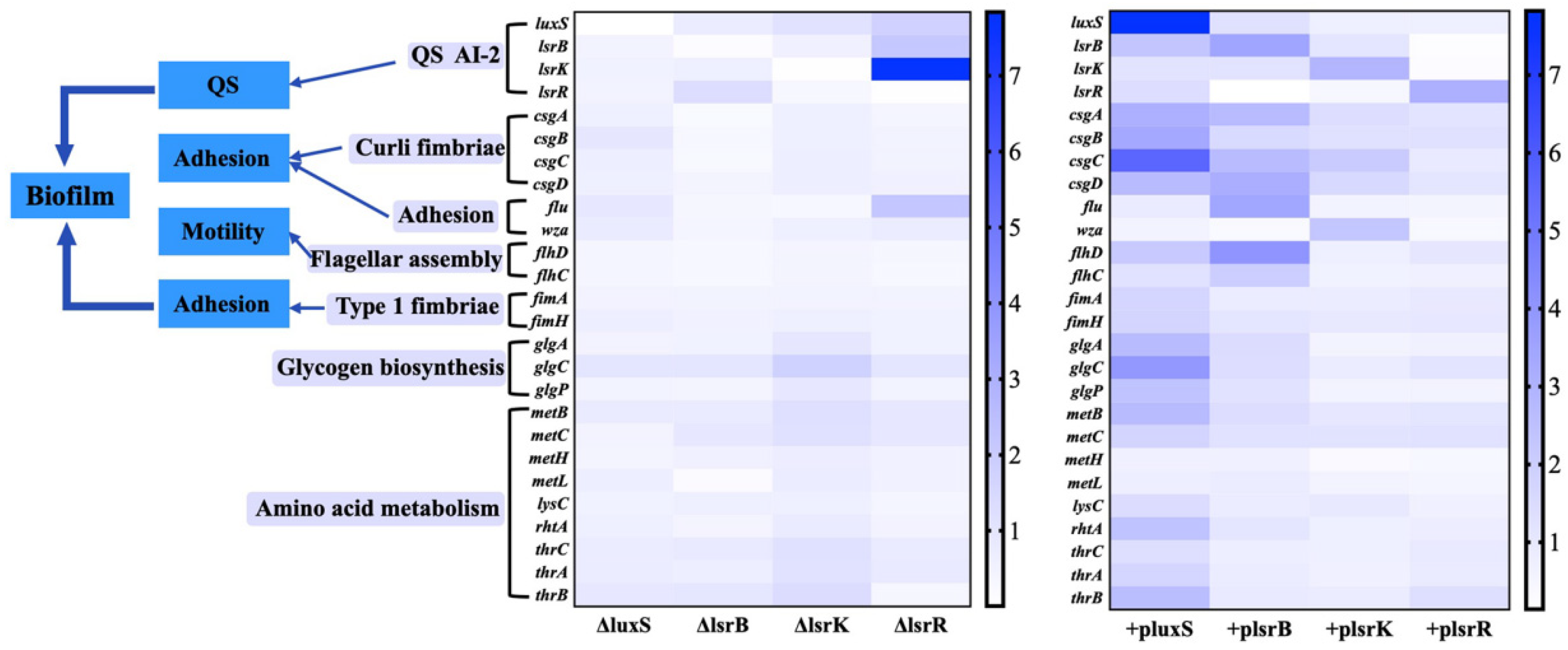
2.6. qRT-PCR Analysis of the Free-Cell Fermentation of luxS, lsrB, lsrK, and lsrR Mutants
2.7. Immobilized Repeated-Batch Fermentation in +pluxS and +plsrB
2.8. SEM Analysis of Biofilm on Carrier
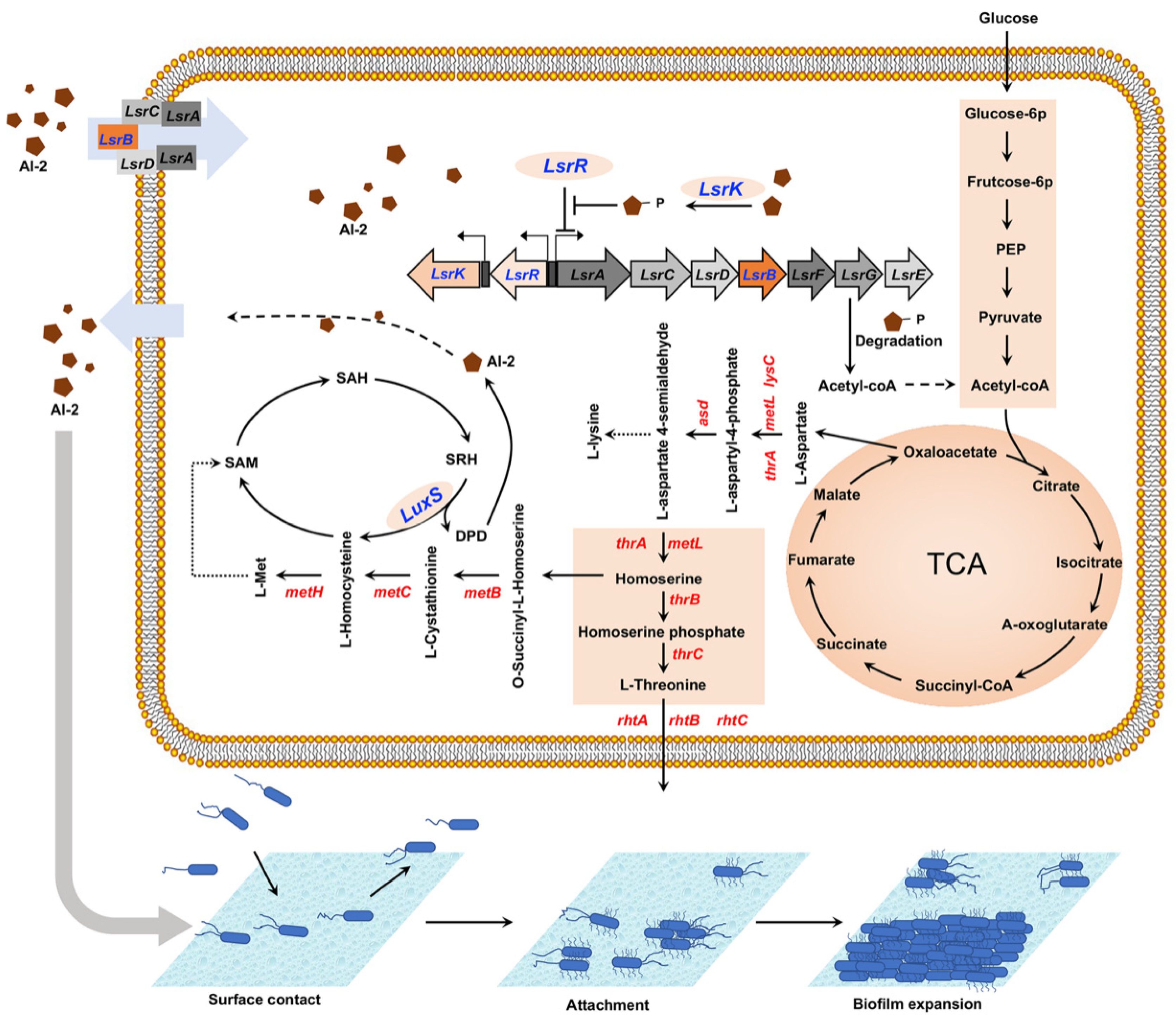
3. Discussion
4. Materials and Methods
4.1. Media and Growth Conditions
4.2. Construction of luxS, lsrB, lsrK, and lsrR Mutants
4.3. Growth Curves
4.4. Biofilm Forming Capacity
4.5. Curli Stain on YESCA-CR Plates
4.6. Motility Assay
4.7. AI-2 Detection
4.8. qRT-PCR Analysis
4.9. Free-Cell Fermentation, Immobilized Fermentation, and SEM
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fang, K.; Jin, X.; Hong, S.H. Probiotic Escherichia coli inhibits biofilm formation of pathogenic E. coli via extracellular activity of DegP. Sci. Rep. 2018, 8, 4939. [Google Scholar] [CrossRef]
- Miquel, S.; Lagrafeuille, R.; Souweine, B.; Forestier, C. Anti-biofilm activity as a health issue. Front. Microbiol. 2016, 7, 592. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T.-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Sharma, G.; Sharma, S.; Sharma, P.; Chandola, D.; Dang, S.; Gupta, S.; Gabrani, R. Escherichia coli biofilm: Development and therapeutic strategies. J. Appl. Microbiol. 2016, 121, 309–319. [Google Scholar] [CrossRef]
- Lister, J.L.; Horswill, A.R. Staphylococcus aureus biofilms: Recent developments in biofilm dispersal. Front. Cell Infect. Microbiol. 2014, 4, 178. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liu, Y.; Zhang, X.; Gao, H.; Mou, L.; Wu, M.; Zhang, W.; Xin, F.; Jiang, M. Biofilm application in the microbial biochemicals production process. Biotechnol. Adv. 2021, 48, 107724. [Google Scholar] [CrossRef]
- Chen, T.; Liu, N.; Ren, P.; Xi, X.; Yang, L.; Sun, W.; Yu, B.; Ying, H.; Ouyang, P.; Liu, D.; et al. Efficient Biofilm-Based Fermentation Strategies for L-Threonine Production by Escherichia coli. Front. Microbiol. 2019, 10, 1773. [Google Scholar] [CrossRef]
- Yang, L.; Zheng, C.; Chen, Y.; Shi, X.; Ying, Z.; Ying, H. Nitric oxide increases biofilm formation in Saccharomyces cerevisiae by activating the transcriptional factor Mac1p and thereby regulating the transmembrane protein Ctr1. Biotechnol. Biofuels 2019, 12, 30. [Google Scholar] [CrossRef]
- Yu, B.; Zhang, X.; Sun, W.; Xi, X.; Zhao, N.; Huang, Z.; Ying, Z.; Liu, L.; Liu, D.; Niu, H.; et al. Continuous citric acid production in repeated-fed batch fermentation by Aspergillus niger immobilized on a new porous foam. J. Biotechnol. 2018, 276, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Chen, Y.; Li, A.; Ding, F.; Zhou, T.; He, Y.; Li, B.; Niu, H.; Lin, X.; Xie, J.; et al. Enhanced butanol production by modulation of electron flow in Clostridium acetobutylicum B3 immobilized by surface adsorption. Bioresour. Technol. 2013, 129, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Shi, S.; Chen, J.; Zhao, W.; Chen, T.; Li, G.; Zhang, K.; Yu, B.; Liu, D.; Chen, Y.; et al. Blue Light Signaling Regulates Escherichia coli W1688 Biofilm Formation and l-Threonine Production. Microbiol. Spectr. 2022, 10, e0246022. [Google Scholar] [CrossRef]
- Berlanga, M.; Guerrero, R. Living together in biofilms: The microbial cell factory and its biotechnological implications. Microb. Cell Fact. 2016, 15, 165. [Google Scholar] [CrossRef]
- Pan, Y.; Breidt, F., Jr.; Gorski, L. Synergistic effects of sodium chloride, glucose, and temperature on biofilm formation by Listeria monocytogenes serotype 1/2a and 4b strains. Appl. Environ. Microbiol. 2010, 76, 1433–1441. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yu, B.; Sun, W.; Liang, C.; Ying, H.; Zhou, S.; Niu, H.; Wang, Y.; Liu, D.; Chen, Y. Calcineurin signaling pathway influences Aspergillus niger biofilm formation by affecting hydrophobicity and cell wall integrity. Biotechnol. Biofuels 2020, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef] [PubMed]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yi, L.; Zhang, Z.; Fan, H.; Cheng, X.; Lu, C. Overexpression of luxS cannot increase autoinducer-2 production, only affect the growth and biofilm formation in Streptococcus suis. Sci. World J. 2013, 2013, 924276. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.; Sun, L.; Grenier, D.; Yi, L. The LuxS/AI-2 system of Streptococcus suis. Appl. Microbiol. Biotechnol. 2018, 102, 7231–7238. [Google Scholar] [CrossRef]
- Brackman, G.; Coenye, T. Quorum sensing inhibitors as anti-biofilm agents. Curr. Pharm. Des. 2015, 21, 5–11. [Google Scholar] [CrossRef]
- Banerjee, G.; Ray, A.K. Quorum-sensing network-associated gene regulation in Gram-positive bacteria. Acta Microbiol. Immunol. Hung. 2017, 64, 439–453. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.; Wang, J.H.; Swatton, J.E.; Davenport, P.; Price, B.; Mikkelsen, H.; Stickland, H.; Nishikawa, K.; Gardiol, N.; Spring, D.R.; et al. Variations on a theme: Diverse N-acyl homoserine lactone-mediated quorum sensing mechanisms in gram-negative bacteria. Sci. Prog. 2006, 89, 167–211. [Google Scholar] [CrossRef]
- Xavier, K.B.; Bassler, B.L. Interference with AI-2-mediated bacterial cell–cell communication. Nature 2005, 437, 750–753. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Attila, C.; Wang, L.; Wood, T.K.; Valdes, J.J.; Bentley, W.E. Quorum sensing in Escherichia coli is signaled by AI-2/LsrR: Effects on small RNA and biofilm architecture. J. Bacteriol. 2007, 189, 6011–6020. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.S.; de Regt, A.K.; Brito, P.H.; Miller, S.T.; Xavier, K.B. Identification of functional LsrB-like autoinducer-2 receptors. J. Bacteriol. 2009, 191, 6975–6987. [Google Scholar] [CrossRef]
- De Keersmaecker, S.C.; Sonck, K.; Vanderleyden, J. Let LuxS speak up in AI-2 signaling. Trends Microbiol. 2006, 14, 114–119. [Google Scholar] [CrossRef]
- Schauder, S.; Shokat, K.; Surette, M.G.; Bassler, B.L. The LuxS family of bacterial autoinducers: Biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 2001, 41, 463–476. [Google Scholar] [CrossRef]
- Stephens, K.; Zargar, A.; Emamian, M.; Abutaleb, N.; Choi, E.; Quan, D.N.; Payne, G.; Bentley, W.E. Engineering Escherichia coli for enhanced sensitivity to the autoinducer-2 quorum sensing signal. Biotechnol. Prog. 2019, 35, e2881. [Google Scholar] [CrossRef]
- Torres-Cerna, C.E.; Morales, J.A.; Hernandez-Vargas, E.A. Modeling quorum sensing dynamics and interference on Escherichia coli. Front. Microbiol. 2019, 10, 1835. [Google Scholar] [CrossRef]
- Rezzonico, F.; Duffy, B. Lack of genomic evidence of AI-2 receptors suggests a non-quorum sensing role for luxS in most bacteria. BMC Microbiol. 2008, 8, 154. [Google Scholar] [CrossRef]
- Laganenka, L.; Sourjik, V. Autoinducer 2-dependent Escherichia coli biofilm formation is enhanced in a dual-species coculture. Appl. Environ. Microbiol. 2018, 84, e02638-17. [Google Scholar] [CrossRef]
- Kapuscinski, J. DAPI: A DNA-specific fluorescent probe. Biotech. Histochem. 1995, 70, 220–233. [Google Scholar] [CrossRef]
- Partridge, J.D.; Nhu, N.T.; Dufour, Y.S.; Harshey, R.M. Escherichia coli remodels the chemotaxis pathway for swarming. mBio 2019, 10, e00316-19. [Google Scholar] [CrossRef] [PubMed]
- Xavier, K.B.; Bassler, B.L. LuxS quorum sensing: More than just a numbers game. Curr. Opin. Microbiol. 2003, 6, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Fang, Y.; Wang, X.; Zhao, L.; Wang, J.; Li, Y. Increasing L-threonine production in Escherichia coli by engineering the glyoxylate shunt and the L-threonine biosynthesis pathway. Appl. Microbiol. Biotechnol. 2018, 102, 5505–5518. [Google Scholar] [CrossRef]
- Miwa, K.; Tsuchida, T.; Kurahashi, O.; Nakamori, S.; Sano, K.; Momose, H. Construction of L-threonine overproducing strains of Escherichia coli K-12 using recombinant DNA techniques. Agric. Biol. Chem. 1983, 47, 2329–2334. [Google Scholar] [CrossRef]
- Lee, K.H.; Park, J.H.; Kim, T.Y.; Kim, H.U.; Lee, S.Y. Systems metabolic engineering of Escherichia coli for L-threonine production. Mol. Syst. Biol. 2007, 3, 149. [Google Scholar] [CrossRef]
- Liu, S.; Liang, Y.; Liu, Q.; Tao, T.; Lai, S.; Chen, N.; Wen, T. Development of a two-stage feeding strategy based on the kind and level of feeding nutrients for improving fed-batch production of l-threonine by Escherichia coli. Appl. Microbiol. Biotechnol. 2013, 97, 573–583. [Google Scholar] [CrossRef]
- Liu, J.; Li, H.; Xiong, H.; Xie, X.; Chen, N.; Zhao, G.; Caiyin, Q.; Zhu, H.; Qiao, J. Two-stage carbon distribution and cofactor generation for improving L-threonine production of Escherichia coli. Biotechnol. Bioeng. 2019, 116, 110–120. [Google Scholar] [CrossRef]
- DeLisa, M.P.; Valdes, J.J.; Bentley, W.E. Quorum signaling via AI-2 communicates the "Metabolic Burden" associated with heterologous protein production in Escherichia coli. Biotechnol. Bioeng. 2001, 75, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Pratt, L.A.; Kolter, R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998, 30, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Walters, M.; Sircili, M.P.; Sperandio, V. AI-3 synthesis is not dependent on luxS in Escherichia coli. J. Bacteriol. 2006, 188, 5668–5681. [Google Scholar] [CrossRef]
- Galloway, W.R.; Hodgkinson, J.T.; Bowden, S.D.; Welch, M.; Spring, D.R. Quorum sensing in Gram-negative bacteria: Small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem. Rev. 2011, 111, 28–67. [Google Scholar] [CrossRef]
- Bassler, B.L.; Greenberg, E.P.; Stevens, A.M. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 1997, 179, 4043–4045. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, B.; Duan, C.; Sun, B.; Yang, J.; Yang, S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015, 81, 2506–2514. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wu, R.; Zhang, J.; Li, P. Overexpression of luxS promotes stress resistance and biofilm formation of Lactobacillus paraplantarum L-ZS9 by regulating the expression of multiple genes. Front. Microbiol. 2018, 9, 2628. [Google Scholar] [CrossRef]
- Collinson, S.; Doig, P.; Doran, J.; Clouthier, S.; Trust, T.; Kay, W. Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J. Bacteriol. 1993, 175, 12–18. [Google Scholar] [CrossRef]
- Greenberg, E.; Hastings, J.W.; Ulitzur, S. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch. Microbiol. 1979, 120, 87–91. [Google Scholar] [CrossRef]



| Strain | Time (h) | L-Threonine (g/L) | Productivity (g/L/h) | Conversion Rate (g/g) | Reference |
|---|---|---|---|---|---|
| Batch/shake-flash | |||||
| E. coli W1688 | 36 | 10.16 | 0.282 | 0.339 | This study |
| E. coli W1688+pluxS | 28 | 15.27 | 0.545 | 0.509 | This study |
| E. coli W1688+plsrB | 30 | 13.38 | 0.446 | 0.446 | This study |
| E. coli TWF006/pFW01 | 36 | 15.9 | 0.44 | 0.53 | [35] |
| E. coli βIM4 | 72 | 13.4 | 0.186 | 0.447 | [36] |
| Fed-batch | |||||
| E. coli TH28C | 50 | 82.4 | 1.648 | 0.393 | [37] |
| E. coli EC125 | 48 | 105.3 | 2.194 | 0.405 | [38] |
| E. coli THPE5 | 40 | 70.8 | 1.77 | 0.404 | [39] |
| Strains/Plasmids | Relevant Characteristics | Culture Temperature | Sources |
|---|---|---|---|
| Strains | |||
| E. coli W1688 | L-threonine producing strain | 37 °C | Stored in our lab |
| ⊗luxS | E. coli W1688 with the deletion of luxS | 37 °C | This study |
| ⊗lsrB | E. coli W1688 with the deletion of lsrB | 37 °C | This study |
| ⊗lsrK | E. coli W1688 with the deletion of lsrK | 37 °C | This study |
| ⊗lsrR | E. coli W1688 with the deletion of lsrR | 37 °C | This study |
| +pluxS | E. coli W1688 harbouring plasmid pET28a-luxS | 37 °C | This study |
| +plsrB | E. coli W1688 harbouring plasmid pET28a-lsrB | 37 °C | This study |
| +plsrK | E. coli W1688 harbouring plasmid pET28a-lsrK | 37 °C | This study |
| +plsrR | E. coli W1688 harbouring plasmid pET28a-lsrR | 37 °C | This study |
| V. harveyi BB170 | BB120luxN:Tn5 (sensor 1-, sensor 2+) | 30 °C | [44] |
| Plasmids | |||
| pET28a | Kan resistance | 37 °C | Stored in our lab |
| pCas | Kan resistance | 30 °C | [45] |
| pTarget | Streptomycin resistance | 37 °C | [45] |
| Primer Name | Primer Sequence (5′ to 3′) |
|---|---|
| luxS-sg-F | TGTTGCTGATGCCTGGAAAGGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGT |
| luxS-sg-R | CTTTCCAGGCATCAGCAACAACTAGTATTATACCTAGGACTGAGCTAGCTG |
| luxS-up-F | GGAACCGGGTGATCCTCGA |
| luxS-up-R | GTGCAGTTCCTGCGCTATCTAACAACGGCATTTAGCCAC |
| luxS-dn-F | TTGTTAGATAGCGCAGGAACTGCACATCTAGTCAG |
| luxS-dn-R | CCGTCTGCTTCTACTGGCG |
| lsrB-sg-F | TGGTGTGAGAGTGCTGACCTGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGT |
| lsrB-sg-R | AGGTCAGCACTCTCACACCAACTAGTATTATACCTAGGACTGAGCTAGCTG |
| lsrB-up-F | CTGGCTCTGGCTGCATAAAAC |
| lsrB-up-R | CGCTCCGGGAGCGCTAAGTAAGGCGATTTTCT |
| lsrB-dn-F | AGCGCTCCCGGAGCGCGTGATATTCAAC |
| lsrB-dn-R | CCGATATAAACCTGCGCCGC |
| lsrK-sg-F | TGGCTGGCCTATATGCTCAGGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCT |
| lsrK-sg-R | CTGAGCATATAGGCCAGCCAACTAGTATTATACCTAGGACTGAGCTAGCTG |
| lsrK-up-F | GCAGCGGATGTGGCGA |
| lsrK-up-R | CTGATCCTGCCGGTGCCTGCATCC |
| lsrK-dn-F | AGGCACCGGCAGGATCAGCTGGGGCT |
| lsrK-dn-R | GTGTTAGTTGGAGGTGGGAAGGT |
| lsrR-sg-F | GGGAATCGGGCAGCTTAACGGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCT |
| lsrR-sg-R | CGTTAAGCTGCCCGATTCCCACTAGTATTATACCTAGGACTGAGCTAGCTG |
| lsrR-up-F | TTGGGAAAAGCAGCGGTTCCT |
| lsrR-up-R | TGCAGCGGCGTCGTGATAGTAAAACCACGCG |
| lsrR-dn-F | TCACGACGCCGCTGCAATGAAAGGC |
| lsrR-dn-R | GTTCGCTAACTTCGCGTGCC |
| luxS-F | AGGAGATATACCATGCCGTTGTTAGATAGCTTCACAG |
| luxS-R | GGTGGTGGTGCTCGAGGATGTGCAGTTCCTGCAACTTCT |
| lsrB-F | AGGAGATATACCATGACACTTCATCGCTTTAAGAAAATCGC |
| lsrB-R | GGTGGTGGTGCTCGAGGAAATCGTATTTGCCGATATTCTCTTTGTTGAA |
| lsrK-F | AGGAGATATACCATGGCTCGACTCTTTACCCTTTCAG |
| lsrK-R | GGTGGTGGTGCTCGAGTAACCCAGGCGCTTTCCATAACGAC |
| lsrR-F | AGGAGATATACCATGATGACAATCAACGATTCGGCAAT |
| lsrR-R | GGTGGTGGTGCTCGAGACTACGTAAAATCGCCGCTG |
| Gene | Forward Primer Sequence (5′-3′) | Reverse Primer Sequence (5′-3′) |
|---|---|---|
| luxS | GAACGTCTACCAGTGTGGCA | GTGCCAGTTCTTCGTTGCTG |
| lsrB | ATACTAAACCGGAGTGCCGC | AACGCGACTTTGGCTTTGTC |
| lsrK | TGGCCTACGTGCCGATATTC | TTGTGAACTTACCACGCCCA |
| lsrR | ATTGGTTTTGGCGAGGCAAC | TATAAGAACCGACGCCACCG |
| fimA | GGCAATCGTTGTTCTGTCGG | TCCCACCATTAACCGTCGTG |
| fimH | GATGCGGGCAACTCGATT | CGCCCTGTGCAGGTGAA |
| flhD | CGTCTGGTGGCTGTCAAAAC | CGTTAGCGGCACTGACTCTT |
| flhC | GCTTGTGGGCACTGTTCAAG | CCGGTTTGTGTAATGGCGTC |
| csgA | AGTACGGTGGCGGTAACTCT | TGTCATCTGAGCCCTGACCA |
| csgB | TAGTGCTCAGTTACGGCAGG | TGGTCAATCTTTGCCCGGTT |
| csgC | TTCAGGGCAAAGTCAGACGA | TAGCAGGCAATGAAAGGGTCT |
| csgD | CGTAAAGTAGCATTCGCCGC | GATTACCCGTACCGCGACAT |
| flu | ACGGTAAATGGCGGACTGTT | CACGGATGGTCAGGGTATCG |
| wza | CCCGCAGGTGGACGTTAATA | TAGTCAGTGGCACGTTGGTG |
| glgA | CAAAGACAAACCCACTGGCG | TGGGCTTGCTGATACGGTTT |
| glgP | AGTGCAGTTCCGATACACCG | GATACCGATCTGCTGGGACG |
| glgC | CATAACGCCAAATGCGGAGG | GCGGGCGACCATATCTACAA |
| thrA | CTTCACCCCCGCACCAT | ATCAGGCAAGGGATCTGGAA |
| thrB | CGAGCTGGAAGGCCGTATC | AACACGGTGCCACGTTGTC |
| thrC | AAGCGACTCAGGCGACGTTA | CACACGCGGCCAGTTGT |
| lysC | GCACAGCCTGAATATGCTGC | GCGAGGATGCCGAAAACTTC |
| rhtA | TCGTCGCCCGGTAGATTTC | GCAGGAACCACAGACCAAGAA |
| metB | CCCGGCATTACAAAATCCGC | GAATGCAACACCAGATCGGC |
| metC | ACTCTCGGCGCGGTAAATAG | CAAAGACCAGCGAAGAAGCG |
| metH | GTCAACGATCCGGCATTTCG | CGCTTTGGTGGACTCTCGAT |
| metL | TACACGCCGGATCAAGGTTC | GACATCATCGTGGCTGGTGA |
| 16S rna | TCGGGAACCGTGAGACAGG | CCGCTGGCAACAAAGGATAAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, H.; Zhang, K.; Li, G.; Yu, Y.; Shi, S.; Liang, C.; Niu, H.; Zhuang, W.; Liu, D.; Yang, P.; et al. Autoinducer-2: Its Role in Biofilm Formation and L-Threonine Production in Escherichia coli. Fermentation 2023, 9, 916. https://doi.org/10.3390/fermentation9100916
Han H, Zhang K, Li G, Yu Y, Shi S, Liang C, Niu H, Zhuang W, Liu D, Yang P, et al. Autoinducer-2: Its Role in Biofilm Formation and L-Threonine Production in Escherichia coli. Fermentation. 2023; 9(10):916. https://doi.org/10.3390/fermentation9100916
Chicago/Turabian StyleHan, Hui, Kaijie Zhang, Guoxiong Li, Ying Yu, Shuqi Shi, Caice Liang, Huanqing Niu, Wei Zhuang, Dong Liu, Pengpeng Yang, and et al. 2023. "Autoinducer-2: Its Role in Biofilm Formation and L-Threonine Production in Escherichia coli" Fermentation 9, no. 10: 916. https://doi.org/10.3390/fermentation9100916
APA StyleHan, H., Zhang, K., Li, G., Yu, Y., Shi, S., Liang, C., Niu, H., Zhuang, W., Liu, D., Yang, P., Chen, T., Sun, W., & Chen, Y. (2023). Autoinducer-2: Its Role in Biofilm Formation and L-Threonine Production in Escherichia coli. Fermentation, 9(10), 916. https://doi.org/10.3390/fermentation9100916






