Selection of Yeast and Lactic Acid Bacteria Strains, Isolated from Spontaneous Raw Milk Fermentation, for the Production of a Potential Probiotic Fermented Milk
Abstract
:1. Introduction
2. Materials and Methods
2.1. Raw Milk Spontaneous Fermentation
2.2. Microorganism Enumeration
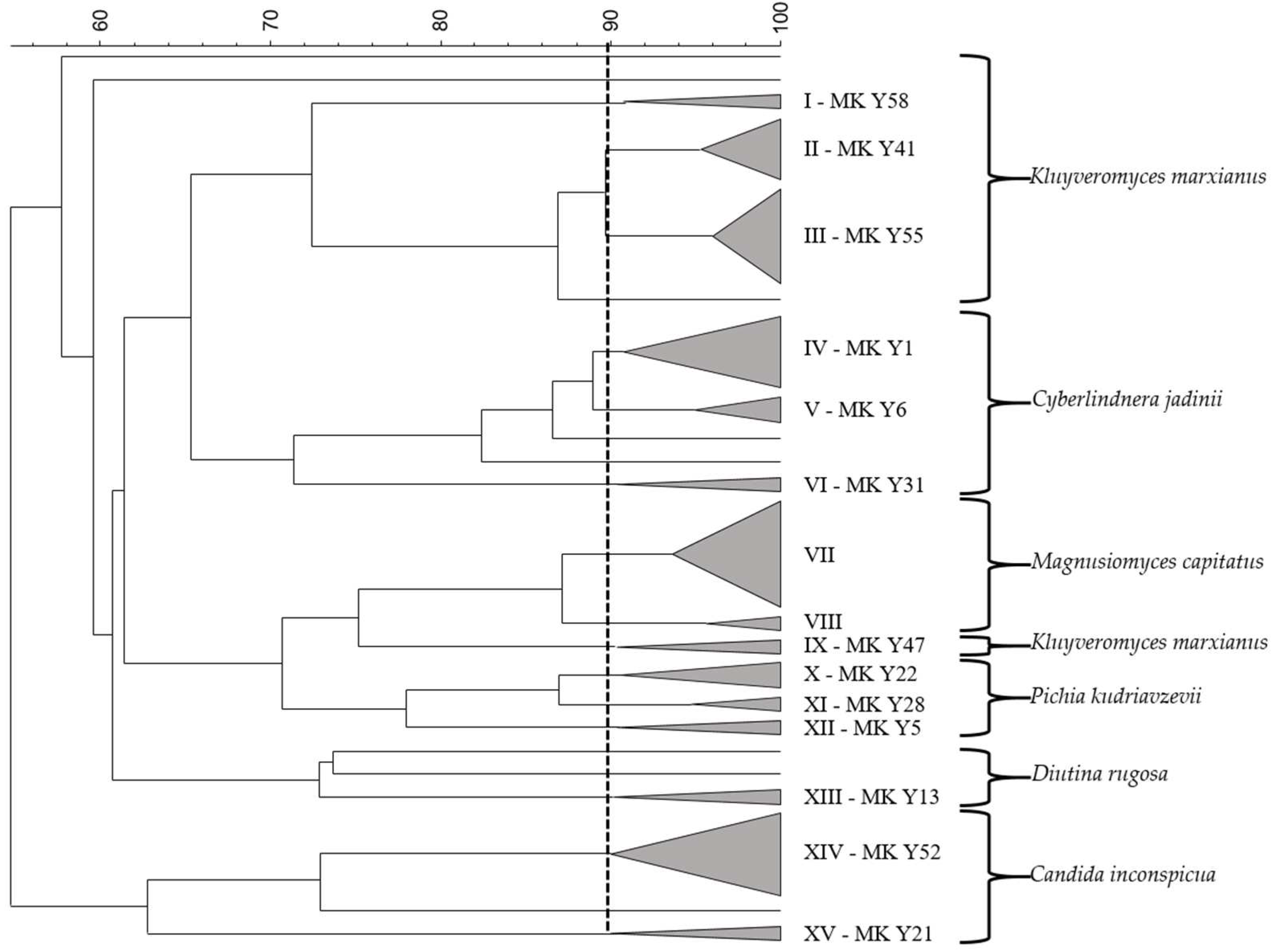
2.3. Identification and Typing of Yeasts
2.4. Microorganisms and Culture Condition
2.5. Yeast and Lactic Acid Bacteria Probiotic Features Assessment
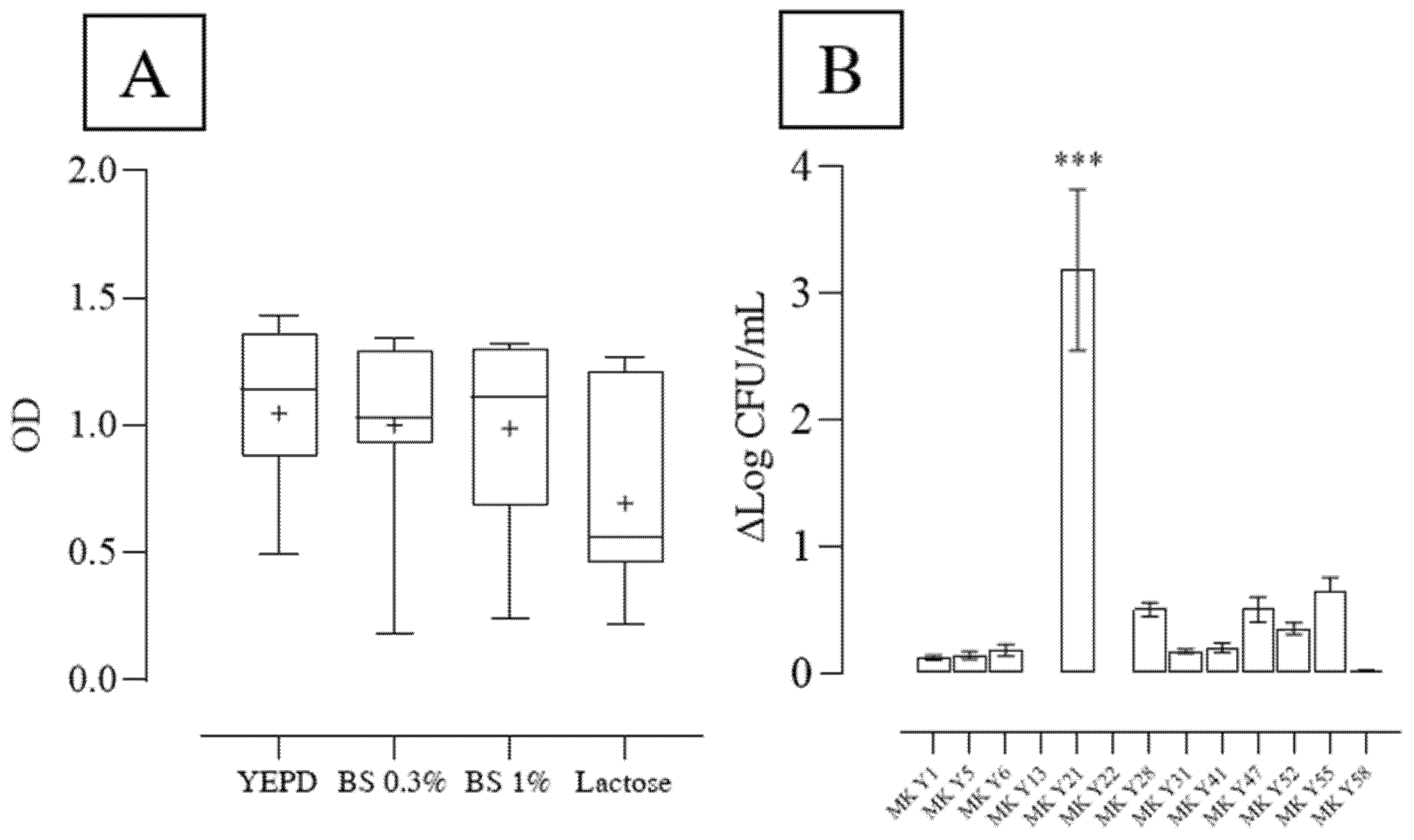
2.5.1. Bile Salt Tolerance
2.5.2. Tolerance to Acidic Condition
2.6. Lactose Utilization
2.7. Resistance to Simulated Human Gastrointestinal (GI) Conditions
2.8. Manufacture of Fermented Milk and Microbial Viability during Storage
2.9. Resistance to Simulated Gastrointestinal Conditions of Microorganisms in Fermented Milk
2.10. Organic Acid Determination by High-Pressure Liquid Chromatography (HPLC)
2.11. Statistical Analysis
3. Results and Discussion
3.1. Raw Cow Milk Spontaneous Fermentation and Yeast Identification
3.2. Yeast Screening for Probiotic Features
3.3. Resistance of Yeasts to Simulated Gastrointestinal (GI) Conditions
3.4. Lactic Acid Bacteria Screening for Probiotic Features
3.5. Resistance of Lactic Acid Bacteria to Simulated Gastrointestinal Conditions
3.6. Fermented Milk Production
3.7. Microbial Survival during Simulated Gastrointestinal Conditions in Fermented Milk
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- CXS 243–2003; Codex Standard for Fermented Milks. WHO: Geneva, Switzerland, 2003.
- Tamang, J.P.; Watanabe, K.; Holzapfel, W.H. Review: Diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 2016, 7, 377. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.V.; Aryana, K.J.; Prinyawiwatkul, W.; Ordonez, K.C.; Boeneke, C.A. The effects of frozen storage on the survival of probiotic microorganisms found in traditionally and commercially manufactured kefir. J. Dairy Sci. 2016, 99, 7043–7048. [Google Scholar] [CrossRef] [PubMed]
- FAO/WHO. Food and Agriculture Organization and World Health Organization Expert Consultation. In Evaluation of Health and Nutritional Properties of Powder Milk and Live Lactic Acid Bacteria; Food and Agriculture Organization of the United Nations and World Health Organization: Córdoba, Argentina, 2001; Available online: http://ftp.fao.org/es/esn/food/probio_report_en.pdf (accessed on 13 May 2022).
- Islam, S.U. Clinical uses of probiotics. Medicine 2016, 95, e2658. [Google Scholar] [CrossRef] [PubMed]
- Martoni, C.J.; Srivastava, S.; Leyer, G.J. Lactobacillus acidophilus DDS-1 and Bifidobacterium lactis UABla-12 improve abdominal pain severity and symptomology in irritable bowel syndrome: Randomized controlled trial. Nutrients 2020, 12, 363. [Google Scholar] [CrossRef]
- Pesce, M.; Suella, L.; Del Re, A.; Lu, J.; Palenca, I.; Corpetti, C.; Rurgo, S.; Sanseverino, W.; Sarnelli, G.; Esposito, G. Next-Generation Probiotics for Inflammatory Bowel Disease. Int. J. Mol. Sci. 2022, 23, 5466. [Google Scholar] [CrossRef]
- Tripathi, M.K.; Giri, S.K. Probiotic functional foods: Survival of probiotics during processing and storage. J. Funct. Foods 2014, 9, 225–241. [Google Scholar] [CrossRef]
- Abid, R.; Waseem, H.; Ali, J.; Ghazanfar, S.; Muhammad Ali, G.; Elasbali, A.M.; Alharethi, S.H. Probiotic Yeast Saccharomyces: Back to Nature to Improve Human Health. J. Fungi. 2022, 8, 444. [Google Scholar] [CrossRef]
- Pais, P.; Almeida, V.; Yılmaz, M.; Teixeira, M.C. Saccharomyces boulardii: What makes it tick as successful probiotic? J. Fungi. 2020, 6, 78. [Google Scholar] [CrossRef]
- Shruthi, B.; Deepa, N.; Somashekaraiah, R.; Adithi, G.; Divyashree, S.; Sreenivasa, M.Y. Exploring biotechnological and functional characteristics of probiotic yeasts: A review. Biotechnol. Rep. 2022, 34, e00716. [Google Scholar] [CrossRef]
- Karim, A.; Gerliani, N.; Aïder, M. Kluyveromyces marxianus: An emerging yeast cell factory for applications in food and biotechnology. Int. J. Food Microbiol. 2020, 333, 108818. [Google Scholar] [CrossRef]
- Lane, M.M.; Morrissey, J.P. Kluyveromyces marxianus: A yeast emerging from its sister’s shadow. Fungal Biol. Rev. 2010, 24, 17–26. [Google Scholar] [CrossRef]
- Diosma, G.; Romanin, D.E.; Rey-Burusco, M.F.; Londero, A.; Garrote, G.L. Yeasts from kefir grains: Isolation, identification, and probiotic characterization. World J. Microbiol. Biotechnol. 2014, 30, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Gethins, L.; Rea, M.C.; Stanton, C.; Ross, R.P.; Kilcawley, K.; O’Sullivan, M.; Crotty, S.; Morrissey, J.P. Acquisition of the yeast Kluyveromyces marxianus from unpasteurised milk by a kefir grain enhances kefir quality. FEMS Microbiol. Lett. 2016, 363, fnw165. [Google Scholar] [CrossRef] [PubMed]
- Tofalo, R.; Fasoli, G.; Schirone, M.; Perpetuini, G.; Pepe, A.; Corsetti, A.; Suzzi, G. The predominance, biodiversity and biotechnological properties of Kluyveromyces marxianus in the production of Pecorino di Farindola cheese. Int. J. Food Microbial. 2014, 187, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Panel, E.B.; Herman, L. Scientific Opinion on the update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA (2017–2019). EFSA J. 2020, 18, e05966. [Google Scholar]
- Mari, E.; Guerrini, S.; Granchi, L.; Vincenzini, M. Enumeration and rapid identification of yeasts during extraction processes of extra virgin olive oil in Tuscany. World J. Microbiol. Biotechnol. 2016, 32, 1–10. [Google Scholar] [CrossRef]
- Huey, B.I.N.G.; Hall, J.E.F.F. Hypervariable DNA fingerprinting in Escherichia coli: Minisatellite probe from bacteriophage M13. J. Bacteriol. 1989, 171, 2528–2532. [Google Scholar] [CrossRef]
- Reguant, C.; Bordons, A. Typification of Oenococcus oeni strains by multiplex RAPD-PCR and study of population dynamics during malolactic fermentation. J. Appl. Microbiol. 2003, 95, 344–353. [Google Scholar] [CrossRef]
- Granchi, L.; Bosco, M.; Messini, A.; Vincenzini, M. Rapid detection and quantification of yeast species during spontaneous wine fermentation by PCR–RFLP analysis of the rDNA ITS region. J. Appl. Microbiol. 1999, 87, 949–956. [Google Scholar] [CrossRef]
- Galli, V.; Venturi, M.; Mari, E.; Guerrini, S.; Granchi, L. Gamma-aminobutyric acid (GABA) production in fermented milk by lactic acid bacteria isolated from spontaneous raw milk fermentation. Int. Dairy J. 2022, 127, 105284. [Google Scholar] [CrossRef]
- Fernández, M.F.; Boris, S.; Barbes, C. Probiotic properties of human lactobacilli strains to be used in the gastrointestinal tract. J. Appl. Microbiol. 2003, 94, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, K.; Touchette, M.; St-Gelais, D.; Labrie, S. Characterization of the fungal microflora in raw milk and specialty cheeses of the province of Quebec. Dairy Sci. Technol. 2012, 92, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Brejová, B.; Lichancová, H.; Brázdovič, F.; Hegedűsová, E.; Forgáčová Jakúbková, M.; Hodorová, V.; Džugasová, V.; Baláž, A.; Zeiselová, L.; Cillingová, A.; et al. Genome sequence of the opportunistic human pathogen Magnusiomyces capitatus. Curr. Genet. 2019, 65, 539–560. [Google Scholar] [CrossRef]
- Delavenne, E.; Mounier, J.; Asmani, K.; Jany, J.L.; Barbier, G.; Le Blay, G. Fungal diversity in cow, goat and ewe milk. Int. J. Food Microbiol. 2011, 151, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Merchán, A.V.; Benito, M.J.; Galván, A.I.; de Herrera, S.R.M.S. Identification and selection of yeast with functional properties for future application in soft paste cheese. LW-Food Sci. Technol. 2020, 124, 109173. [Google Scholar] [CrossRef]
- Cardoso, V.M.; Borelli, B.M.; Lara, C.A.; Soares, M.A.; Pataro, C.; Bodevan, E.C.; Rosa, C.A. The influence of seasons and ripening time on yeast communities of a traditional Brazilian cheese. Food Res. Int. 2015, 69, 331–340. [Google Scholar] [CrossRef]
- Coloretti, F.; Chiavari, C.; Luise, D.; Tofalo, R.; Fasoli, G.; Suzzi, G.; Grazia, L. Detection and identification of yeasts in natural whey starter for Parmigiano Reggiano cheese-making. Int. Dairy J. 2017, 66, 13–17. [Google Scholar] [CrossRef]
- EFSA Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Herman, L. Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 15: Suitability of taxonomic units notified to EFSA until September 2021. EFSA J. 2022, 20, e07045. [Google Scholar]
- Gil-Rodríguez, A.M.; Carrascosa, A.V.; Requena, T. Yeasts in foods and beverages: In vitro characterisation of probiotic traits. LWT-Food Sci. Technol. 2015, 64, 1156–1162. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, H.; Du, H.; Wang, F.; Li, H.; Zhao, X. Identification and characterization of Diutina rugosa SD-17 for potential use as a probiotic. LWT-Food Sci. Technol. 2019, 109, 283–288. [Google Scholar] [CrossRef]
- Helmy, E.A.; Soliman, S.A.; Abdel-Ghany, T.M.; Ganash, M. Evaluation of potentially probiotic attributes of certain dairy yeast isolated from buffalo sweetened Karish cheese. Heliyon 2019, 5, e01649. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xie, Y.H.; Xiong, L.X.; Dong, R.T.; Pan, C.L.; Teng, G.X.; Zhang, H.X. Effect and mechanism of cholesterol-lowering by Kluyveromyces from tibetan kefir. Adv. Mat. Res. 2012, 343, 1290–1298. [Google Scholar] [CrossRef]
- Lucena, R.M.; Dolz-Edo, L.; Brul, S.; de Morais, M.A., Jr.; Smits, G. Extreme low cytosolic pH is a signal for cell survival in acid stressed yeast. Genes 2020, 11, 656. [Google Scholar] [CrossRef] [PubMed]
- Maccaferri, S.; Klinder, A.; Brigidi, P.; Cavina, P.; Costabile, A. Potential probiotic Kluyveromyces marxianus B0399 modulates the immune response in Caco-2 cells and peripheral blood mononuclear cells and impacts the human gut microbiota in an in vitro colonic model system. Appl. Environ. Microbiol. 2012, 78, 956–964. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.J.; Kim, D.H.; Jeong, D.; Seo, K.H.; Jeong, H.S.; Lee, H.G.; Kim, H. Characterization of yeasts isolated from kefir as a probiotic and its synergic interaction with the wine byproduct grape seed flour/extract. LWT-Food Sci. Technol. 2018, 90, 535–539. [Google Scholar] [CrossRef]
- Xie, Y.; Zhang, H.; Liu, H.; Xiong, L.; Gao, X.; Jia, H.; Lian, Z.; Tong, N.; Han, T. Hypocholesterolemic effects of Kluyveromyces marxianus M3 isolated from Tibetan mushrooms on diet-induced hypercholesterolemia in rat. Braz. J. Microbiol. 2015, 46, 389–395. [Google Scholar] [CrossRef]
- Kondrotiene, K.; Lauciene, L.; Andruleviciute, V.; Kasetiene, N.; Serniene, L.; Sekmokiene, D.; Malakauskas, M. Safety assessment and preliminary in vitro evaluation of probiotic potential of Lactococcus lactis strains naturally present in raw and fermented milk. Curr. Microbiol. 2020, 77, 3013–3023. [Google Scholar] [CrossRef]
- Hyacinta, M.; Hana, K.S.; Andrea, B.; Barbora, C. Bile tolerance and its effect on antibiotic susceptibility of probiotic Lactobacillus candidates. Folia Microbiol. 2015, 60, 253–257. [Google Scholar] [CrossRef]
- Hernández-Gómez, J.G.; López-Bonilla, A.; Trejo-Tapia, G.; Ávila-Reyes, S.V.; Jiménez-Aparicio, A.R.; Hernández-Sánchez, H. In vitro bile salt hydrolase (BSH) Activity screening of different probiotic microorganisms. Foods 2021, 10, 674. [Google Scholar] [CrossRef]
- Faye, T.; Tamburello, A.; Vegarud, G.E.; Skeie, S. Survival of lactic acid bacteria from fermented milks in an in vitro digestion model exploiting sequential incubation in human gastric and duodenum juice. J. Dairy Sci. 2012, 95, 558–566. [Google Scholar] [CrossRef]
- Garofalo, C.; Ferrocino, I.; Reale, A.; Sabbatini, R.; Milanović, V.; Alkić-Subašić, M.; Boscaino, F.; Aquilanti, L.; Pasquini, M.; Trombetta, M.F.; et al. Study of kefir drinks produced by backslopping method using kefir grains from Bosnia and Herzegovina: Microbial dynamics and volatilome profile. Food Res. Int. 2020, 137, 109369. [Google Scholar] [CrossRef] [PubMed]
- Baschali, A.; Tsakalidou, E.; Kyriacou, A.; Karavasiloglou, N.; Matalas, A.L. Traditional low-alcoholic and non-alcoholic fermented beverages consumed in European countries: A neglected food group. Nutr. Res. Rev. 2017, 30, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Danova, S.; Petrov, K.; Pavlov, P.; Petrova, P. Isolation and characterization of Lactobacillus strains involved in koumiss fermentation. Int. J. Dairy Technol. 2005, 58, 100–105. [Google Scholar] [CrossRef]
- Gul, O.; Mortas, M.; Atalar, I.; Dervisoglu, M.; Kahyaoglu, T. Manufacture and characterization of kefir made from cow and buffalo milk, using kefir grain and starter culture. J. Dairy Sci. 2015, 98, 1517–1525. [Google Scholar] [CrossRef]
- Savastano, M.L.; Pati, S.; Bevilacqua, A.; Corbo, M.R.; Rizzuti, A.; Pischetsrieder, M.; Losito, I. Influence of the production technology on kefir characteristics: Evaluation of microbiological aspects and profiling of phosphopeptides by LC-ESI-QTOF-MS/MS. Food Res. Int. 2020, 129, 108853. [Google Scholar] [CrossRef]
- Chaves-López, C.; Serio, A.; Grande-Tovar, C.D.; Cuervo-Mulet, R.; Delgado-Ospina, J.; Paparella, A. Traditional fermented foods and beverages from a microbiological and nutritional perspective: The Colombian heritage. Compr. Rev. Food Sci. 2014, 13, 1031–1048. [Google Scholar] [CrossRef]
- Beshkova, D.M.; Simova, E.D.; Simov, Z.I.; Frengova, G.I.; Spasov, Z.N. Pure cultures for making kefir. Food Microbiol. 2002, 19, 537–544. [Google Scholar] [CrossRef]
- Kök-Taş, T.; Seydim, A.C.; Özer, B.; Guzel-Seydim, Z.B. Effects of different fermentation parameters on quality characteristics of kefir. J. Dairy Sci. 2013, 96, 780–789. [Google Scholar] [CrossRef]
- Leite, A.M.O.; Leite, D.C.A.; Del Aguila, E.M.; Alvares, T.S.; Peixoto, R.S.; Miguel, M.A.L.; Silva, J.T.; Paschoalin, V.M.F. Microbiological and chemical characteristics of Brazilian kefir during fermentation and storage processes. J. Dairy Sci. 2013, 96, 4149–4159. [Google Scholar] [CrossRef]
- Ryan, J.; Hutchings, S.C.; Fang, Z.; Bandara, N.; Gamlath, S.; Ajlouni, S.; Ranadheera, C.S. Microbial, physico-chemical and sensory characteristics of mango juice-enriched probiotic dairy drinks. Int. J. Dairy Technol. 2020, 73, 182–190. [Google Scholar] [CrossRef]
- Ziarno, M.; Zaręba, D. Effects of milk components and food additives on survival of three bifidobacteria strains in fermented milk under simulated gastrointestinal tract conditions. Microb. Ecol. Health Dis. 2015, 26, 27812. [Google Scholar] [CrossRef] [PubMed]
- Ranadheera, C.S.; Evans, C.A.; Adams, M.C.; Baines, S.K. In vitro analysis of gastrointestinal tolerance and intestinal cell adhesion of probiotics in goat’s milk ice cream and yogurt. Food Res. Int. 2012, 49, 619–625. [Google Scholar] [CrossRef]
- Toscano, M.; De Grandi, R.; Pastorelli, L.; Vecchi, M.; Drago, L. A consumer’s guide for probiotics: 10 golden rules for a correct use. Dig. Liver Dis. 2017, 49, 1177–1184. [Google Scholar] [CrossRef] [PubMed]
- Taverniti, V.; Koirala, R.; Dalla Via, A.; Gargari, G.; Leonardis, E.; Arioli, S.; Guglielmetti, S. Effect of cell concentration on the persistence in the human intestine of four probiotic strains administered through a multispecies formulation. Nutrients 2019, 11, 285. [Google Scholar] [CrossRef] [PubMed]




| FM-S | FM-B | |
|---|---|---|
| Lactic acid bacteria (CFU/mL) | (2.44 ± 0.69) × 109 a | (1.42 ± 0.44) × 109 a |
| Yeasts (CFU/mL) | (2.14 ± 0.55) × 107 a | (2.76 ± 0.34) × 107 a |
| Final pH | 4.21 ± 0.17 b | 4.59 ± 0.01 a |
| ΔpH | −2.22 ± 0.11 b | −1.63 ± 0.04 a |
| Lactose (g/L) | 31.75 ± 0.64 | 31.85 ± 0.64 |
| Lactic acid (g/L) | 7.00 ± 0.49 b | 5.00 ± 0.26 a |
| Acetic acid (g/L) | 0.58 ± 0.05 b | 0.38 ± 0.10 a |
| Ethanol (v/v %) | 0.61 ± 0.06 a | 0.80 ± 0.07 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galli, V.; Venturi, M.; Mari, E.; Guerrini, S.; Granchi, L. Selection of Yeast and Lactic Acid Bacteria Strains, Isolated from Spontaneous Raw Milk Fermentation, for the Production of a Potential Probiotic Fermented Milk. Fermentation 2022, 8, 407. https://doi.org/10.3390/fermentation8080407
Galli V, Venturi M, Mari E, Guerrini S, Granchi L. Selection of Yeast and Lactic Acid Bacteria Strains, Isolated from Spontaneous Raw Milk Fermentation, for the Production of a Potential Probiotic Fermented Milk. Fermentation. 2022; 8(8):407. https://doi.org/10.3390/fermentation8080407
Chicago/Turabian StyleGalli, Viola, Manuel Venturi, Eleonora Mari, Simona Guerrini, and Lisa Granchi. 2022. "Selection of Yeast and Lactic Acid Bacteria Strains, Isolated from Spontaneous Raw Milk Fermentation, for the Production of a Potential Probiotic Fermented Milk" Fermentation 8, no. 8: 407. https://doi.org/10.3390/fermentation8080407
APA StyleGalli, V., Venturi, M., Mari, E., Guerrini, S., & Granchi, L. (2022). Selection of Yeast and Lactic Acid Bacteria Strains, Isolated from Spontaneous Raw Milk Fermentation, for the Production of a Potential Probiotic Fermented Milk. Fermentation, 8(8), 407. https://doi.org/10.3390/fermentation8080407







