Improved Hydrogen Peroxide Stress Resistance of Zymomonas mobilis NADH Dehydrogenase (ndh) and Alcohol Dehydrogenase (adhB) Mutants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Cultivation
2.2. Viability Tests for Hydrogen Peroxide Resistance
2.3. Preparation of Cell-Free Extracts, Cytoplasmic Membranes, and Permeabilized Cells
2.4. Enzymatic Assays
2.5. RT-qPCR
2.6. Analytical Procedures
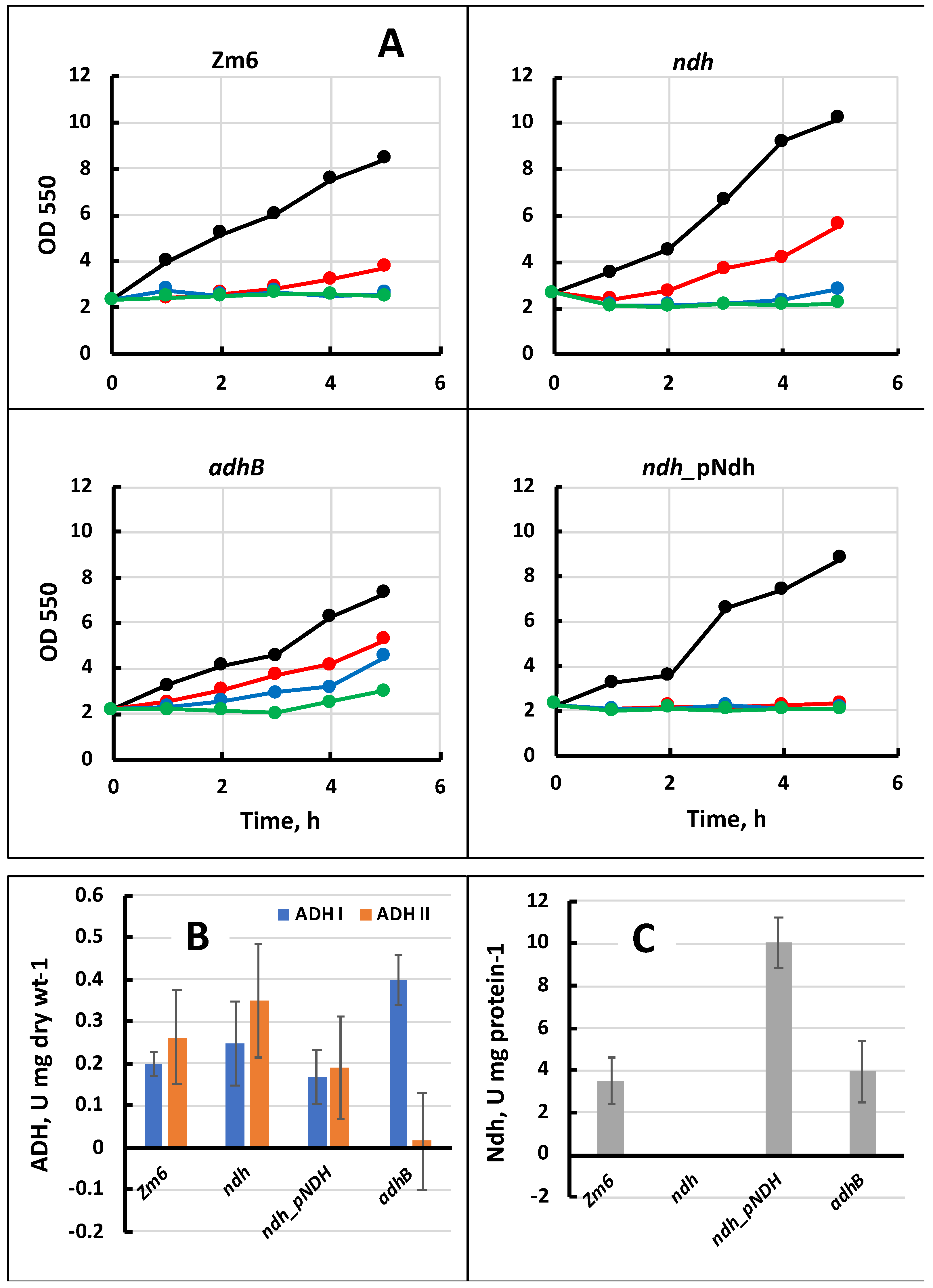
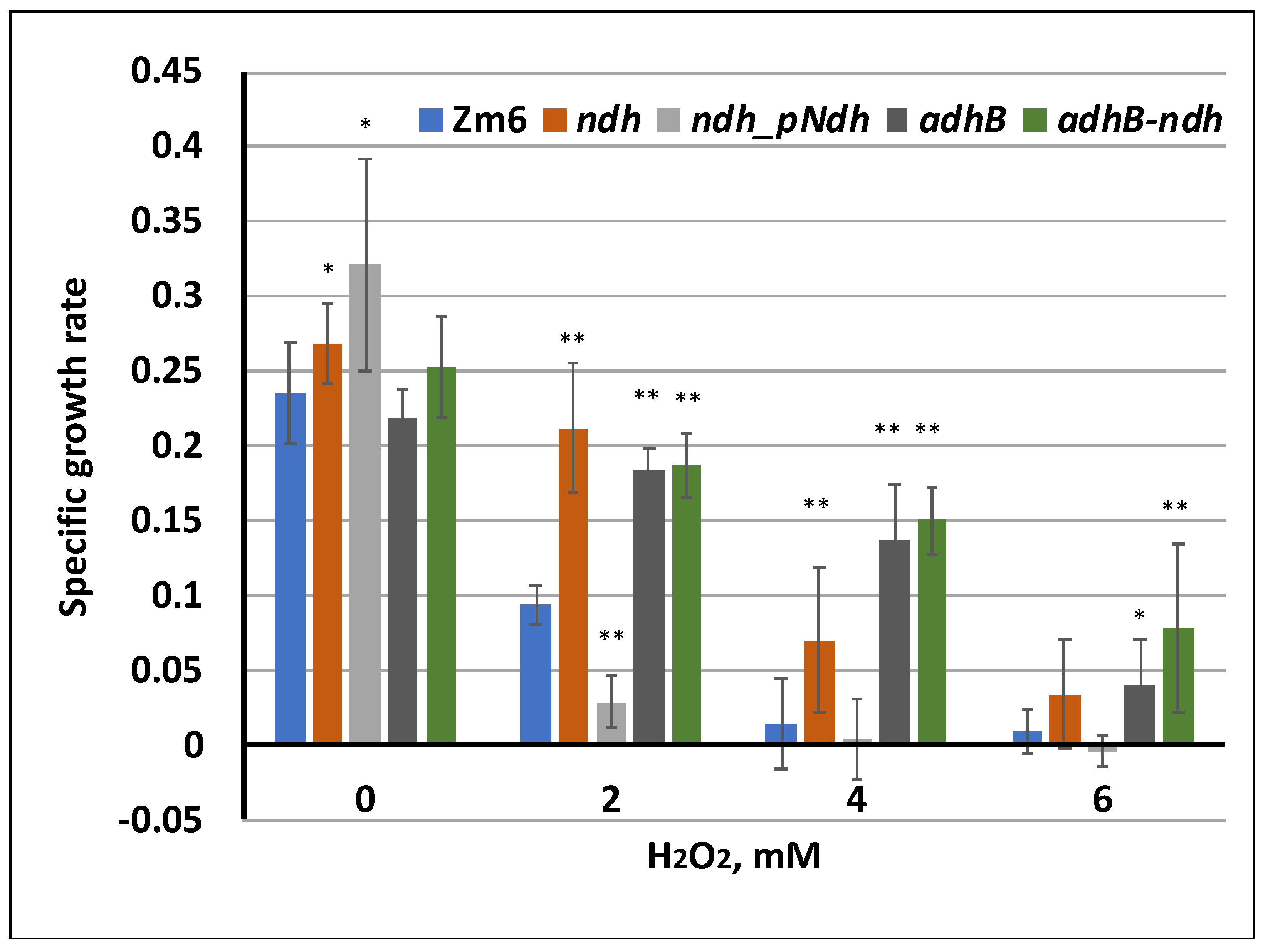
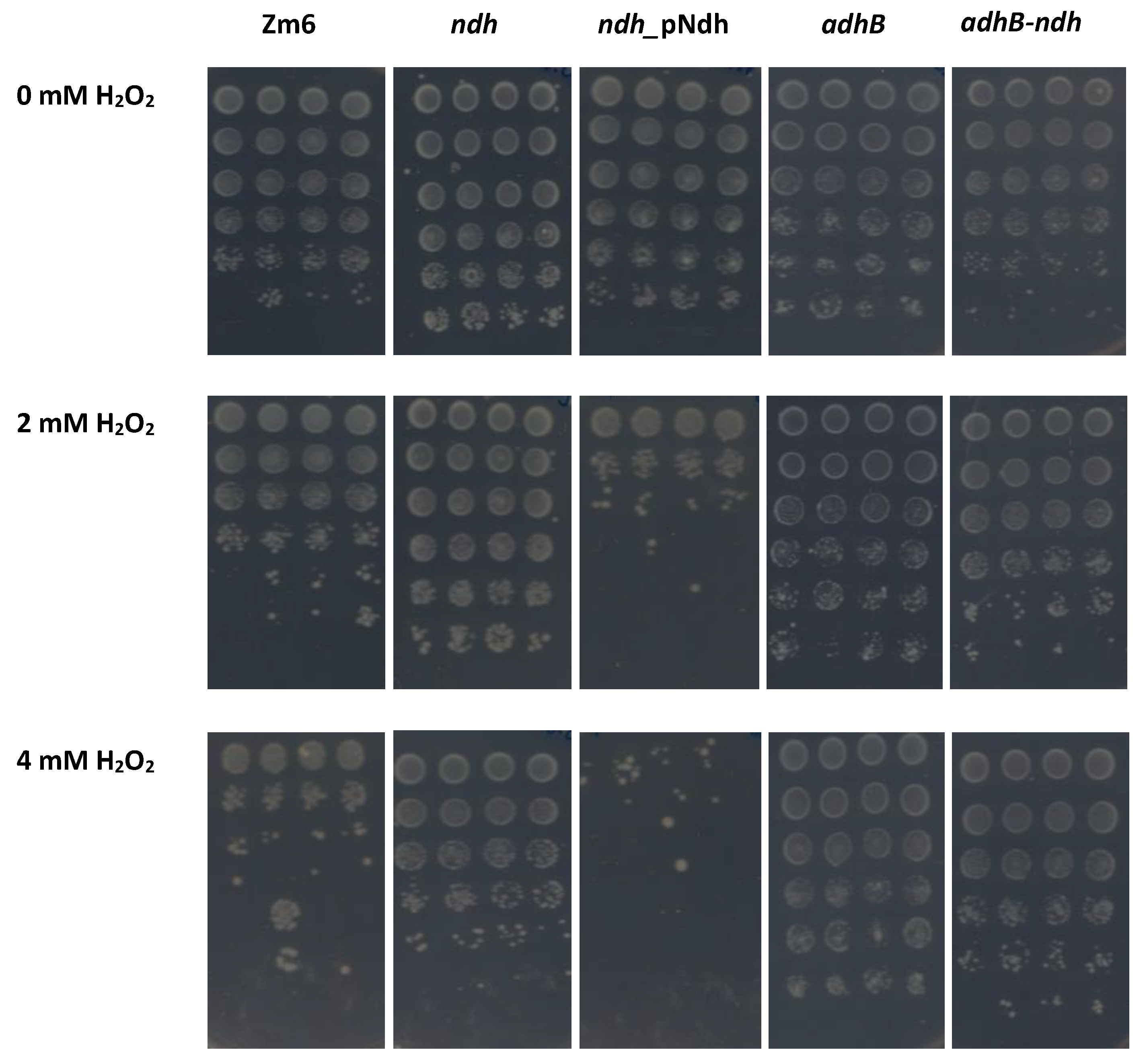
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rogers, P.L.; Lee, K.J.; Skotnicki, M.L.; Tribe, D.E. Ethanol production by Zymomonas mobilis. In Microbial Reactions; Advances in Biochemical Engineering; Springer: Berlin/Heidelberg, Germany, 1982; Volume 23, pp. 37–84. [Google Scholar]
- Sprenger, G. Carbohydrate metabolism in Zymomonas mobilis: A catabolic highway with some scenic routes. FEMS Microbiol. Lett. 1996, 145, 301–307. [Google Scholar] [CrossRef]
- Strohdeicher, M.; Neuß, B.; Bringer-Meyer, S.; Sahm, H. Electron transport chain of Zymomonas mobilis. Interaction with the membrane-bound glucose dehydrogenase and identification of ubiquinone 10. Arch. Microbiol. 1990, 154, 536–543. [Google Scholar]
- Kalnenieks, U.; Galinina, N.; Bringer-Meyer, S.; Poole, R.K. Membrane d-lactate oxidase in Zymomonas mobilis: Evidence for a branched respiratory chain. FEMS Microbiol. Lett. 1998, 168, 91–97. [Google Scholar] [CrossRef]
- Rutkis, R.; Galinina, N.; Strazdina, I.; Kalnenieks, U. The inefficient aerobic energetics of Zymomonas mobilis: Identifying the bottleneck. J. Basic Microbiol. 2014, 54, 1090–1097. [Google Scholar] [CrossRef] [PubMed]
- Rutkis, R.; Strazdina, I.; Balodite, E.; Lasa, Z.; Galinina, N.; Kalnenieks, U. The low energy-coupling respiration in Zymomonas mobilis accelerates flux in the Entner–Doudoroff pathway. PLoS ONE 2016, 11, e0153866. [Google Scholar] [CrossRef] [Green Version]
- Pentjuss, A.; Odzina, I.; Kostromins, A.; Fell, D.A.; Stalidzans, E.; Kalnenieks, U. Biotechnological potential of respiring Zymomonas mobilis: A stoichiometric analysis of its central metabolism. J. Biotechnol. 2013, 165, 1–10. [Google Scholar] [CrossRef]
- Swings, J.; De Ley, J. The biology of Zymomonas. Bacteriol. Rev. 1977, 41, 367–377. [Google Scholar] [CrossRef]
- Cazetta, M.L.; Celligoi, M.A.P.C.; Buzato, J.B.; Scarmino, I.S. Fermentation of molasses by Zymomonas mobilis: Effects of temperature and sugar concentration on ethanol production. Biores. Technol. 2007, 98, 2824–2828. [Google Scholar] [CrossRef]
- Zhang, M.; Eddy, C.; Deanda, K.; Finkelstein, M.; Picataggio, S. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science 1995, 267, 240–243. [Google Scholar] [CrossRef]
- Mohagheghi, A.; Linger, J.G.; Yang, S.H.; Smith, H.; Dowe, N.; Zhang, M.; Pienkos, P.T. Improving a recombinant Zymomonas mobilis strain 8b through continuous adaptation on dilute acid pretreated corn stover hydrolysate. Biotechnol. Biofuels 2015, 8, 55. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Mohagheghi, A.; Franden, M.A.; Chou, Y.C.; Chen, X.; Dowe, N.; Himmel, M.E.; Zhang, M. Metabolic engineering of Zymomonas mobilis for 2,3-butanediol production from lignocellulosic biomass sugars. Biotechnol. Biofuels 2016, 9, 189. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Shen, W.; Yan, X.; He, Q.; Cai, D.; Chen, S.; Wei, H.; Knoshaug, E.P.; Zhang, M.; Himmel, M.E.; et al. Metabolic engineering of Zymomonas mobilis for anaerobic isobutanol production. Biotechnol. Biofuels 2020, 13, 15. [Google Scholar] [CrossRef] [PubMed]
- Kalnenieks, U.; Balodite, E.; Strähler, S.; Strazdina, I.; Rex, J.; Pentjuss, A.; Fuchino, K.; Bruheim, P.; Rutkis, R.; Pappas, K.M.; et al. Improvement of acetaldehyde production in Zymomonas mobilis by engineering of its aerobic metabolism. Front. Microbiol. 2019, 10, 2533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, P.L.; Jeon, Y.J.; Lee, K.J.; Lawford, H.G. Zymomonas mobilis for fuel ethanol and higher value products. Adv. Biochem. Eng./Biotechnol. 2007, 108, 263–288. [Google Scholar]
- Wang, X.; He, Q.; Yang, Y.; Wang, J.; Haning, K.; Hu, Y.; Wu, B.; He, M.; Zhang, Y.; Bao, J.; et al. Advances and prospects in metabolic engineering of Zymomonas mobilis. Metab. Eng. 2018, 50, 57–73. [Google Scholar] [CrossRef]
- Kalnenieks, U.; Pappas, K.M.; Bettenbrock, K. Zymomonas mobilis metabolism: Novel tools and targets for its rational engineering. Adv. Micr. Physiol. 2020, 77, 37–88. [Google Scholar]
- Hayashi, T.; Furuta, Y.; Furukawa, K. Respiration-deficient mutants of Zymomonas mobilis show improved growth and ethanol fermentation under aerobic and high temperature conditions. J. Biosci. Bioeng. 2011, 111, 414–419. [Google Scholar] [CrossRef]
- Hayashi, T.; Kato, T.; Watakabe, S.; Song, W.; Aikawa, S.; Furukawa, K. The respiratory chain provides salt stress tolerance by maintaining a low NADH/NAD+ ratio in Zymomonas mobilis. Microbiology 2015, 161, 2384–2394. [Google Scholar] [CrossRef]
- Strazdina, I.; Balodite, E.; Lasa, Z.; Rutkis, R.; Galinina, N.; Kalnenieks, U. Aerobic catabolism and respiratory lactate bypass in Ndh-negative Zymomonas mobilis. Metabol. Eng. Commun. 2018, 7, e00081. [Google Scholar] [CrossRef]
- Kalnenieks, U.; Galinina, N.; Strazdina, I.; Kravale, Z.; Pickford, J.L.; Rutkis, R.; Poole, R.K. NADH dehydrogenase deficiency results in low respiration rate and improved aerobic growth of Zymomonas mobilis. Microbiology 2008, 154, 989–994. [Google Scholar] [CrossRef] [Green Version]
- Melo, A.M.; Bandeiras, T.M.; Teixeira, M. New Insights into Type II NAD(P)H:Quinone Oxidoreductases. Microbiol. Mol. Biol. Rev. 2004, 68, 603–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalnenieks, U.; Balodite, E.; Rutkis, R. Metabolic engineering of bacterial respiration: High vs. low P/O and the case of Zymomonas mobilis. Front. Bioeng. Biotechnol. 2019, 7, 327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charoensuk, K.; Irie, A.; Lertwattanasakul, N.; Sootsuwan, K.; Thanonkeo, P.; Yamada, M. Physiological importance of cytochrome c peroxidase in ethanologenic thermotolerant Zymomonas mobilis. J. Mol. Microbiol. Biotechnol. 2011, 20, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Balodite, E.; Strazdina, I.; Galinina, N.; McLean, S.; Rutkis, R.; Poole, R.K.; Kalnenieks, U. Structure of the Zymomonas mobilis respiratory chain: Oxygen affinity of electron transport and the role of cytochrome c peroxidase. Microbiology 2014, 160, 2045–2052. [Google Scholar] [CrossRef]
- An, H.; Scopes, R.K.; Rodriguez, M.; Keshav, K.F.; Ingram, L.O. Gel electrophoretic analysis of Zymomonas mobilis glycolytic and fermentative enzymes: Identification of alcohol dehydrogenase II as a stress protein. J. Bacteriol. 1991, 173, 5975–5982. [Google Scholar] [CrossRef] [Green Version]
- Echave, P.; Tamarit, J.; Cabiscol, E.; Ros, J. Novel antioxidant role of alcohol dehydrogenase E from Escherichia coli. J. Biol. Chem. 2003, 278, 30193–30198. [Google Scholar] [CrossRef] [Green Version]
- Beckham, K.S.H.; Connolly, J.P.R.; Ritchie, J.M.; Wang, D.; Gawthorne, J.A.; Tahoun, A.; Gally, D.L.; Burgess, K.; Burchmore, R.J.; Smith, B.O.; et al. The metabolic enzyme AdhE controls the virulence of Escherichia coli O157:H7. Mol. Microbiol. 2014, 93, 199–211. [Google Scholar] [CrossRef]
- Lin, G.H.; Hsieh, M.C.; Shu, H.Y. Role of Iron-Containing Alcohol Dehydrogenases in Acinetobacter baumannii ATCC 19606 Stress Resistance and Virulence. Int. J. Mol. Sci. 2021, 22, 9921. [Google Scholar] [CrossRef]
- Luong, T.T.; Kim, E.H.; Bak, J.P.; Nguyen, C.T.; Choi, S.; Briles, D.E.; Pyo, S.; Rhee, D.K. Ethanol-induced alcohol dehydrogenase E (AdhE) potentiates pneumolysin in Streptococcus pneumoniae. Infect. Immun. 2015, 83, 108–119. [Google Scholar] [CrossRef] [Green Version]
- Kavanaugh, D.W.; Porrini, C.; Dervyn, R.; Ramarao, N. The pathogenic biomarker alcohol dehydrogenase protein is involved in Bacillus cereus virulence and survival against host innate defence. PLoS ONE 2022, 17, e0259386. [Google Scholar] [CrossRef]
- Tamarit, J.; Cabiscol, E.; Aguilar, J.; Ros, J. Differential inactivation of alcohol dehydrogenase isoenzymes in Zymomonas mobilis by oxygen. J. Bacteriol. 1997, 179, 1102–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalnenieks, U.; Galinina, N.; Toma, M.M.; Pickford, J.L.; Rutkis, R.; Poole, R.K. Respiratory behaviour of a Zymomonas mobilis adhB::kanr mutant supports the hypothesis of two alcohol dehydrogenase isoenzymes catalysing opposite reactions. FEBS Lett. 2006, 580, 5084–5088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strazdina, I.; Kravale, Z.; Galinina, N.; Rutkis, R.; Poole, R.K.; Kalnenieks, U. Electron transport and oxidative stress in Zymomonas mobilis respiratory mutants. Arch. Microbiol. 2012, 194, 461–471. [Google Scholar] [CrossRef]
- Osman, Y.A.; Conway, T.; Bonetti, S.J.; Ingram, L.O. Glycolytic flux in Zymomonas mobilis: Enzyme and metabolite levels during batch fermentation. J. Bacteriol. 1987, 169, 3726–3736. [Google Scholar] [CrossRef] [Green Version]
- Kalnenieks, U.; Galinina, N.; Toma, M.M. Physiological regulation of the properties of alcohol dehydrogenase II (ADH II) of Zymomonas mobilis: NADH renders ADH II resistant to cyanide and aeration. Arch. Microbiol. 2005, 183, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Flecha, B.; Demple, B. Intracellular generation of superoxide as a by-product of Vibrio harveyi luciferase expressed in Escherichia coli. J. Bacteriol. 1994, 176, 2293–2299. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, S.; Kakizono, T.; Kadota, K.; Das, K.; Taguchi, H. Purification of two alcohol dehydrogenases from Zymomonas mobilis and their properties. Appl. Microbiol. Biotechnol. 1985, 22, 249–254. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Markwell, M.A.; Haas, S.M.; Bieber, L.L.; Tolbert, N.E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 1978, 87, 206–210. [Google Scholar] [CrossRef]
- Sen, A.; Imlay, J.A. How Microbes Defend Themselves From Incoming Hydrogen Peroxide. Front. Immunol. 2021, 12, 667343. [Google Scholar] [CrossRef]
- Brumaghim, J.L.; Li, Y.; Henle, E.; Linn, S. Effects of hydrogen peroxide upon nicotinamide nucleotide metabolism in Escherichia coli: Changes in enzyme levels and nicotinamide nucleotide pools and studies of the oxidation of NAD(P)H by Fe(III). J. Biol. Chem. 2003, 278, 42495–42504. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Strain/Plasmid | Characteristics | Source |
|---|---|---|
| pGEMndh::cmr | Plasmid pGEM-Zf(+) derivative, carrying a 2.6 kb DNA fragment between the HindIII and BamHI sites of its MCS, containing a PCR-amplified ORF of the Type II NADH dehydrogenase gene ndh (ZMO_RS04970) with a Cmr marker inserted in its AgeI site | [21] |
| pNdh | Plasmid pBBR1MCS-2 derivative, carrying a 1.5 kb a 2.6 kb DNA fragment between the HindIII and BamHI sites of its MCS, containing a PCR-amplified ORF of the Type II NADH dehydrogenase gene ndh (ZMO_RS04970) with a Cmr marker inserted in its AgeI site | [6] |
| Zm6 | Wild type, parent strain | ATCC 29191 |
| Zm6-adhB | Zm6 with a Kanr insert in the ORF of the iron-containing alcohol dehydrogenase gene adhB (ZMO_RS07165) | [33] |
| Zm6-ndh | Zm6 with a Cmr insert in the ORF of respiratory Type II NADH dehydrogenase gene ndh (ZMO_RS04970) | [21] |
| Zm6-adhB-ndh | Zm6-adhB with a Cmr insert in the ORF of ndh | Present work |
| Zm6-ndh_pNdh | Zm6-ndh transformed with the pNdh plasmid | [6] |
| Zm6-cat | Zm6 with a Cmr insert in the ORF of catalase gene cat (ZMO_RS04105) | [34] |
| Zm6-adhB-cat | Zm6-adhB with a Cmr insert in the ORF of cat | [14] |
| Zm6-cat_pNdh | Zm6-cat, transformed with the pNdh plasmid | [14] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kovtuna, K.; Strazdina, I.; Bikerniece, M.; Galinina, N.; Rutkis, R.; Martynova, J.; Kalnenieks, U. Improved Hydrogen Peroxide Stress Resistance of Zymomonas mobilis NADH Dehydrogenase (ndh) and Alcohol Dehydrogenase (adhB) Mutants. Fermentation 2022, 8, 289. https://doi.org/10.3390/fermentation8060289
Kovtuna K, Strazdina I, Bikerniece M, Galinina N, Rutkis R, Martynova J, Kalnenieks U. Improved Hydrogen Peroxide Stress Resistance of Zymomonas mobilis NADH Dehydrogenase (ndh) and Alcohol Dehydrogenase (adhB) Mutants. Fermentation. 2022; 8(6):289. https://doi.org/10.3390/fermentation8060289
Chicago/Turabian StyleKovtuna, Kristiana, Inese Strazdina, Mara Bikerniece, Nina Galinina, Reinis Rutkis, Jekaterina Martynova, and Uldis Kalnenieks. 2022. "Improved Hydrogen Peroxide Stress Resistance of Zymomonas mobilis NADH Dehydrogenase (ndh) and Alcohol Dehydrogenase (adhB) Mutants" Fermentation 8, no. 6: 289. https://doi.org/10.3390/fermentation8060289
APA StyleKovtuna, K., Strazdina, I., Bikerniece, M., Galinina, N., Rutkis, R., Martynova, J., & Kalnenieks, U. (2022). Improved Hydrogen Peroxide Stress Resistance of Zymomonas mobilis NADH Dehydrogenase (ndh) and Alcohol Dehydrogenase (adhB) Mutants. Fermentation, 8(6), 289. https://doi.org/10.3390/fermentation8060289





