Apple Pomace Modulates the Microbiota and Increases the Propionate Ratio in an In Vitro Piglet Gastrointestinal Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Equipment, Inocula, Culture Media and Sample Collection
2.2. Microbial Metabolites
2.3. 16S rRNA Gene Sequencing
2.4. Statistics
3. Results
3.1. SCFA Results
3.2. 16S rRNA Gene Sequencing
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, Y.; Foo, L.Y. Identification and quantification of major polyphenols in apple pomace. Food Chem. 1997, 59, 187–194. [Google Scholar] [CrossRef]
- Boyer, J.; Liu, R.H. Apple phytochemicals and their health benefits. Nutr. J. 2004, 3, 5. [Google Scholar] [CrossRef] [PubMed]
- Grigoras, C.G.; Destandau, E.; Fougère, L.; Elfakir, C. Evaluation of apple pomace extracts as a source of bioactive compounds. Ind. Crops Prod. 2013, 49, 794–804. [Google Scholar] [CrossRef]
- Reis, S.F.; Rai, D.K.; Abu-Ghannam, N. Apple pomace as a potential ingredient for the development of new functional foods. Int. J. Food Sci. Technol. 2014, 49, 1743–1750. [Google Scholar] [CrossRef]
- Waldbauer, K.; McKinnon, R.; Kopp, B. Apple pomace as potential source of natural active compounds. Planta Med. 2017, 83, 994–1010. [Google Scholar] [CrossRef]
- Okoro, O.V.; Nie, L.; Hobbi, P.; Shavandi, A. Valorization of waste apple pomace for production of platform biochemicals: A multi-objective optimization study. Waste Biomass Valorization 2021, 12, 6887–6901. [Google Scholar] [CrossRef]
- Awasthi, M.K.; Ferreira, J.A.; Sirohi, R.; Sarsaiya, S.; Khoshnevisan, B.; Baladi, S.; Sindhu, R.; Binod, P.; Pandey, A.; Juneja, A.; et al. A critical review on the development stage of biorefinery systems towards the management of apple processing-derived waste. Renew. Sustain. Energy Rev. 2021, 143, 110972. [Google Scholar] [CrossRef]
- Lyu, F.; Luiz, S.F.; Azeredo, D.R.P.; Cruz, A.G.; Ajlouni, S.; Ranadheera, C.S. Apple pomace as a functional and healthy ingredient in food products: A review. Processes 2020, 8, 319. [Google Scholar] [CrossRef]
- Sabater, C.; Calvete-Torre, I.; Villamiel, M.; Moreno, F.J.; Margolles, A.; Ruiz, L. Vegetable waste and by-products to feed a healthy gut microbiota: Current evidence, machine learning and computational tools to design novel microbiome-targeted foods. Trends Food Sci. Technol. 2021, 118, 399–417. [Google Scholar] [CrossRef]
- Juśkiewicz, J.; Zary-Sikorska, E.; Zduńczyk, Z.; Król, B.; Jarosławska, J.; Jurgoński, A. Effect of dietary supplementation with unprocessed and ethanol-extracted apple pomaces on caecal fermentation, antioxidant and blood biomarkers in rats. Br. J. Nutr. 2012, 107, 1138–1146. [Google Scholar] [CrossRef]
- Sehm, J.; Treutter, D.; Lindermayer, H.; Meyer, H.H.D.; Pfaffl, M.W. The influence of apple- or red-grape pomace enriched piglet diet on blood parameters, bacterial colonisation, and marker gene expression in piglet white blood cells. Food Nutr. Sci. 2011, 2, 366–376. [Google Scholar] [CrossRef]
- Sehm, J.; Lindermayer, H.; Dummer, C.; Treutter, D.; Pfaffl, M.W. The influence of polyphenol rich apple pomace or red-wine pomace diet on the gut morphology in weaning piglets. J. Anim. Physiol. Anim. Nutr. 2007, 91, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Uerlings, J.; Schroyen, M.; Bautil, A.; Courtin, C.; Richel, A.; Sureda, E.A.; Bruggeman, G.; Tanghe, S.; Willems, E.; Bindelle, J.; et al. In vitro prebiotic potential of agricultural by-products on intestinal fermentation, gut barrier and inflammatory status of piglets. Br. J. Nutr. 2020, 123, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Martinov, J.; Krsti, M.; Spasic, S.; Miletic, S.; Stefanovic-Kojic, J.; Nikolic-Kokic, A.; Blagojevic, D.; Spasojevic, I.; Spasic, M.B. Apple pectin-derived oligosaccharides produce carbon dioxide radical anion in Fenton reaction and prevent growth of Escherichia coli and Staphylococcus aureus. Food Res. Int. 2017, 100, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Weary, D.M.; Jasper, J.; Hötzel, M.J. Understanding weaning distress. Appl. Anim. Behav. Sci. 2008, 110, 24–41. [Google Scholar] [CrossRef]
- Pluske, J.R.; Le Dividich, J.; Verstegen, M.W.A. Weaning the Pig. Concepts and Consequences; Wageningen Academic Publishers: Wageningen, The Netherlands, 2003. [Google Scholar]
- Campbell, J.M.; Crenshaw, J.D.; Polo, J. The biological stress of early weaned piglets. J. Anim. Sci. Biotechnol. 2013, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Niewold, T. Intestinal Health. Key to Maximise Growth Performance in Livestock; Wageningen Academic Publishers: Wageningen, The Netherlands, 2015. [Google Scholar]
- Rhouma, M.; Fairbrother, J.M.; Beaudry, F.; Letellier, A. Post weaning diarrhea in pigs: Risk factors and non-colistin-based control strategies. Acta Vet. Scand. 2017, 59, 31. [Google Scholar] [CrossRef] [PubMed]
- Gresse, R.; Chaucheyras-Durand, F.; Fleury, M.A.; Van de Wiele, T.; Forano, E.; Blanquet-Diot, S. Gut microbiota dysbiosis in postweaning piglets: Understanding the keys to health. Trends Microbiol. 2017, 25, 851–873. [Google Scholar] [CrossRef]
- Dufourny, S.; Everaert, N.; Lebrun, S.; Douny, C.; Scippo, M.L.; Li, B.; Taminiau, B.; Marzorati, M.; Wavreille, J.; Froidmont, E.; et al. Baby-SPIME: A dynamic in vitro piglet model mimicking gut microbiota during the weaning process. J. Microbiol. Methods 2019, 167, 105735. [Google Scholar] [CrossRef]
- Douny, C.; Dufourny, S.; Brose, F.; Verachtert, P.; Rondia, P.; Lebrun, S.; Marzorati, M.; Everaert, N.; Delcenserie, V.; Scippo, M.-L. Development of an analytical method to detect short-chain fatty acids by SPME-GC–MS in samples coming from an in vitro gastrointestinal model. J. Chromatogr. B 2019, 1124, 188–196. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef]
- Aguirre, M.; Ramiro-Garcia, J.; Koenen, M.E.; Venema, K. To pool or not to pool? Impact of the use of individual and pooled fecal samples for in vitro fermentation studies. J. Microbiol. Methods 2014, 107, 1–7. [Google Scholar] [CrossRef]
- Karasova, D.; Crhanova, M.; Babak, V.; Jerabek, M.; Brzobohaty, L.; Matesova, Z.; Rychlik, I. Development of piglet gut microbiota at the time of weaning influences development of postweaning diarrhea—A field study. Res. Vet. Sci. 2021, 135, 59–65. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, K.; Chen, H.; Su, Y.; Zhu, W. Caecal infusion of the short-chain fatty acid propionate affects the microbiota and expression of inflammatory cytokines in the colon in a fistula pig model. Microb. Biotechnol. 2018, 11, 859–868. [Google Scholar] [CrossRef]
- Skinner, R.C.; Gigliotti, J.C.; Ku, K.-M.; Tou, J.C. A comprehensive analysis of the composition, health benefits, and safety of apple pomace. Nutr. Rev. 2018, 76, 893–909. [Google Scholar] [CrossRef]
- Sembries, S.; Dongowski, G.; Jacobasch, G.; Mehrländer, K.; Will, F.; Dietrich, H. Effects of dietary fibre-rich juice colloids from apple pomace extraction juices on intestinal fermentation products and microbiota in rats. Br. J. Nutr. 2003, 90, 607–615. [Google Scholar] [CrossRef]
- Dufourny, S.; Antoine, N.; Pitchugina, E.; Delcenserie, V.; Godbout, S.; Douny, C.; Scippo, M.-L.; Froidmont, E.; Rondia, P.; Wavreille, J.; et al. Apple Pomace and Performance, Intestinal Morphology and Microbiota of Weaned Piglets—A Weaning Strategy for Gut Health? Microorganisms 2021, 9, 572. [Google Scholar] [CrossRef]
- Li, D.; Wang, P.; Wang, P.; Hu, X.; Chen, F. Targeting the gut microbiota by dietary nutrients: A new avenue for human health. Crit. Rev. Food Sci. Nutr. 2017, 59, 181–195. [Google Scholar] [CrossRef]
- Chénard, T.; Prévost, K.; Dubé, J.; Massé, E. Immune system modulations by products of the gut microbiota. Vaccines 2020, 8, 461. [Google Scholar] [CrossRef]
- Sehm, J.; Lindermayer, H.; Meyer, H.H.D.; Pfaffl, M.W. The influence of apple- and red-wine pomace rich diet on mRNA expression of inflammatory and apoptotic markers in different piglet organs. Anim. Sci. 2006, 82, 877–887. [Google Scholar] [CrossRef]
- Hosseini, E.; Grootaert, C.; Verstraete, W.; Van de Wiele, T. Propionate as a health-promoting microbial metabolite in the human gut. Nutr. Rev. 2011, 69, 245–258. [Google Scholar] [CrossRef]
- De Vadder, F.; Kovatcheva-Datchary, P.; Goncalves, D.; Vinera, J.; Zitoun, C.; Duchampt, A.; Bäckhed, F.; Mithieux, G. Microbiota-Generated Metabolites Promote Metabolic Benefits via Gut-Brain Neural Circuits. Cell 2014, 156, 84–96. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Lan, C.; Xie, K.; Li, H.; Devillard, E.; He, J.; Liu, L.; Cai, J.; Tian, G.; Wu, A.; et al. Active or autoclaved Akkermansia muciniphila relieves TNF-α-induced inflammation in intestinal epithelial cells through distinct pathways. Front. Immunol. 2021, 12, 788638. [Google Scholar] [CrossRef] [PubMed]
- Langella, P.; Guarner, F.; Martín, R. Editorial: Next-generation probiotics: From commensal bacteria to novel drugs and food supplements. Front. Microbiol. 2019, 10, 1973. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.J.; Lin, T.L.; Tsai, Y.L.; Wu, T.R.; Lai, W.F.; Lu, C.C.; Lai, H.C. Next generation probiotics in disease amelioration. J. Food Drug Anal. 2019, 27, 615–622. [Google Scholar] [CrossRef]
- Cani, P.D.; de Vos, W.M. Next-generation beneficial microbes: The case of Akkermansia muciniphila. Front. Microbiol. 2017, 8, 1765. [Google Scholar] [CrossRef]
- El Hage, R.; Hernandez-Sanabria, E.; Arroyo, M.C.; Van de Wiele, T. Supplementation of a propionate-producing consortium improves markers of insulin resistance in an in vitro model of gut-liver axis. Am. J. Physiol. Endocrinol. Metab. 2020, 318, E742–E749. [Google Scholar] [CrossRef]
- Barbosa, J.C.; Machado, D.; Almeida, D.; Andrade, J.C.; Brandelli, A.; Gomes, A.M.; Freitas, A.C. Next-generation probiotics. In Probiotics, 1st ed.; Brandelli, A., Ed.; Academic Press: Cambridge, MA, USA, 2022; ISBN 9780323851701. [Google Scholar]
- Roopchand, D.E.; Carmody, R.N.; Kuhn, P.; Moskal, K.; Rojas-Silva, P.; Turnbaugh, P.J.; Raskin, I. Dietary polyphenols promote growth of the gut bacterium akkermansia muciniphila and attenuate high-fat diet-induced metabolic syndrome. Diabetes 2015, 64, 2847–2858. [Google Scholar] [CrossRef]
- Kumari, M.; Singh, P.; Nataraj, B.H.; Kokkiligadda, A.; Naithani, H.; Azmal Ali, S.; Behare, P.V.; Nagpal, R. Fostering next-generation probiotics in human gut by targeted dietary modulation: An emerging perspective. Food Res. Int. 2021, 150, 110716. [Google Scholar] [CrossRef]
- Luo, Y.; Ren, W.; Smidt, H.; Wright, A.G.; Yu, B.; Schyns, G.; McCormack, U.M.; Cowieson, A.J.; Yu, J.; He, J.; et al. Dynamic distribution of gut microbiota in pigs at different growth stages: Composition and contribution. Microbiol. Spectr. 2022, 10, e00688-21. [Google Scholar] [CrossRef]
- Wallenborn, J.T.; Vonaesch, P. Intestinal microbiota research from a global perspective. Gastroenterol. Rep. 2022, 10, goac010. [Google Scholar] [CrossRef]
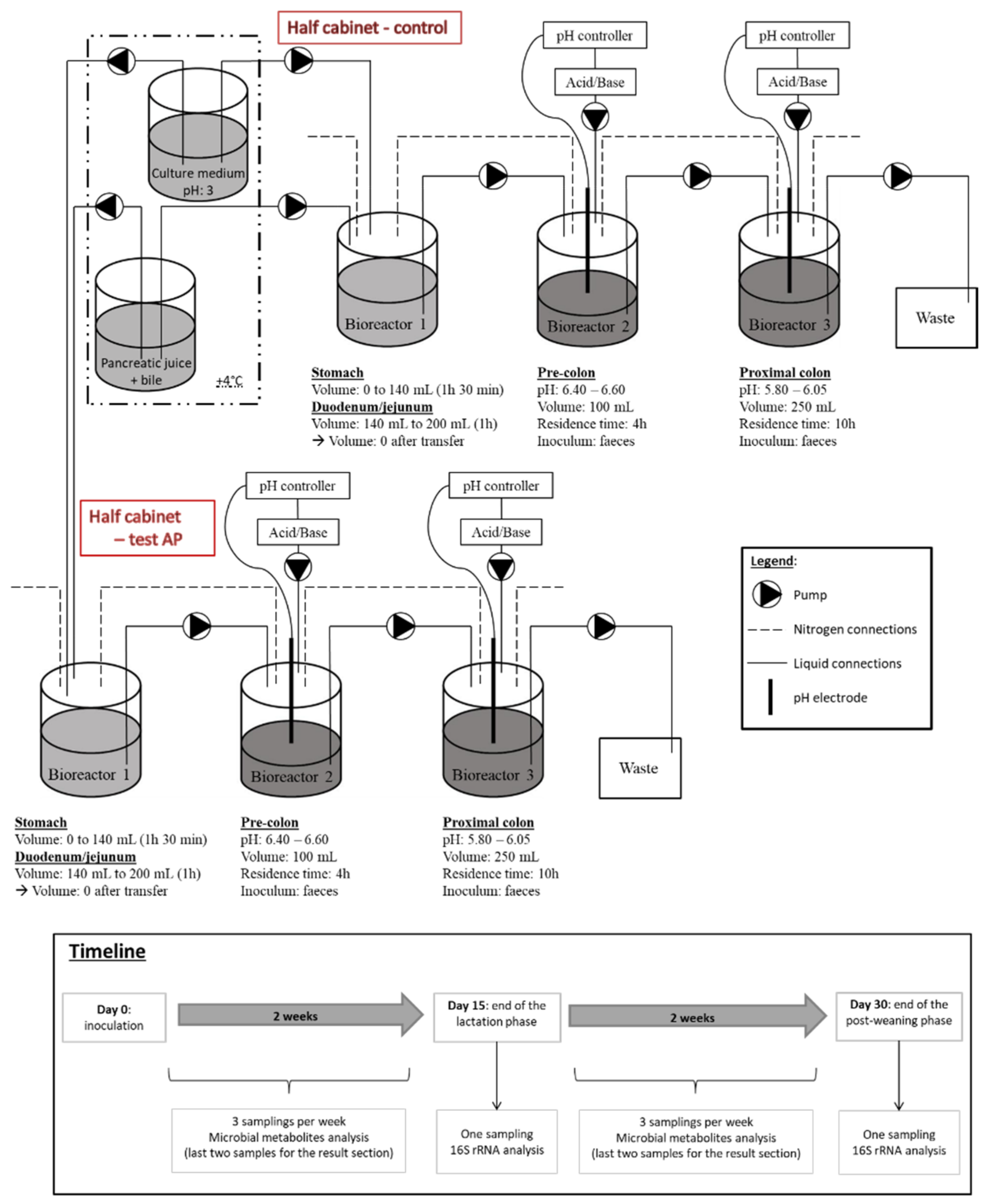

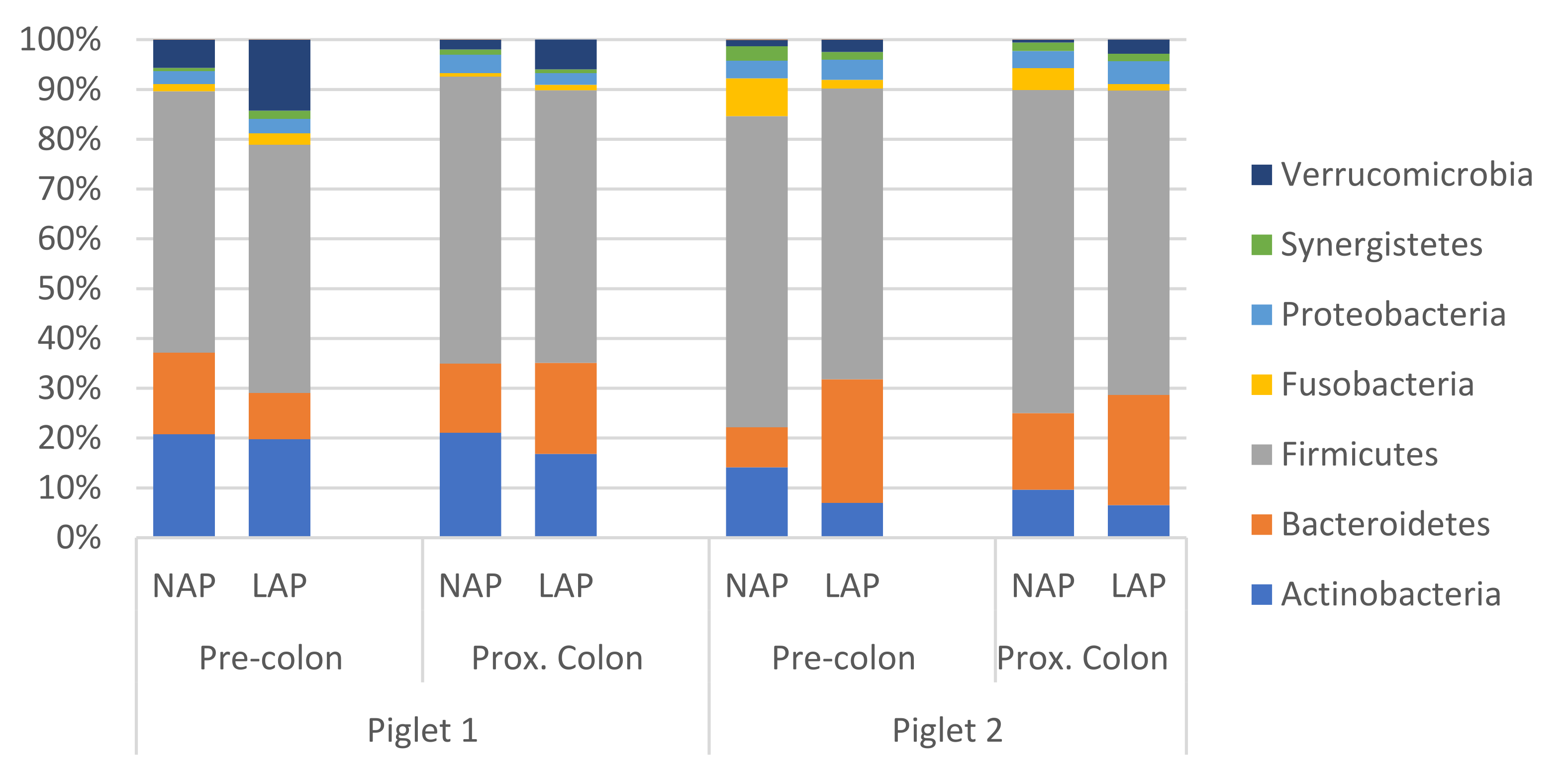
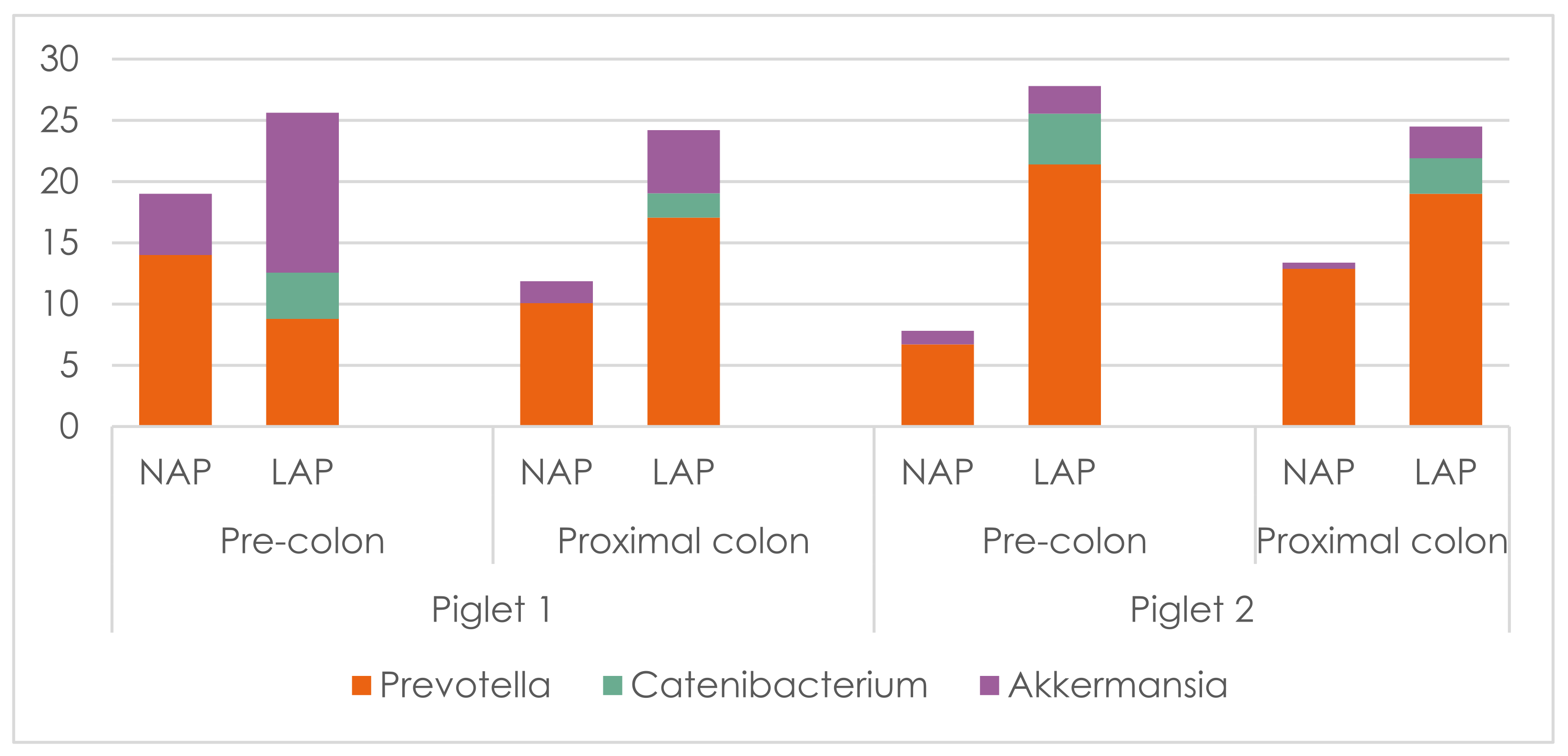
| Ingredients | Lactation CM | PW Control CM | PW AP CM |
|---|---|---|---|
| Mucin (Sigma-Aldrich, St-Louis, MO, USA) | 6.0 g/L | 6.0 g/L | 6.0 g/L |
| Proteose-Peptone n°3 (BD Bacto Biosciences, Franklin Lakes, NJ, USA) | 1.0 g/L | 1.0 g/L | 1.0 g/L |
| Potato starch (Sigma-Aldrich, St-Louis, MO, USA) | 1.0 g/L | 1.0 g/L | 1.0 g/L |
| L-Cysteine hydrochloride (Merck, Darmstadt, Germany) | 0.2 g/L | 0.2 g/L | 0.2 g/L |
| Nuklospray Yoghurt 1 (Dumoulin, Andenne, Belgium) | 8.0 g/L | 0.0 g/L | 0.0 g/L |
| Post-weaning diet for piglets 2 (ABZDiervoeding, Nijkerk, The Netherlands) | 0.0 g/L | 8.0 g/L | 8.0 g/L |
| Apple pomace (Extr’Apple SAS, Pleudihen-sur-Rance, France) | 0.0 g/L | 0.0 g/L | 0.65 g/L |
| SCFA | Pre-Colon Bioreactors | Proximal Colon Bioreactors | p-Value | ||||
|---|---|---|---|---|---|---|---|
| Control (n = 2) | AP (n = 2) | Control (n = 2) | AP (n = 2) | Bioreactor (B) | Medium (M) | B * M | |
| C2 | 2019 | 2146 | 2157 | 1685 | ns | ns | ns |
| C3 | 1126 | 1352 | 1200 | 1165 | ns | ns | ns |
| iC4 | 38.1 | 44.5 | 36.1 | 44.6 | ns | ns | ns |
| C4 | 692 | 579 | 714 | 556 | ns | ns | ns |
| iC5 | 103 | 98 | 111 | 92 | ns | ns | ns |
| C5 | 556 | 531 | 591 | 494 | ns | ns | ns |
| C6 | 110 | 137 | 166 | 118 | ns | ns | ns |
| Sum | 4644 | 4887 | 4975 | 4155 | ns | ns | ns |
| Ratio C2 (%) | 59.3 | 58.8 | 59.6 | 56.1 | ns | ns | ns |
| Ratio C3 (%) | 26.9 | 30.4 | 27.0 | 31.4 | ns | 0.021 | ns |
| Ratio C4 (%) | 13.8 | 10.8 | 13.5 | 12.6 | ns | 0.009 | ns |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dufourny, S.; Lebrun, S.; Douny, C.; Dubois, B.; Scippo, M.-L.; Wavreille, J.; Rondia, P.; Everaert, N.; Delcenserie, V. Apple Pomace Modulates the Microbiota and Increases the Propionate Ratio in an In Vitro Piglet Gastrointestinal Model. Fermentation 2022, 8, 408. https://doi.org/10.3390/fermentation8080408
Dufourny S, Lebrun S, Douny C, Dubois B, Scippo M-L, Wavreille J, Rondia P, Everaert N, Delcenserie V. Apple Pomace Modulates the Microbiota and Increases the Propionate Ratio in an In Vitro Piglet Gastrointestinal Model. Fermentation. 2022; 8(8):408. https://doi.org/10.3390/fermentation8080408
Chicago/Turabian StyleDufourny, Sandrine, Sarah Lebrun, Caroline Douny, Benjamin Dubois, Marie-Louise Scippo, José Wavreille, Pierre Rondia, Nadia Everaert, and Véronique Delcenserie. 2022. "Apple Pomace Modulates the Microbiota and Increases the Propionate Ratio in an In Vitro Piglet Gastrointestinal Model" Fermentation 8, no. 8: 408. https://doi.org/10.3390/fermentation8080408
APA StyleDufourny, S., Lebrun, S., Douny, C., Dubois, B., Scippo, M.-L., Wavreille, J., Rondia, P., Everaert, N., & Delcenserie, V. (2022). Apple Pomace Modulates the Microbiota and Increases the Propionate Ratio in an In Vitro Piglet Gastrointestinal Model. Fermentation, 8(8), 408. https://doi.org/10.3390/fermentation8080408






