Rational Metabolic Engineering Combined with Biosensor-Mediated Adaptive Laboratory Evolution for l-Cysteine Overproduction from Glycerol in Escherichia coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Plasmids
2.2. Growth Medium and Culture Conditions
2.3. Gene Deletion with CRISPR/Cas9
2.4. Gene Overexpression
2.5. Construction and Verification of the Serine-Biosensor
2.6. Biosensor-Driven Evolution Experiment
2.7. Genome Sequencing
2.8. Analytical Methods
3. Results
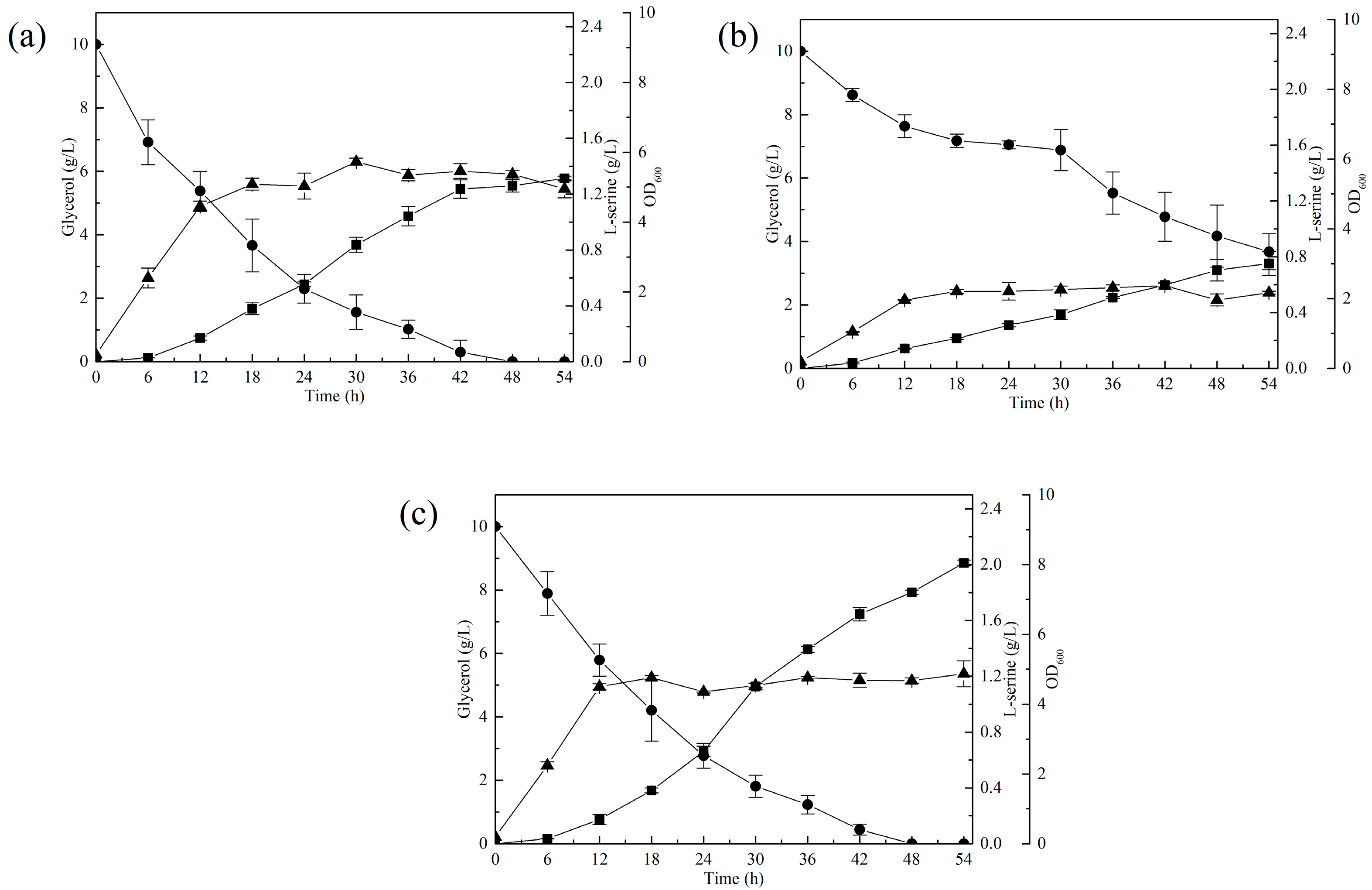
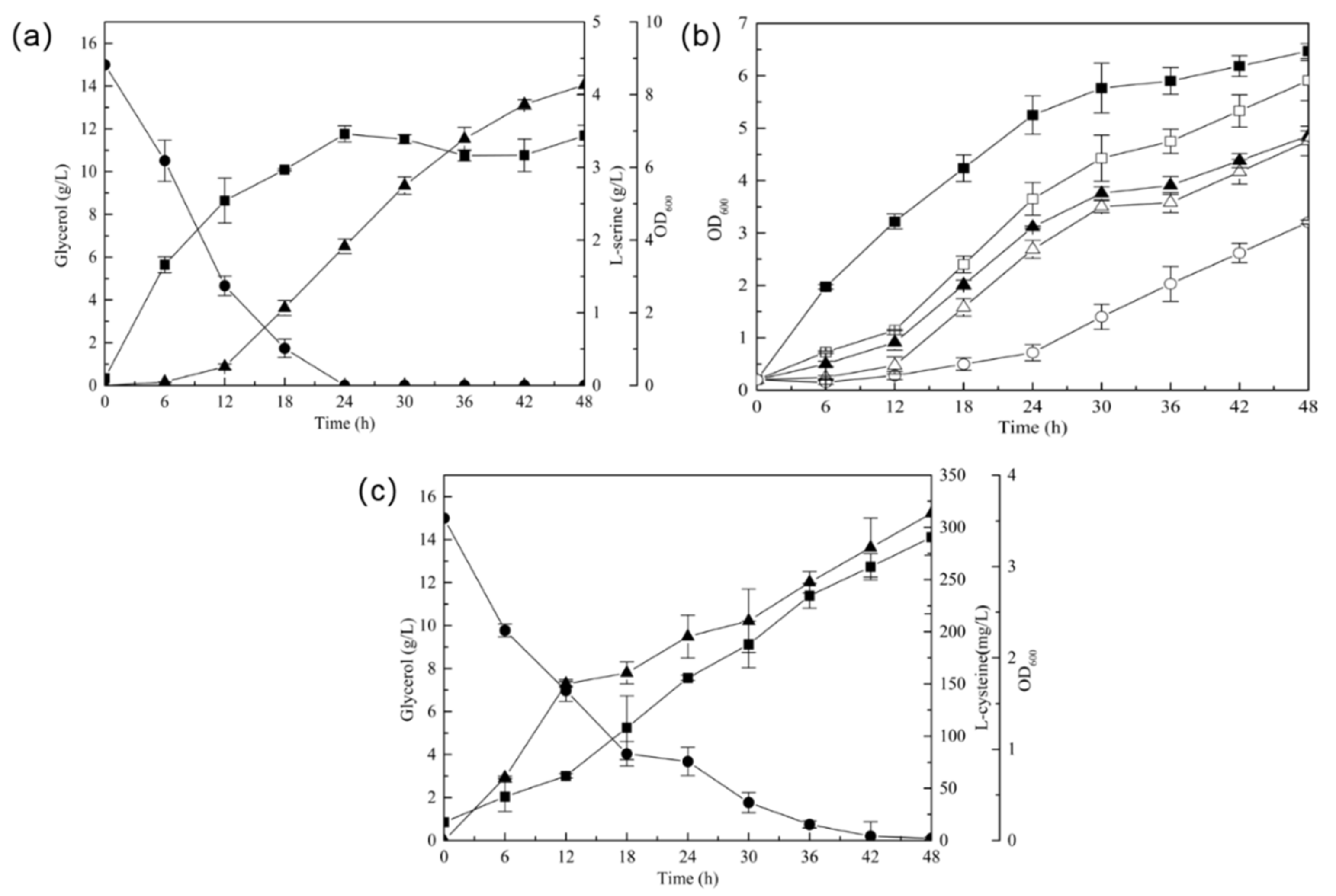
3.1. Improved the Precursor l-Serine Accumulation by Decreasing l-Serine Degradation in E. coli
3.2. Increased l-Serine Production through ALE Combined with a Serine-Biosensor
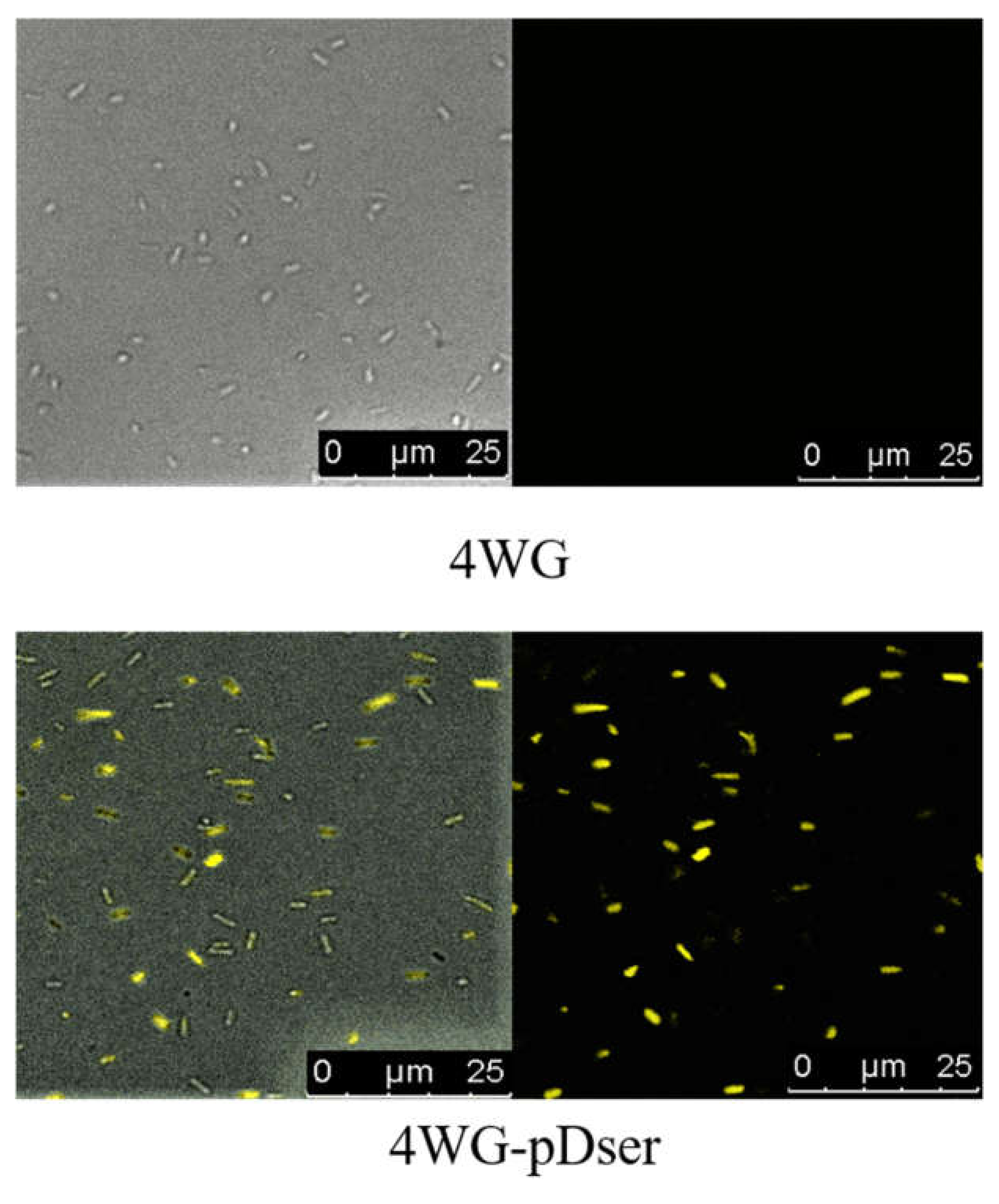
3.2.1. Construction and Verification of a Serine-Biosensor in E. coli
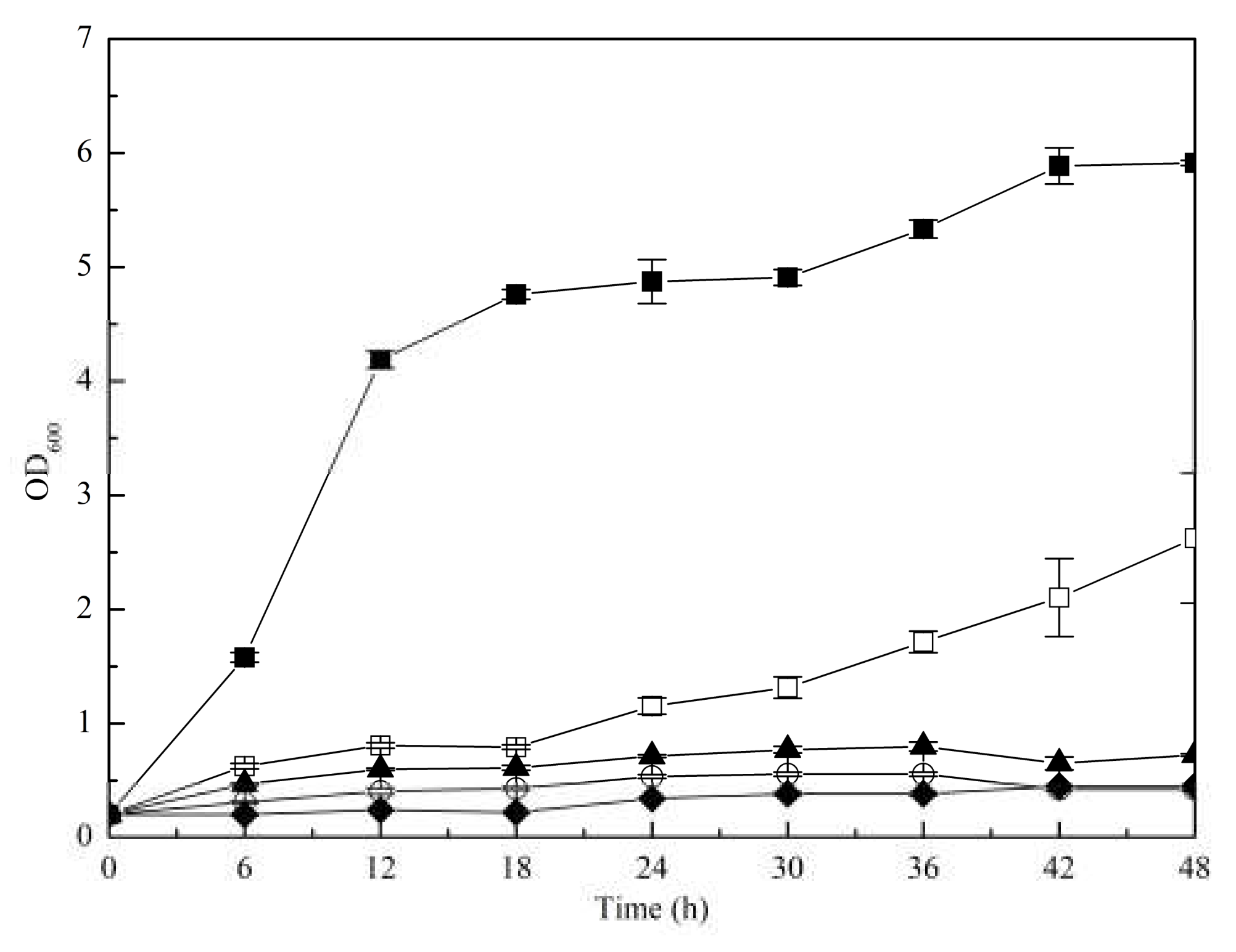
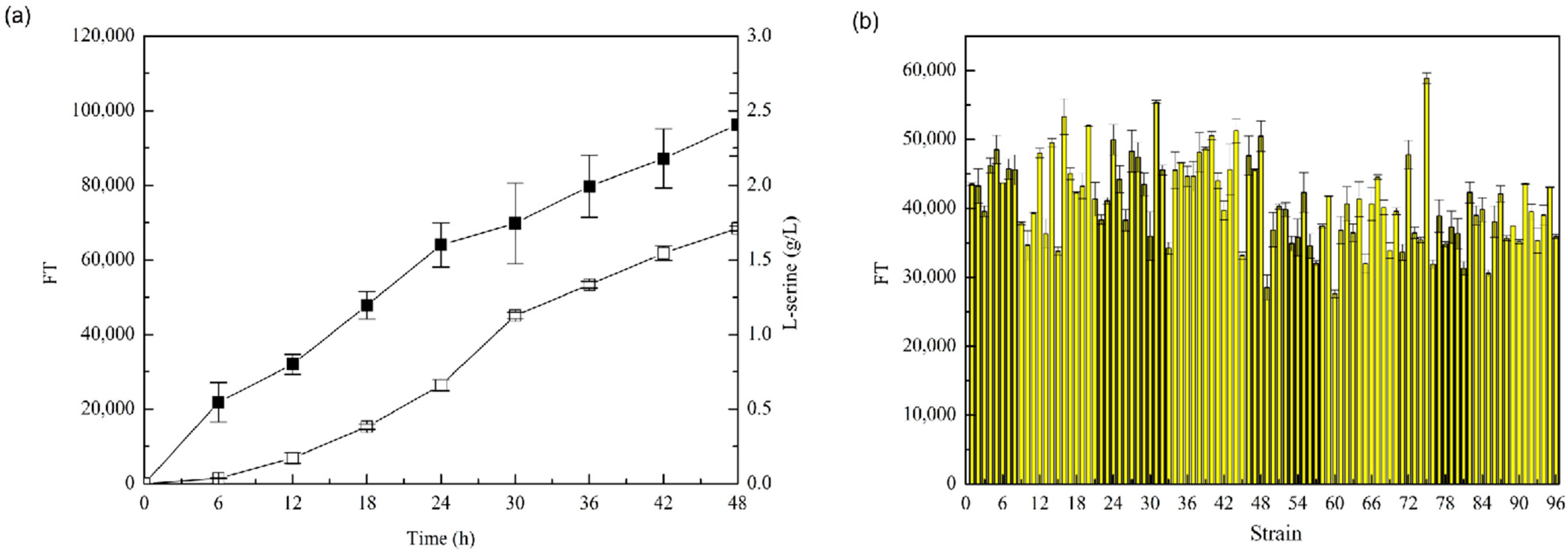
3.2.2. Increased l-Serine Yield Achieved by Biosensor-Driven Evolution
3.3. Construction of l-Cysteine-Producing Recombinant Strain
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Takagi, H.; Ohtsu, I. l-cysteine metabolism and fermentation in microorganisms. Adv. Biochem. 2017, 159, 129–151. [Google Scholar]
- Wendisch, V.F. Metabolic engineering advances and prospects for amino acid production. Metab. Eng. 2020, 58, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, Y.; Hou, Y.; Li, Z. Fitness of chassis cells and metabolic pathways for l-cysteine overproduction in Escherichia coli. J. Agric. Food. Chem. 2020, 68, 14928–14937. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Ren, W.; Yang, G.; Duan, J.; Huang, X.; Fang, R.; Li, C.; Li, T.; Yin, Y.; Hou, Y.; et al. l-Cysteine metabolism and its nutritional implications. Mol. Nutr. Food. Res. 2016, 60, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Kishino, K.; Kondoh, M.; Takagi, H. Enhanced l-cysteine production by overexpressing potential l-cysteine exporter genes in an l-cysteine-producing recombinant strain of Corynebacterium glutamicum. Biosci. Biotechnol. Biochem. 2019, 83, 2390–2393. [Google Scholar] [CrossRef]
- Kawano, Y.; Ohtsu, I.; Takumi, K.; Tamakoshi, A.; Nonaka, G.; Funahashi, E.; Ihara, M.; Takagi, H. Enhancement of l-cysteine production by disruption of yciW in Escherichia coli. J. Biosci. Bioeng. 2015, 119, 176–179. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Wang, H.; Xu, N.; Zhou, W.; Ju, J.; Liu, J.; Ma, Y. Metabolic engineering of Corynebacterium glutamicum for l-cysteine production. Appl. Microbiol. Biotechnol. 2019, 103, 1325–1338. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Fang, G.; Wu, H.; Li, Z.; Ye, Q. l-cysteine production in Escherichia coli based on rational metabolic engineering and modular strategy. Biotechnol. J. 2018, 13, 1–6. [Google Scholar] [CrossRef]
- Kawano, Y.; Onishi, F. Improved fermentative l-cysteine overproduction by enhancing a newly identified thiosulfate assimilation pathway in Escherichia coli. Appl. Microbiol. Biotechnol. 2017, 101, 6879–6889. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, K.; Albermann, C.; Sprenger, G.A. Improvement of l-phenylalanine production from glycerol by recombinant Escherichia coli strains: The role of extra copies of glpK, glpX, and tktA genes. Microb. Cell. Fact. 2014, 13, 16. [Google Scholar] [CrossRef]
- Nguyen-Vo, T.P.; Liang, Y.; Sankaranarayanan, M.; Seol, E.; Chun, A.Y.; Ashok, S.; Chauhan, A.S.; Kim, J.R.; Park, S. Development of 3-hydroxypropionic-acid-tolerant strain of Escherichia coli W and role of minor global regulator yieP. Metab. Eng. 2019, 53, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Sprenger, G.A. Engineering of microorganisms for the production of chemicals and fuels from renewable resources. Springer Nat. Verl. 2017, 4, 93–123. [Google Scholar]
- Luo, X.; Ge, X.; Cui, S.; Li, Y. Value-added processing of crude glycerol into chemicals and polymers. Bioresour. Technol. 2016, 215, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Trondle, J.; Trachtmann, N.; Sprenger, G.A.; Weuster-Botz, D. Fed-batch production of l-tryptophan from glycerol using recombinant Escherichia coli. Biotechnol. Bioeng. 2018, 115, 2881–2892. [Google Scholar] [CrossRef]
- Yang, F.X.; Hanna, M.A.; Sun, R.C. Value-added uses for crude glycerol-a byproduct of biodiesel production. Biotechnol. Biofuels. 2012, 5, 10. [Google Scholar] [CrossRef]
- Li, Q.; Wu, H.; Li, Z.; Ye, Q. Enhanced succinate production from glycerol by engineered Escherichia coli strains. Bioresour. Technol. 2016, 218, 217–223. [Google Scholar] [CrossRef]
- Litsanov, B.; Brocker, M.; Bott, M. Glycerol as a substrate for aerobic succinate production in minimal medium with Corynebacterium glutamicum. Microb. Biotechnol. 2013, 6, 189–195. [Google Scholar] [CrossRef]
- Mazumdar, S.; Blankschien, M.D.; Clomburg, J.M.; Gonzalez, R. Efficient synthesis of l-lactic acid from glycerol by metabolically engineered Escherichia coli. Microb. Cell. Fact. 2013, 12, 11. [Google Scholar] [CrossRef]
- Weiner, M.; Albermann, C.; Gottlieb, K.; Sprenger, G.A.; Weuster-Botz, D. Fed-batch production of l-phenylalanine from glycerol and ammonia with recombinant Escherichia coli. Biochem. Eng. J. 2014, 83, 62–69. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Zhu, J.F.; Liu, W.; Xu, G.Q.; Zhang, X.M.; Shi, J.S.; Xu, Z.H. High-yield production of l-serine from glycerol by engineered Escherichia coli. J. Ind. Microbiol. Biotechnol. 2019, 46, 883–885. [Google Scholar] [CrossRef]
- Rennig, M.; Mundhada, H.; Wordofa, G.G.; Gerngross, D.; Wulff, T.; Worberg, A.; Nielsen, A.T.; Nørholm, M.H.H. Industrializing a bacterial strain for l-serine production through translation initiation optimization. ACS. Synth. Biol. 2019, 8, 2347–2358. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.M.; Zhang, J.; Ji, X.M.; Fang, Z.; Wu, Z.M.; Chen, J.; Du, G.C. Evolutionary engineering of industrial microorganisms-strategies and applications. Appl. Microbiol. Biotechnol. 2018, 102, 4615–4627. [Google Scholar] [CrossRef] [PubMed]
- Leavitt, J.M.; Wagner, J.M.; Tu, C.C.; Tong, A.; Liu, Y.Y.; Alper, H.S. Biosensor-enabled directed evolution to improve muconic acid production in Saccharomyces cerevisiae. Biotechnol. J. 2017, 12, 9. [Google Scholar] [CrossRef]
- Sandberg, T.E.; Salazar, M.J.; Weng, L.L.; Palsson, B.O.; Feist, A.M. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab. Eng. 2019, 56, 1–16. [Google Scholar] [CrossRef]
- Della, C.D.; Van, B.H.L.; Syberg, F.; Schallmey, M.; Tobola, F.; Cormann, K.U.; Schlicker, C.; Baumann, P.T.; Krumbach, K.; Sokolowsky, S.; et al. Engineering and application of a biosensor with focused ligand specificity. Nat. Commun. 2020, 11, 4851. [Google Scholar] [CrossRef] [PubMed]
- Binder, S.; Schendzielorz, G.; Stabler, N.; Krumbach, K.; Hoffmann, K.; Bott, M.; Eggeling, L. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome. Biol. 2012, 13, 12. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, S.; Jiang, P.; Yao, J.; He, Y.; Chen, L.; Gui, X.; Dong, Z.; Tang, S.Y. Design of an ectoine-responsive AraC mutant and its application in metabolic engineering of ectoine biosynthesis. Metab. Eng. 2015, 30, 149–155. [Google Scholar] [CrossRef]
- Cress, B.F.; Trantas, E.A.; Ververidis, F.; Linhardt, R.J.; Koffas, M.A.G. Sensitive cells: Enabling tools for static and dynamic control of microbial metabolic pathways. Curr. Opin. Biotechnol. 2015, 36, 205–214. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Q.; Zheng, P.; Zhang, Z.; Liu, Y.; Sun, C.; Cao, G.; Zhou, W.; Wang, X.; Zhang, D.; et al. Developing a high-throughput screening method for threonine overproduction based on an artificial promoter. Microb. Cell. Fact. 2015, 14, 11. [Google Scholar] [CrossRef]
- Schulte, J.; Baumgart, M.; Bott, M. Development of a single-cell GlxR-based cAMP biosensor for Corynebacterium glutamicum. J. Biotechnol. 2017, 258, 33–40. [Google Scholar] [CrossRef]
- Wang, Q.Z.; Tang, S.Y.; Yang, S. Genetic biosensors for small-molecule products: Design and applications in high-throughput screening. Front. Chem. Sci. Eng. 2017, 11, 15–26. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, X.M.; Xu, G.Q.; Shi, J.S.; Xu, Z.H. Integration of ARTP mutagenesis with biosensor-mediated high-throughput screening to improve l-serine yield in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2018, 102, 5939–5951. [Google Scholar] [CrossRef]
- Martinez, A.; Grabar, T.B.; Shanmugam, K.T.; Yomano, L.P.; York, S.W.; Ingram, L.O. Low salt medium for lactate and ethanol production by recombinant Escherichia coli. B. Biotechnol. Lett. 2007, 29, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Feist, A.M.; Zielinski, D.C.; Orth, J.D.; Schellenberger, J.; Herrgard, M.J.; Palsson, B.O. Model-driven evaluation of the production potential for growth-coupled products of Escherichia coli. Metab. Eng. 2010, 12, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Chu, B.C.; Otten, R.; Krewulak, K.D.; Mulder, F.A.; Vogel, H.J. The solution structure, binding properties, and dynamics of the bacterial siderophore-binding protein FepB. J. Biol. Chem. 2014, 289, 29219–29234. [Google Scholar] [CrossRef]
- Storek, K.M.; Vij, R.; Sun, D.; Smith, P.A.; Koerber, J.T.; Rutherford, S.T. The Escherichia coli β-barrel assembly machinery is sensitized to perturbations under high membrane fluidity. J. Bacteriol. 2018, 201, e00517-18. [Google Scholar] [CrossRef] [PubMed]
- Doyle, M.T.; Bernstein, H.D. BamA forms a translocation channel for polypeptide export across the bacterial outer membrane. Mol. Cell. 2021, 81, 2000–2012. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.C.; Cottrill, M.A.; Forsberg, C.W.; Jia, Z. Functional insights revealed by the crystal structures of Escherichia coli glucose-1-phosphatase. J. Biol. Chem. 2003, 278, 31412–31418. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Corsi, I.D.; Bier, N.; Koehler, T.M. BrnQ-type branched-chain amino acid transporters Influence Bacillus anthracis growth and virulence. mBio 2022, 25, e03640-21. [Google Scholar] [CrossRef]
- Eggeling, L.; Bott, M. A giant market and a powerful metabolism: l-lysine provided by Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2015, 99, 3387–3394. [Google Scholar] [CrossRef]
- Johnson, A.O.; Gonzalez-Villanueva, M.; Wong, L.; Steinbuchel, A.; Tee, K.L.; Xu, P.; Wong, T.S. Design and application of genetically-encoded malonyl-CoA biosensors for metabolic engineering of microbial cell factories. Metab. Eng. 2017, 44, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Xu, P. Production of chemicals using dynamic control of metabolic fluxes. Curr. Opin. Biotechnol. 2018, 53, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Mahr, R.; Von Boeselager, R.F.; Wiechert, J.; Frunzke, J. Screening of an Escherichia coli promoter library for a phenylalanine biosensor. Appl. Microbiol. Biotechnol. 2016, 100, 6739–6753. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.Y.; Lee, Y.J.; Park, J.W.; Kim, S.N.; Kim, E.Y.; Kim, Y.; Kim, O.B. An Escherichia coli FdrA Variant Derived from Syntrophic Coculture with a Methanogen Increases Succinate Production Due to Changes in Allantoin Degradation. mSphere 2021, 6, e0065421. [Google Scholar] [CrossRef] [PubMed]
- Volpon, L.; Lievre, C.; Osborne, M.J.; Gandhi, S.; Iannuzzi, P.; Larocque, R.; Cygler, M.; Gehring, K.; Ekiel, I. The solution structure of YbcJ from Escherichia coli reveals a recently discovered alphal motif involved in RNA binding. J. Bacteriol. 2003, 185, 4204–4210. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tagliabue, L.; Antoniani, D.; Maciąg, A.; Bocci, P.; Raffaelli, N.; Landini, P. The diguanylate cyclase YddV controls production of the exopolysaccharide poly-N-acetylglucosamine (PNAG) through regulation of the PNAG biosynthetic pgaABCD operon. Microbiology 2010, 156, 2901–2911. [Google Scholar] [CrossRef]
- Masulis, I.S.; Sukharycheva, N.A.; Kiselev, S.S.; Andreeva, Z.S.; Ozoline, O.N. Between computational predictions and high-throughput transcriptional profiling: In depth expression analysis of the OppB trans-membrane subunit of Escherichia coli OppABCDF oligopeptide transporter. Res. Microbiol. 2020, 171, 55–63. [Google Scholar] [CrossRef]
- Minamino, T.; Inoue, Y.; Kinoshita, M.; Namba, K. FliK-Driven Conformational Rearrangements of FlhA and FlhB Are Required for Export Switching of the Flagellar Protein Export Apparatus. J. Bacteriol. 2020, 202, e00637-19. [Google Scholar] [CrossRef]
- Lolkema, J.S. Domain structure and pore loops in the 2-hydroxycarboxylate transporter family. J. Mol. Microbiol. Biotechnol. 2006, 11, 318–325. [Google Scholar] [CrossRef]
- Poyner, R.R.; Larsen, T.M.; Wong, S.W.; Reed, G.H. Functional and structural changes due to a serine to alanine mutation in the active-site flap of enolase. Arch. Biochem. Biophys. 2002, 401, 155–163. [Google Scholar] [CrossRef]
 ) indicated genes that were deleted. cysE encoded serine acetyltransferase, the red line indicated gene that were overexpressed. The starting strain E. coli 4W had been constructed in our previous study with deletion of sdaA, sdaB, and tdcG (The three genes encoded l-serine deaminases), and the removal of feedback inhibition of serA (serA encoded 3-phosphoglycerate dehydrogenase).
) indicated genes that were deleted. cysE encoded serine acetyltransferase, the red line indicated gene that were overexpressed. The starting strain E. coli 4W had been constructed in our previous study with deletion of sdaA, sdaB, and tdcG (The three genes encoded l-serine deaminases), and the removal of feedback inhibition of serA (serA encoded 3-phosphoglycerate dehydrogenase).
 ) indicated genes that were deleted. cysE encoded serine acetyltransferase, the red line indicated gene that were overexpressed. The starting strain E. coli 4W had been constructed in our previous study with deletion of sdaA, sdaB, and tdcG (The three genes encoded l-serine deaminases), and the removal of feedback inhibition of serA (serA encoded 3-phosphoglycerate dehydrogenase).
) indicated genes that were deleted. cysE encoded serine acetyltransferase, the red line indicated gene that were overexpressed. The starting strain E. coli 4W had been constructed in our previous study with deletion of sdaA, sdaB, and tdcG (The three genes encoded l-serine deaminases), and the removal of feedback inhibition of serA (serA encoded 3-phosphoglycerate dehydrogenase).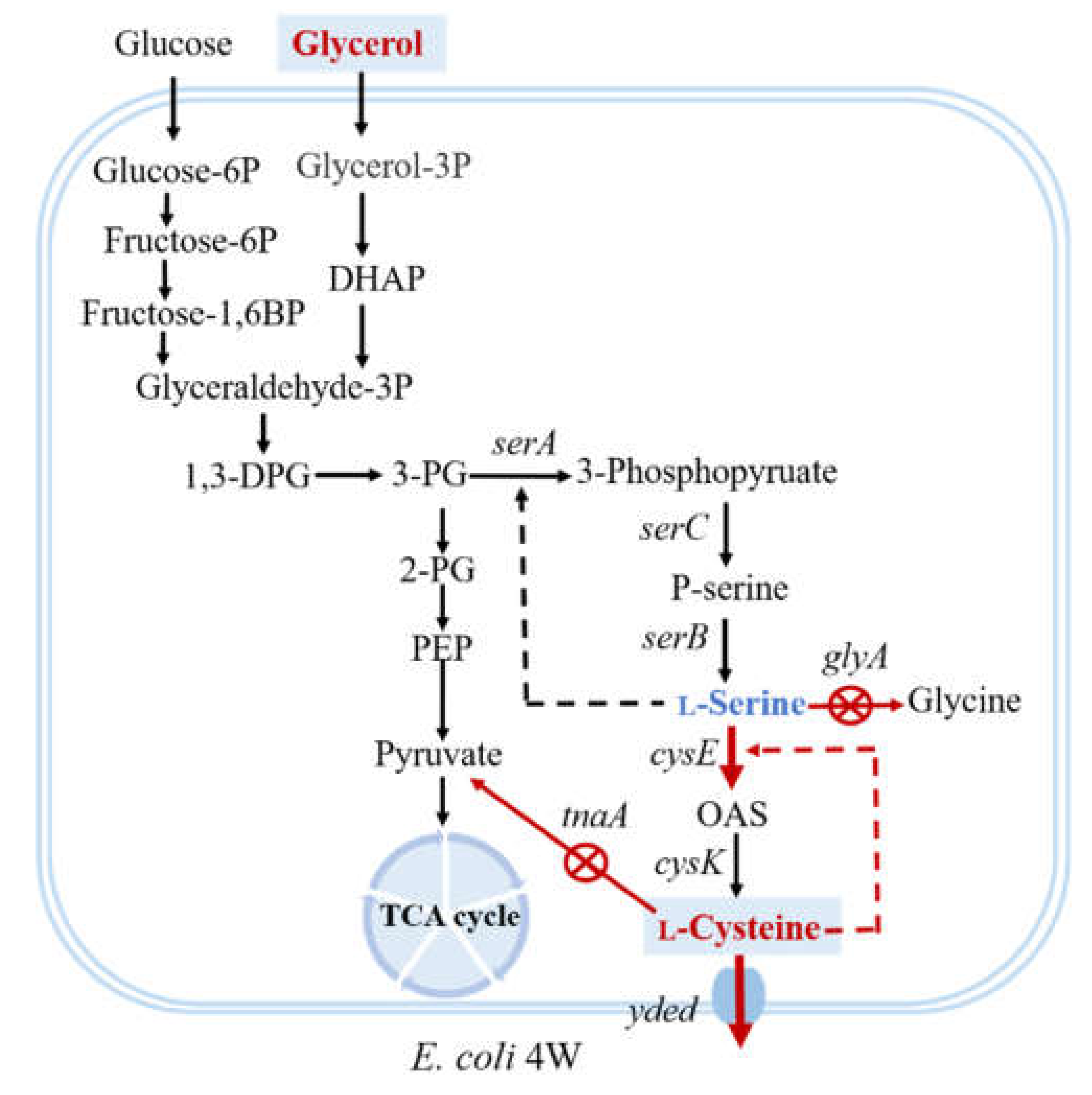
| Strains or Plasmids | Description | Sources |
| Strains E. coli JM109 | recA1, endA1, gyrA96, thi-1, hsd R17(rk- mk+) supE44 | Invitrogen |
| 4W | W3110△tdcG△sdaA△sdaB serAdr | Invitrogen |
| 4WG | 4W with glyA deletion | This study |
| 4W-pDer | 4W harboring serine-biosensor pDser | This study |
| 4WGX | A mutant derived from 4W | This study |
| 4WG-pDer | 4WG harboring serine- biosensor pDser | This study |
| 4WG-cysE | 4WG harboring pEtac-cysE | This study |
| 4WG-cysE-ydeD | 4WG harboring pEtac-cysE-ydeD | This study |
| 4WG-∆tnaA | 4WG with tnaA deletion | This study |
| 4WG-∆tnaA-cysE | 4WG-∆tnaA harboring pEtac-cysE | This study |
| 4WG-∆tnaA-cysE-ydeD | 4WG-∆tnaA harboring pEtac-cyE-ydeD | This study |
| Plasmids | ||
| pCas | Carrying Cas9 and λRed System, kan | Invitrogen |
| pTargetF | Carrying N20 sequence, spc or smr | Invitrogen |
| pDser | Biosensor, kan | Invitrogen |
| pEtac | Inducible expression plasmid, tac, kan | This study |
| pEtac-cysE | Carrying cysE gene from E. coli | This study |
| pEtac-cysE-ydeD | Carrying cysE and ydeD gene from E. coli | This study |
| Primers | Sequence |
| pTargetF-△glyA1-F | ACTGTGGCAGGCTATGGAGCGTTTTAGAGCTAGAAATAGCAAGTT |
| pTargetF-△glyA1-R | GCTCCATAGCCTGCCACAGTACTAGTATTATACCTAGGACTGAGC |
| pTargetF-△glyA2-F | AGAAGCCGAAGCGAAAGAACGTTTTAGAGCTAGAAA-TAGCAAGTT |
| pTargetF-△glyA2-R | GTTCTTTCGCTTCGGCTTCTACTAGTATTATACCTAGGACTGAGC |
| glyA-U-F | AGCCCTGCAATGTAAATGGTT |
| glyA-U-R | ACAGCAAATCACCGTTTCGCCCGCATCTCCTGACTCAGCTA |
| glyA-D-F | AGCTGAGTCAGGAGATGCGGGCGAAACGGTGATTTGCTGTC |
| glyA-D-R | TCGCCAGACAGGATTTAACCC |
| pTargetF:1756F23 | CCCTGATTCTGTGGATAACCGTA |
| pTargetF:78R23 | ACATCAGTCGATCATAGCACGAT |
| cysE-F | TTCACACAGGAAACAGAATTCATGTCGTGTGAAGAACTGGAAATTG |
| cysE-R | TGCGGCCGCAAGCTTGTCGACTTAGATCCCATCCCCATACTCAA |
| ydeD-F | GGGATCTAAGTCGACAAGCTTCGCTGAGCAATAACTAGCATAACC |
| ydeD-R | GTGGTGGTGGTGGTGCTCGAGTTAACTTCCCACCTTTACCGCT |
| tnaA-U-F | TTGCATATATATCTGGCGAATTAATCGG |
| tnaA-U-R | GCCACTCTGTAGTATTAAGTATCAAAGAAATAGTTAGAGAACGCCA |
| tnaA-D-F | ACTTAATACTACAGAGTGGCTATAAGGATGTT |
| tnaA-D-R | ACGAAAATGGCTGTGCAGAT |
| pTargetF-∆tnaA-F | CGTTCTCTTTCACATGTTTAACTAGTATTATACCTAGGACTG |
| pTargetF-∆tnaA-R | TAAACATGTGAAAGAGAACGTTTTAGAGCTAGAAATAGCAA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Sun, Z.; Bian, J.; Gao, Y.; Zhang, D.; Xu, G.; Zhang, X.; Li, H.; Shi, J.; Xu, Z. Rational Metabolic Engineering Combined with Biosensor-Mediated Adaptive Laboratory Evolution for l-Cysteine Overproduction from Glycerol in Escherichia coli. Fermentation 2022, 8, 299. https://doi.org/10.3390/fermentation8070299
Zhang X, Sun Z, Bian J, Gao Y, Zhang D, Xu G, Zhang X, Li H, Shi J, Xu Z. Rational Metabolic Engineering Combined with Biosensor-Mediated Adaptive Laboratory Evolution for l-Cysteine Overproduction from Glycerol in Escherichia coli. Fermentation. 2022; 8(7):299. https://doi.org/10.3390/fermentation8070299
Chicago/Turabian StyleZhang, Xiaomei, Zhenhang Sun, Jinyu Bian, Yujie Gao, Dong Zhang, Guoqiang Xu, Xiaojuan Zhang, Hui Li, Jinsong Shi, and Zhenghong Xu. 2022. "Rational Metabolic Engineering Combined with Biosensor-Mediated Adaptive Laboratory Evolution for l-Cysteine Overproduction from Glycerol in Escherichia coli" Fermentation 8, no. 7: 299. https://doi.org/10.3390/fermentation8070299
APA StyleZhang, X., Sun, Z., Bian, J., Gao, Y., Zhang, D., Xu, G., Zhang, X., Li, H., Shi, J., & Xu, Z. (2022). Rational Metabolic Engineering Combined with Biosensor-Mediated Adaptive Laboratory Evolution for l-Cysteine Overproduction from Glycerol in Escherichia coli. Fermentation, 8(7), 299. https://doi.org/10.3390/fermentation8070299






