1. Introduction
Rumen fermentation plays an important role in feed efficiency, productivity, and health for ruminants. With the help of colonized microbes, the rumen can transfer low-quality plant protein and nonprotein nitrogen to high-quality microbial protein and can hydrolyze plant fibers to volatile fatty acid (VFA), thus improving the feed digestibility [
1]. Some studies have concluded that microbial protein synthesis in the rumen provides the majority of required protein (accounting for 50 to 80% of the total absorbable protein) [
2] and that VFAs produced in the rumen provide about 75% of the total metabolic energy for ruminants [
3]. Therefore, targeting rumen fermentation is an efficient way to enhance feed digestion for ruminants.
Understanding the main factor affecting rumen fermentation is the key step in enhancing rumen feed digestion. Previous studies observed that the feed composition [
4], the feed type, feed processing, and additives could affect rumen fermentation [
5,
6], but these feed factors could not explain the feed digestibility differences among different species of ruminants, even with similar feed, as observed in our previous studies. Some studies also demonstrated that the composition of rumen bacteria, fungi, and protozoa, as well as their cooperative and competitive relationships, affect rumen fermentation [
7,
8,
9], such as the feed degradation rate of cattle, which can be improved by transferring the rumen contents of buffalo to the rumens of cattle [
10]. The degradation of crude fiber increased after the removal of protozoa [
11], and supplementation with
Bacillus licheniformis in feed increased the digestibility of neutral detergent fiber (NDF), acid detergent fiber (ADF), and organic matter (OM) [
12]. Some studies have also shown that the rumen microbial community of different species of ruminants differed [
13,
14]. Thus, we speculated that the microbiota composition difference of different species of ruminants might contribute to their feed digestibility variation and that some bacterial taxa might play key roles.
To test this hypothesis, in this study in vitro fermentation, which is a widely used method to evaluate rumen fermentation, was performed [
15], and goats (Taihang White cashmere goat), sheep (Hu sheep), and cows (Holstein cow) were used as experimental animals [
16,
17] to establish the correlation between their rumen bacterial communities and fermentation parameters with a random forest machine learning algorithm.
4. Discussion
Gas production (GP) is produced by rumen microorganisms, and it is positively correlated with both the feed quality and the activity of rumen microorganisms [
28,
29]. In this study, even with the same feed substrate, we observed different GP and GP parameters between different species of ruminants, and the sheep group had the highest GP and theoretical maximum GP (a + b). Correspondingly, we also observed that the nutrient digestibility of OM, DM, CP, and GE in the goat and cow groups were lower than that of the sheep group, although no difference was observed in the nutrient digestibility of NDF and ADF between the goat, sheep, and cow groups. Li et al. previously found that the anaerobic fungi population in the sheep rumen were greater than that of the goat, but the bacteria and ciliates populations were smaller than those of the goat [
30]. The ability of gas production and the degradation of plant fragments for different microbes differed [
31], which might be the reason why gas production and feed digestibility differed between the goat, sheep, and cow groups, even when the same feed was provided.
Ruminal pH is an important indicator of the homeostasis of the rumen metabolism and environment and usually varies from 5.0 to 7.5 [
32]. In this study, the ruminal pH values of the goat, sheep, and cow groups were all within the normal range, but the ruminal pH of the sheep group was lower than those of the goat and cow groups, and there were no significant differences between the goat and cow groups. This might be caused by the greater quantities of TVFA produced during the rumen microbial fermentation of the sheep, but no significant difference in the TVFA produced by goats and cows was observed. Domngue et al. [
33] also found that goats and sheep with the same forage feed also had different VFA contents in the rumen, indicating the microbes in different species of ruminants might contribute to the ruminal pH and VFA content variations. Isac et al. [
34] found that the A:P of goats was significantly higher than that of sheep, which is the same as our findings. Some studies inferred that methyl malonyl CoA mutase activity played an important role in changing propionic acid production in the rumen and causing the dysregulation of glucose homeostasis for ruminants, which were mainly produced by
Prevotella, which could be affected by multiple factors [
35,
36]. We also observed that the relative abundances of
Prevotella in the sheep and cow groups were higher than that of the goat group as well as low propionic acid production in the rumens of goats, which may be the main reason for the increased A:P. NH
3-N can be used to synthesize MCP and is the intermediate product of rumen microbial decomposition of nitrogen-containing substances, The optimal concentration range of NH
3-N is 2.37–27.3 mg/dL [
37], and MCP is the most important nitrogen source for ruminants, providing 40%−80% of their protein requirements. In this study, the content of NH
3-N and MCP in the sheep group was also higher than in the goat and cow groups.
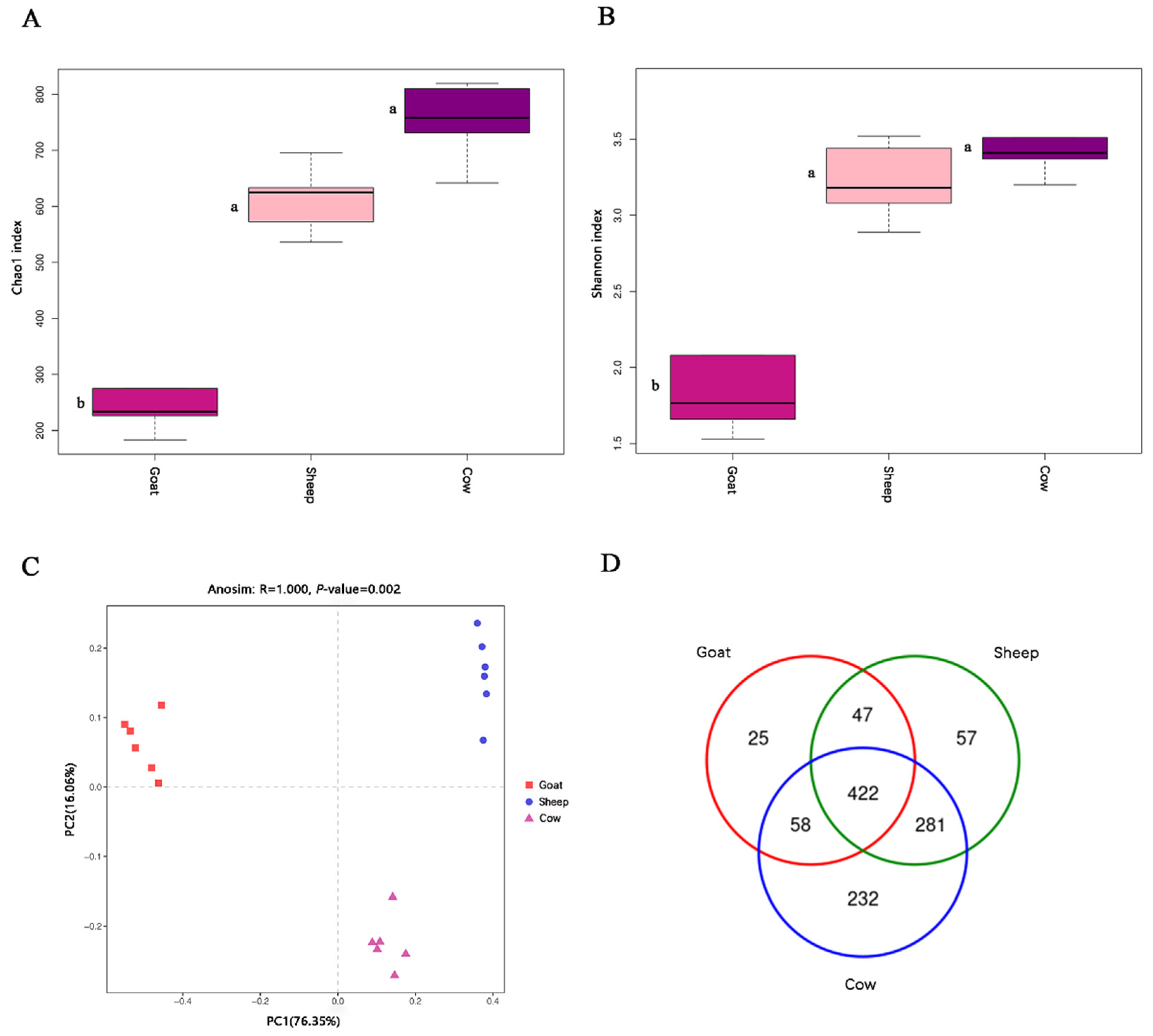
We found that there were significant differences in the bacterial communities of the goat, sheep, and cow. Firmicutes were the main dominant phyla of the sheep and cow, but Proteobacteria was the main dominant phylum of the goat. Firmicutes could improve the abundance of genes encoding enzymes related to energy metabolism [
38], promote the digestibility of oligosaccharides, starch, and cellulose [
39], and improve the enzyme activity and rumen fermentation. Proteobacteria are Gram-negative bacteria, mainly including
escherichia coli,
vibrio,
spirobacter, and other pathogens, most of which are the main factors threatening the health of livestock [
40]. At the genus level,
Prevotella and
Streptococcus were the main dominant genera of the sheep and cow.
Streptococcus could obtain glucose through the phosphotransferase system and produce lactic acid through glycolysis or the hexose diphosphate pathway [
41]. It was found that starch-based high-concentrate feed can cause the rapid proliferation and growth of
Streptococcus in the rumens of ruminants, which generate large amounts of lactic acid by fermentation with easily fermentable carbohydrates [
42], resulting in lactic acid accumulation and accelerating the process of rumen acidosis. Therefore, maintaining rumen
Streptococcus in the normal range was of great significance for preventing rumen acidosis in ruminants. The results of this study suggest that the high abundance of
Streptococcus in the sheep and cow rumens might induce a greater susceptibility to rumen acidosis than the goat.
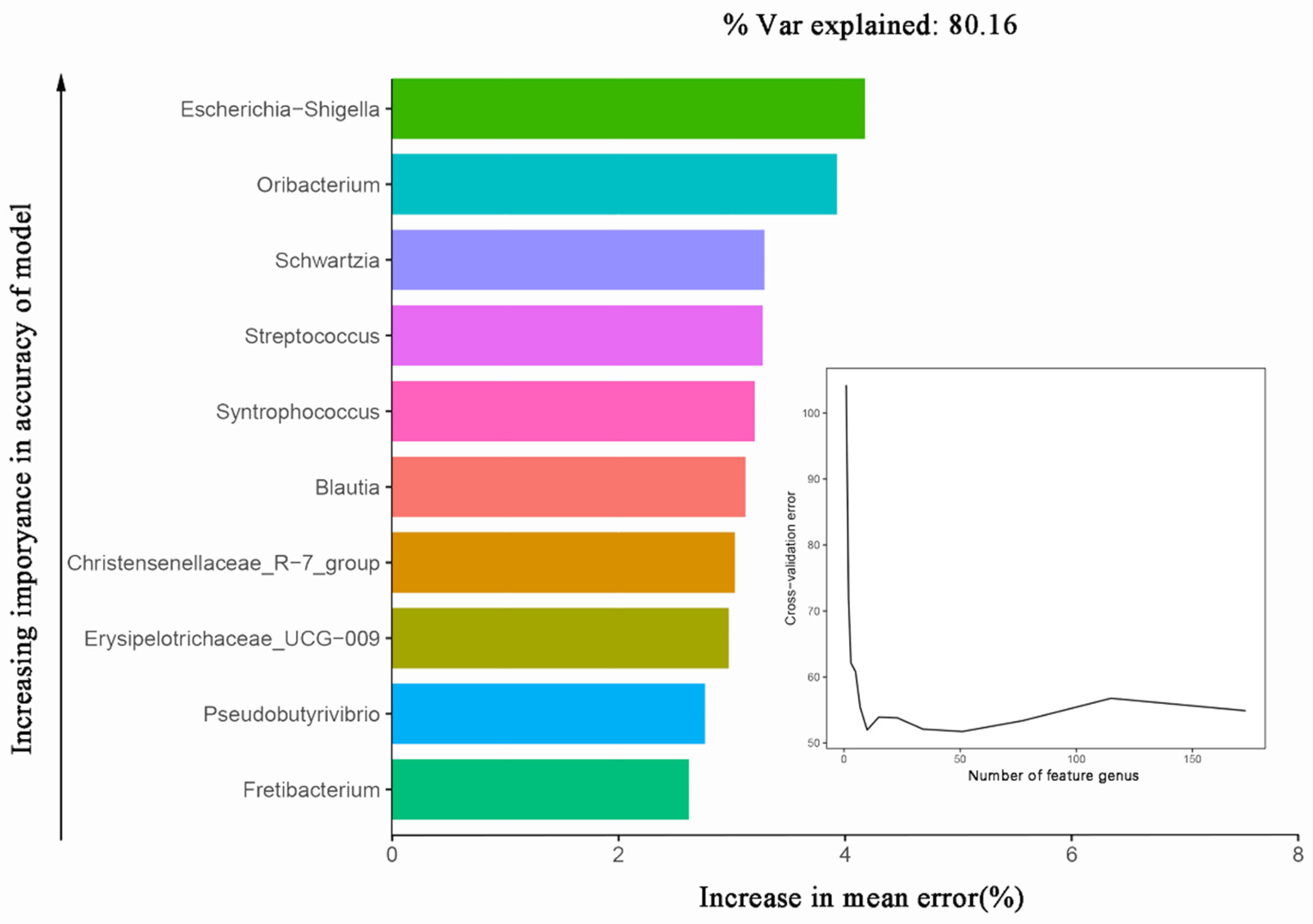
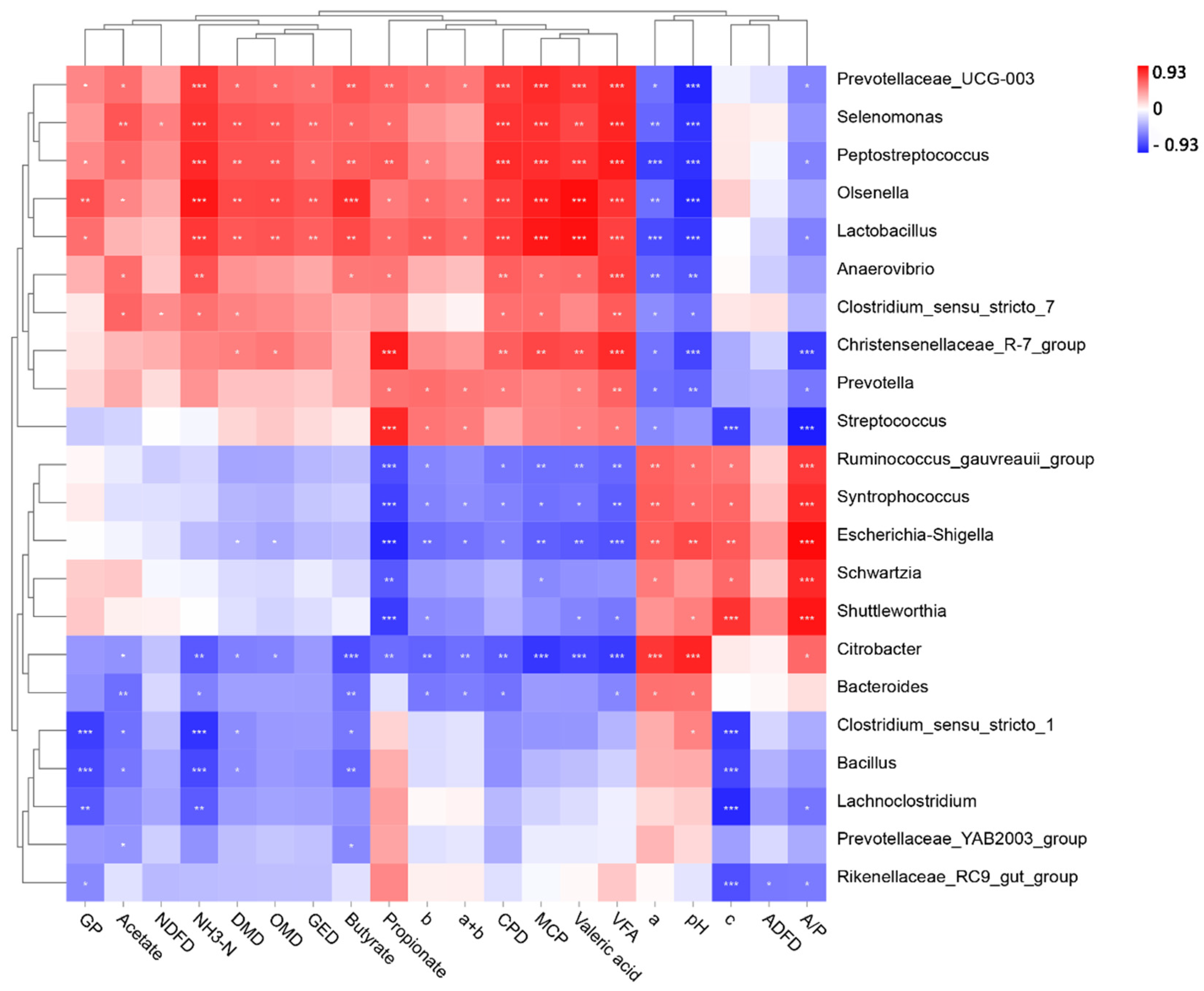
Multiple differences in bacterial composition and fermentation parameters were observed between goats, sheep, and cows, and we inferred that the bacterial community differences between goats, sheep, and cows contributed to the fermentation parameter alterations. With the random forest machine learning algorithm, we found that most of the in vitro fermentation parameters could be accurately predicted by rumen bacterial composition. In addition, we made a correlation heat map based on the bacteria selected from the random forest model to determine the main bacteria that affect the fermentation parameters. The results showed that
Prevotellaceae_UCG-003,
Lactobacillus,
Olsenella,
Selenomonas, and
Peptostreptococcus were the main bacteria that affected the in vitro fermentation parameters of ruminants.
Prevotellaceae_UCG-003 belongs to the family Prevotellaceae, which could increase rumen propionic acid content and promote feed digestion [
35]. A high abundance of
Lactobacillus could reduce ruminal pH and restrain the aerobic consumption caused by harmful bacteria growth [
43], so
Lactobacillus abundance was also significantly positively correlated with most fermentation parameters.
Olsenella and
Peptostreptococcus are anaerobic bacteria that produce lactic acid from glucose fermentation, similar to the metabolites of
Lactobacillus [
44].
Selenomonas is a very important lactic acid utilization bacterium in the rumen, which could reduce rumen lactic acid accumulation, stabilize ruminal pH, provide a suitable environment for rumen microorganisms, and accelerate the production of propionic acid [
45], thus enhancing rumen fermentation.
We could accurately predict the fermentation parameters of GP, a, b, c, a + b, pH, NH
3-N, propionic acid, valeric acid, and A:P by rumen bacteria (explanation > 55% of variations), but the predictive effect for DMD, OMD, GED, CPD, NDFD, ADFD, MCP, acetic acid, butyric acid, and TVFA were inadequate, especially for feed digestibility. The rumen microbial community is composed of bacteria, fungi, and protozoa, and they also have complex relationships in predation, competition, and even interdependence, which could also affect rumen fermentation. Some studies have demonstrated that rumen fungi could infiltrate plant cell walls through abundant rhizoids, erode plant tissues, and decrease the tension of plant fragments [
46]. Rumen protozoa could affect the digestibility of carbohydrates and protein and also have the ability to survive phagocytosis activity to conserve the relative abundance of rumen bacteria [
47]. Thus, although the rumen bacteria were the main rumen microbial community of ruminants, rumen fungi and protozoa performed an important role in the process of feed degradation, which might be the main reason for the insufficient prediction effect of feed digestibility in the current experiment. In the future, further studies on the effects of rumen fungi and protozoa on ruminal fermentation parameters and adding the main rumen fungi and protozoa that cause the differences in feed digestibility among goats, sheep, and cows in the prediction model might contribute to a better prediction.
Although the bacterial community analysis based on the 16S rRNA genes did not allow for species level comparisons, our findings from bacterial ecology at the phylum and genus levels indicated that the in vitro fermentation parameter differences between the goat, sheep, and cow were due to their different bacterial communities and hinted that targeting rumen bacteria might be an efficient way to manipulate rumen fermentation. The strategy of rumen bacteria regulation is of great promise and needs to be further studied.
5. Conclusions
Our results showed that the rumen fermentation parameters and bacterial composition differed in goats, sheep, and cows. At the phylum level, Firmicutes were the main dominant phyla of the sheep and cow, but Proteobacteria was the main dominant phylum of the goat; at the genus level, Prevotella and Streptococcus were the main dominant genera of the sheep and cow, while Escherichia-Shigella was the main dominant genus of the goat. The fermentation parameters of GP, a, b, c, a + b, pH, NH3-N, propionic acid, valeric acid, and A:P could be accurately predicted by rumen bacteria (explanation > 55% of variations), and Lactobacillus, Prevotellaceae_UCG-003, Olsenella, Selenomonas, and Peptostreptococcus were the main bacteria that affected in vitro rumen fermentation parameters.