Abstract
Nisin is a green, safe and natural food preservative. With the expansion of nisin application, the demand for nisin has gradually increased, which equates to increased requirements for nisin production. In this study, Lactococcus lactis subsp. lactis lxl was used as the original strain, and the compound mutation method was applied to induce mutations. A high-yielding and genetically stable strain (Lactobacillus lactis A32) was identified, with the nisin titre raised by 332.2% up to 5089.29 IU/mL. Genome and transcriptome sequencing was used to analyse A32 and compare it with the original lxl strain. The comparative genomics results show that 107 genes in the A32 genome had mutations and most base mutations were not located in the four well-researched nisin-related operons, nisABTCIPRK, nisI, nisRK and nisFEG: 39 single-nucleotide polymorphisms (SNPs), 34 insertion mutations and 34 deletion mutations. The transcription results show that the expression of 92 genes changed significantly, with 27 of these differentially expressed genes upregulated, while 65 were downregulated. Our findings suggest that the output of nisin increased in L. lactis strain A32, which was accompanied by changes in the DNA replication-related gene dnaG, the ABC-ATPase transport-related genes patM and tcyC, the cysteine thiometabolism-related gene cysS, and the purine metabolism-related gene purL. Our study provides new insights into the traditional genetic mechanisms involved nisin production in L. lactis, which could provide clues for a more efficient metabolic engineering process.
1. Introduction
Nisin is a small-molecule polypeptide with broad-spectrum antibacterial effects, and is produced by Lactococcus ssp. Nisin is internationally recognized as a safe and nontoxic natural food additive. It has no side effects, and has been used as a food preservative for more than 60 years [1,2]. Due to the high biosafety of nisin, research on its application value has long expanded past the field of food preservation. In recent years, nisin applications, including its use as a packaging material [3,4,5], as a medical antibacterial agent [6,7,8,9], as a biosensor [10], in immune regulation [11,12], as an anticancer drug [13,14] and in other aspects, have been explored.
In the process of fermentation, lactic acid-producing bacteria (LAB) produce a variety of metabolites, and nisin is a byproduct of the growth process of Lactococcus lactis subspecies. The nisin synthesis process is very complex, and its synthesis efficiency is relatively low [1]. (1) Nisin is generated to inhibit other microorganisms that compete with L. lactis ssp. for living space [15,16]. To avoid self-inhibition, L. lactis also exerts a series of immune mechanisms. (2) Nisin is produced by ribosomal synthesis and is a post-translationally modified peptide (RiPP); its production process involves precursor peptide-related genes and modified protein synthesis genes. Nisin biosynthesis genes are transcribed in four operons, nisABTCIPRK, nisI, nisRK and nisFEG [16,17,18]. (3) From a production environment standpoint, during their growth processes, LAB produce a variety of acids, such as carboxylic acid, lactic acid and other acidic substances, decreasing the pH of the media [19]. A low pH inhibits the growth of LAB and the production of nisin, requiring production strains that tolerate acidic environments.
The complexity of nisin synthesis makes it difficult to reconstruct the production pathway in other heterogenic hosts; thus, optimizing nisin production by L. lactis strains is important and useful. At present, two main methods are used to modify bacterial strains, traditional mutagenesis-type breeding and genetic engineering. Genetic engineering of bacteria can alter the expression of several genes; however, the choice of operation sites is based on the clear understanding of the intracellular pathway and the corresponding regulatory mechanism. Furthermore, it is difficult to achieve the coexpression of a series of genes, and accordingly, the corresponding increase in production is limited. Although a greater effort is needed to induce traditional mutations, a series of genes can be changed, which is more favourable than changes in the whole strain. To screen the strains with high nisin yields, in this study, compound mutation combined with high-concentration-nisin plate screening was used to modify and quickly screen high-nisin-yielding strains. Compound mutagenesis involves the successive or simultaneous use of one or more mutagens on microorganisms, or the same mutagen acting on microorganisms repeatedly. It is a commonly used method to improve the production of microorganisms. Atmospheric room temperature plasma (ARTP) can change the structure and permeability of the cell wall membrane, causing gene damage and mutation when plasma jets with highly active particles are discharged toward microbial cells. The method is simple to use, is highly pure and pollution-free, is harmless to the human body and causes various mutations [11]. Ultraviolet (UV) mutation is the most traditional and useful mutation method, and is often used as the first choice for microbial breeding. It can directly affect double-stranded DNA and improve mutation efficiency. Therefore, we chose the use of ARTP and UV light, which are two traditional and modern methods for compound mutagenesis. A high-nisin-concentration screening method can be used to identify strains with increased nisin yields. Together, these methods can effectively mutate and screen mutant strains. In this study, a high-yielding mutant strain (A32) was screened, and then, by comparing the differences at the transcriptome level between the original strain and the mutant strain, we hoped to find a series of genes related to increasing nisin production, which is very important for future genetic engineering of the producing strain.
A high-yielding and genetically stable mutant strain was obtained, which was named L. lactis A32. The yield of the mutant strain was three times that of the original strain L. lactis lxl, and its resistance to nisin was twofold greater. By comparing the whole-genome sequence of L. lactis A32 with that of the original strain lxl, 107 mutated genes were found, including 39 single-nucleotide polymorphisms, 34 insertion mutations and 34 deletion mutations. Through comparative analysis of the transcriptome sequencing results, it was found that the transcription of 92 genes changed significantly, with 27 of these genes significantly upregulated, while 65 genes were significantly downregulated. Analysis of the metabolic pathways of these genes can provide clues for the further study of the molecular mechanism underlying the nisin synthesis pathway.
2. Materials and Methods
2.1. Strains, Media and Growth Conditions
Lactococcus lactis subsp. lactis lxl is a store strain in our laboratory. It was screened from raw milk, with 40 mg/L bromocresol purple and 1000 IU/mL nisin standards in modified M17 medium (10 g/L sucrose as the only carbon source). After morphological and physiological analysis and 16S rDNA sequencing, it was identified as Lactococcus lactis subsp. lactis. The titre test strain was Micrococcus luteus NCIB 8166, which was purchased from China General Microbiological Culture Collection Center, and preserved at −80 °C in 20% (v/v) glycerin in our laboratory.
The composition of the media used is as follows.
GM17 medium (a seed medium): M17 broth medium (soy peptone 5 g/L, peptone 2.5 g/L, casein peptone 2.5 g/L, yeast extract powder 2.5 g/L, beef extract powder 5 g/L, lactose 5 g/L, sodium ascorbate 0.5 g/L, β-sodium glycerophosphate 19 g/L, magnesium sulfate 0.25 g/L) containing glucose (5 g/L). CM1 medium (a fermentation medium): sucrose (20 g/L), peptone (20 g/L), yeast extract (20 g/L), KH2PO4 (10 g/L), NaCl (2 g/L) and MgCl2 (0.2 g/L). S1 medium (a detection medium): peptone (8 g/L), yeast extract (5 g/L), glucose (5 g/L), NaCl (5 g/L) and NaH2PO4·12H2O (2 g/L).
2.2. Fermentation in Flasks and Nisin Titre Assays
A nisin standard (Maclin, China) was prepared in a series of standard solutions of 50, 100, 200, 400, 800, 1000, 2000, 3000 and 4000 IU/mL. For this procedure, a sterilized Oxford cup was placed on the test plate, the culture media containing the tested bacteria was added to the plate, and the Oxford cup was removed after cooling to form a uniform small hole. Then, 200 μL of standard solutions containing a series of nisin concentrations were added to the small holes, after which the plates were cultured at 37 °C until there was an obvious bacteriostatic circle around each small hole, and the diameter of the bacteriostatic circle was measured. A nisin titre standard curve was constructed, with the diameter of the bacteriostatic circle representing the abscissa and the logarithm of the titre representing the ordinate.
As a seed solution, a single colony of the original strain lxl was inoculated into GM17 media and cultured overnight at 30 °C and 200 rpm. Then, the seed liquid was inoculated into CM1 media at a concentration of 5%, fermented at 30 °C and centrifuged 200 rpm for 24 h. Samples were collected every 2 h during the fermentation period, and a bacteriostatic circle experiment was carried out. The nisin titre was calculated according to the nisin standard curve (Figure S1) [20].
2.3. Compound Mutation and Strain Breeding
The starting strain was cultured by scribing, and a single colony was picked and cultured in GM17 liquid medium for 6–8 h. The culture was appropriately diluted with normal saline instead of the mutagenic strain solution, and the optical density (OD600) of the mutagenic strain solution was maintained between 0.8 and 1.0 to ensure the proper colony number and growth state of each mutagenic strain. The lethality of the ARTP and UV mutations was determined, and the time when the lethality was 80–90% was selected as the mutation time to ensure mutation efficiency. The nisin tolerance range of the original strain was determined, and a screening plate containing high concentrations of nisin was constructed. The solution of bacteria with different mutations was poured onto the screening plate, which was subsequently cultured at 30 °C. The grown colonies were picked out and cultured overnight in an Erlenmeyer flask containing 50 mL of GM17 medium. The cultured bacterial solution was then inoculated into a triangular flask containing 50 mL of CM1 medium for fermentation at 30 °C and centrifugation at 200 rpm for 24 h. The fermentation broth was subsequently centrifuged at 8000 rpm for 5 min, and the supernatant was collected for a bacteriostatic circle experiment to determine the titre of the fermentation broth. The strain with the highest titre from this batch of colonies was selected as the starting strain for the next mutation, as shown in Figure 1. The abovementioned process was repeated until the high-yielding strains whose parameters were within the target range were screened.
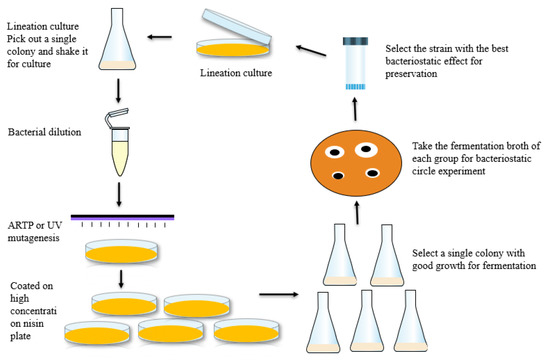
Figure 1.
Compound mutation and breeding process.
2.4. Determination of the Genetic Stability of Mutants
To investigate whether the titre decreased and whether the mutant phenotype was reversible, we determined the genetic stability of the mutants. The strains with high nisin titre were screened out. They were transferred onto plates and cultured at 30 °C for 24 h to form the F1 generation. The colonies were then removed from the F1 plate and cultured on another plate to form the F2 generation. This process was repeated to obtain 20 generations in total. Single colonies were picked every three generations from seed liquid culture, then inoculated into liquid fermentation medium CM1 and incubated at 30 °C and 200 rpm for 24 h, and the supernatant of the fermentation broth was collected to determine the nisin titre.
2.5. Comparison of the Fermentation Process between the Mutant Strain and Original Strain
The mutant with the highest titre that was genetically stable and the original strain were inoculated into CM1 fermentation media at the same time and fermented at 30 °C and 200 rpm for 48 h, and samples were collected every 3 h. The biomass of the sample was measured at 600 nm by a spectrometer (Metash, Shanghai, China), and the nisin titre was tested via bacteriostatic circles.
2.6. DNA Library Construction, DNA Sequencing, Assembly and Annotation
Single colonies of L. lactis A32 and lxl were selected and cultured in CM1 media at a suitable temperature overnight, and the bacteria were collected during the logarithmic growth period. Genomic DNA of L. lactis lxl and A32 was extracted using TIAMP Bacterial DNA Kits (Sparkjade, Jinan, China) according to the manufacturer’s instructions. Then, the quality of the DNA was checked to ensure that the quality was sufficient for subsequent sequencing. The concentration of the standard DNA was measured of by a Qubit instrument (Thermo Fisher Scientific, Waltham, MA, USA) (including the following process of establishing the whole library). A Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) was used to measure the DNA concentration (between OD260/280 and between 1.8 and 2.0), and the DNA integrity was verified by 1% agarose gel electrophoresis. Library construction, Oxford Nanopore Technologies (ONT) sequencing, Illumina sequencing and base determination were carried out by Gene Denovo (Guangzhou, China). The bacterial genome was sequenced by a combination of third-generation ONT sequencing and second-generation Illumina sequencing. This method relies on the long reading characteristics of third-generation sequencers to ensure a more complete genome assembly, and uses second-generation sequencing data for correction to ensure more accurate and reliable assembly results. Fastp software [21] was used to control the ONT and Illumina sequencing data and obtain effective data (clean data). The third -generation sequencing reads were spliced and assembled with Flye (version 2.8.1-b1676) [22]. Pilon (version 1.23) [23] was then used to compare the second-generation sequencing reads to the assembled genome sequence, correct the genome results according to the default parameters of the software, and output the corrected genome sequence and corrected site information. The National Center for Biotechnology Information (NCBI) database was used to predict gene functions. Through BLAST, the predicted gene sequences were compared to the information within the nonredundant (Nr), Swiss-Prot, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Clusters of Orthologous Genes (COG) databases to determine the proteins with the highest sequence similarity to those encoded by a given gene, to obtain the protein functional annotation information of the genes. We used Blast2GO to obtain the GO annotation information of the genes, and performed GO functional classification analysis for all of the genes. Diamond was used to compare all the gene sequences with the sequences in the COG database to obtain the annotation results corresponding to the genes and to classify the functions of proteins according to the annotation results. In addition, the gene function databases Antimicrobial Resistance Genes Database (ARDB) and the Virulence Factors of Pathogenic Bacteria Database (VFDB), within which information is subdivided into an increased number of fields, were used to predict secondary metabolic gene clusters and to annotate virulence factor-related genes and drug resistance-related genes. Moreover, the Protein Family (Pfam) database, the Pathogen Host Interactions (PHI)-base database, the Carbohydrate-Active Enzymes (CAZy) database, the Comprehensive Antibiotic Resistance Database (CARD) and other databases were used for secretory protein prediction, type-N secretion system (TNSS) effector protein prediction and two-component system prediction.
2.7. Whole-Genome Comparisons
L. lactis lxl and A32 bacteria were subjected to comparative genomic analysis. MUMmer [24] software was used to detect the SNPs and insertion deletions (indels) in the assembled genomes of the individuals, as well as to perform statistics based on the positional relationships between the mutant genes and the mutation results, and compare the results with information in databases to assess the functions of the mutated genes.
2.8. RNA Extraction, Transcriptome Sequencing and Analysis
Single colonies of L. lactis A32 and L. lactis lxl were selected and cultured in CM1 media at a suitable temperature overnight, and the bacteria were collected during their logarithmic growth period. An appropriate amount of the cultured bacterial solution was transferred into a suitable centrifuge tube, after which the bacteria were collected by high-speed centrifugation (12,000 rpm). The culture medium in the upper part of the tube was discarded. Total RNA was extracted by the TRIzol (Sevencyd, Jinan, China) method.
The RNA degradation degree and potential contamination were monitored on 1% agarose gels. RNA purity (OD260/OD280, OD260/OD230) was determined using a NanoPhotometer® spectrophotometer (IMPLEN, CA, USA). RNA integrity was measured using a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). An Illumina MRZB12424 Ribo Zero rRNA Removal Kit (Bacteria) (Illumina, San Diego, CA, USA) was used to remove rRNA from 1 μg of total RNA. The quality of the RNA samples was detected using an Agilent 2100 Biological Analyser (Agilent Technologies, Santa Clara, CA, USA). The library construction, transcriptome sequencing and RNA sequencing (RNA-seq) analysis of L. lactis A32 were carried out in Gene Denovo (Guangzhou, China). FASTP (version 0.18.0) [21] was used to filter the original data produced via the Illumina platform. The filtering criteria were as follows: (1) reads with ≥10% unidentified nucleotides (N); (2) reads in which >50% of the bases had Phred quality scores of ≤20 and (3) reads aligned to the barcode adapter. Bowtie2 (version 2.2.8) [25] was used to compare the retained reads to the reference genome, and the reads associated with rRNA in the comparison were deleted. RSEM [26] was used to identify known genes and calculate their expression. Library construction and transcriptome sequencing of L. lactis lxl were carried out by Gene Denovo (Guangzhou, China). The gene expression level was further normalized by using the fragments per kilobase of transcript per million (FPKM) mapped reads method to eliminate the influences of different gene lengths and amounts of sequencing data on the calculation of gene expression. The edgeR package [27] (http://www.r-project.org/ Accessed on 1 January 2022) was then used to identify differentially expressed genes (DEGs) across the samples. Genes were considered differentially expressed if their expression fold-change was ≥2 and their false discovery rate-adjusted P (q value) was <0.05. The DEGs were then subjected to GO functional enrichment and KEGG pathway analyses, and q values <0.05 were used as thresholds. The GO functional terms of the DEGs were analysed, the GO functional classification annotations of the DEGs were obtained, and the pathways in which the DEGs were enriched were identified. Using Rockhopper [28] software, the sequencing results were compared to the reference genome sequence and to the annotated genes. Unknown transcript-coding regions were considered new transcript-coding regions. The newly encoded transcripts were compared to the information in the Nr database for annotation, and the annotated transcripts were regarded as new transcripts with coding potential.
2.9. Statistical Analysis
Student’s t-test was conducted to determine the significant differences in nisin titres between the original L. lactis lxl strain and the mutant A32 strain. The data were statistically analysed using Origin 9.1 (OriginLab, Northampton, MA, USA).
3. Results and Discussion
3.1. Titres of Mutant Strains
As the duration of ARTP and UV treatment increased, the mortality of bacteria also increased. When the bacteria were treated with ARTP and UV light for 120 s and 60 s, respectively, the lethality rates were 90% and 92%, respectively, as shown in Figure 2A,B. In the recent literature on mutagenesis, a treatment time with a lethal rate of approximately 90% is usually used to ensure mutation efficiency [17]. In the present study, ARTP and UV light were applied for 120 s and 60 s, respectively.
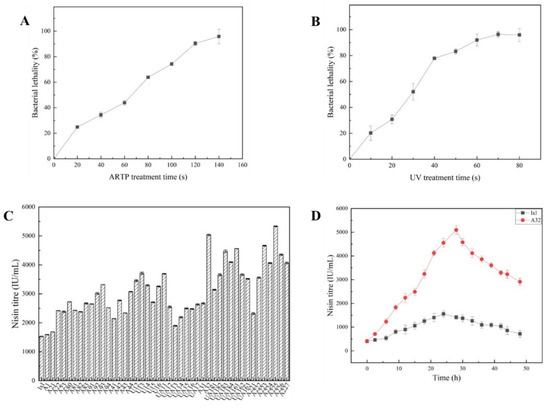
Figure 2.
Mutagenesis preparation and results. (A) Lethality to L. lactis lxl of ARTP mutagenesis; (B) lethality to L. lactis lxl of UV mutagenesis; (C) mutagenic strain titre; (D) titre curves of L. lactis A32 and lxl with increasing fermentation times.
Through a series of compound mutagenesis and step-by-step screening, some mutants that could grow on plates supplemented with high nisin concentration were selected, fermented, and their nisin production titres were determined. The results are shown in Figure 2C. Among all examined mutants, mutants A32 and A225 had higher titres, and their titre peaks were 5089.29 IU/mL and 5361.16 IU/mL, respectively.
3.2. Evaluation of Mutant Stability
The experimental results regarding the genetic stability of the mutants were shown in Table S2. We observe that the ability of the mutant L. lactis A32 to produce nisin was stable, while the ability of A225 to produce nisin was not stable. In the third generation, the nisin titre of A225 was only half that of A32. A32 was also twice as resistant to nisin as the original strain. These results show that the A32 mutant generated by ARTP and UV radiation yielded high amounts of nisin and was genetically stable. Therefore, the mutant strain L. lactis A32 was selected for use in the following experiments.
3.3. Comparison of the Fermentation of L. lactis A32 and Lxl
There was a significant difference in growth curves and nisin titres between L. lactis A32 and lxl. Mutant A32 took significantly longer to reach the stationary phase than the original lxl strain. The A32 biomass peaked at 12 h, and the lxl biomass peaked in the stationary phase at 9 h, as shown in Figure S2. Since nisin is a growth-coupled secondary metabolite, hypothetically, A32 would accumulate more nisin than lxl during this process. This hypothesis is verified in Figure 2D. As shown in Figure 2D, the nisin titre continued to increase within 0–28 h of A32 fermentation, the peak nisin production intensity of A32 reached up to 181.76 IU/(mL·h), which is 5089.29 IU/mL, and lxl yielded a maximum of 1529.79 IU/mL at 24 h. These results show that the correlation between product synthesis and thallus growth was not shifted, but the regulation around the synthetic pathways flux was improved.
3.4. Genome Assembly and Annotation
The PromethION (Oxford Nanopore Technologies, Oxford, UK) and Agilent 2100 sequencing platforms were used to sequence the whole genome of L. lactis A32. A total of 7,244,702 bp of clean reads and 1,086,215,124 bp from A32 were generated. The genome sequencing data of L. lactis A32 have been submitted to the Sequence Read Archive database of the NCBI under accession number PRJNA799214. After data filtering, the genome of L. lactis A32 was assembled, with a total genome length of 2.40 Mb (Table S2). The GC contents (lxl, 34.44%; A32, 34.57%) were similar (Table 1). The complete genome sequences of L. lactis A32 and lxl have been submitted to the GenBank database of the NCBI (accession number for A32, SAMN25149786; accession number for lxl, SAMN25148929). According to the comparative results of five gene function databases (Nr, Swiss-Prot, GO, KEGG and COG), 2278 DNA sequences (CDSs) in lxl and 2285 CDSs in A32 were predicted in the genome (Table 1), and five genes encoded proteins with no predicted function in lxl and A32.

Table 1.
Genome characteristics of L. lactis A32 and lxl.
3.5. Functional Classification and Comparison
All annotated genes of L. lactis A32 and lxl were classified into GO functional categories (Table S3). The GO functional classification system is based on three ontologies, including biological processes, cell components and molecular processes. It includes 58 gene functional classifications, such as cellular processes, metabolic processes, single-organism processes, responses to stimuli, cellular component organization or biogenesis, biological regulation and biological processes. There were some differences in GO cluster analysis between A32 and lxl. Further analysis showed that, compared with lxl, A32 had six more genes related to molecular processes, four more genes related to metabolic processes, four more genes related to catalytic activity, two more genes related to single biological processes, two fewer genes related to responses to stimuli and one less gene related to development processes.
The COG functional categories of all annotated proteins of L. lactis A32 and lxl are shown in Table 2, and were divided into 25 main functional categories. The main functional categories were related to translation; ribosomal structure and biogenesis; replication, recombination and repair; cell wall/membrane/envelope biogenesis; carbohydrate transport; and amino acid transport and metabolism. The distribution patterns of the protein clusters of homologous genes in the two strains were very similar. There were four fewer CDSs in the genome of A32 compared with the original lxl strain. In addition, compared with lxl, A32 had one more CDS related to transcription- and signal transduction-related proteins; two fewer CDSs related to replication, recombination and repair; two fewer CDSs related to cell cycle control, cell division and chromosome distribution; two fewer CDSs related to carbohydrate transport and metabolism; and one less CDS related to inorganic ion transport and metabolism.

Table 2.
GO functional categories.
3.6. Comparison of Genomes between L. lactis lxl and A32
To reveal the relationships between phenotypes and genetic variations, and provide clues for the further study of the molecular mechanism of the nisin synthesis pathway, a genome-wide comparison was performed between the high-nisin-yielding mutant A32 and the original lxl strain. The cyclic images from the genome comparison show the microstructural mutations between L. lactis A32 and lxl (Figure 3). A total of 107 genes in the A32 genome were mutated compared with the lxl genome, including 39 single-base mutations, 34 insertion mutations and 34 deletion mutations. The sequences of the mutated genes were compared with the reference genome sequence in the NCBI database of COG functional annotations. While the functions of some genes were not clear, some genes with base mutations had clear functions (Table 3), and some mutated genes, such as rexB, ftsH, gntP and yfmR, were related to energy metabolism. Additionally, some mutated genes were related to ion transport, such as yfmR, rbcR, zitR, adcA and copB; to DNA replication, transcription and translation, such as dnaG, rpsI, rex and arlR; and to amino acid transport and metabolism, such as patM, tcyC, tcyJ, cysS and brnQ. However, a vast improvement in the nisin titre was achieved, except for one mutation in the four nisin-related operons, nisK; most base mutations were not located in the well-researched nisin-related four operons, nisABTCIPRK, nisI, nisRK and nisFEG. Sequence analysis of the nisin genes nisA, nisB, nisC and nisP before and after mutation showed that these genes were not mutated, thus the amino acid sequence of nisin and the mechanism of action of the modified protein did not change. Based on these analyses, it is reasonable to infer that the nisin produced by the mutant is structurally identical to that of the original bacterium, and thus its stability and other properties remain unchanged.
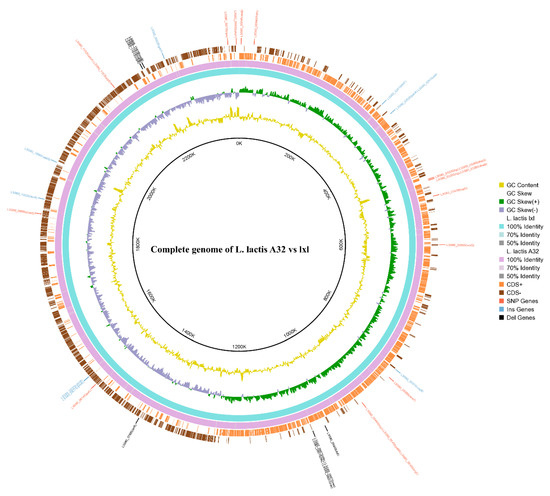
Figure 3.
The cyclic images from the genome comparison.

Table 3.
COG analysis and type of gene mutation.
3.7. Transcriptome Sequencing and Analysis
Transcriptome sequencing was used to detect the differences in gene expression between the original L. lactis lxl strain and A32, the inserted genes were examined to determine the metabolic pathways that they influenced. After quality screening, 19.58 to 24.42 million clean reads and 1.65 to 2.14 Gb of clean data submitted to the NCBI gene expression comprehensive database (BioProject: PRJNA825060) were obtained from the control strain and the mutant strain, respectively (Table S4). The sequencing reads of the original strain and mutant strain were mapped to the A32 genome assembly sequence, and 2231 known mRNAs were identified in A32. The statistics of gene coverage are shown in Figure 4A. Compared with that in the original L. lactis lxl strain, the expression of 92 genes in L. lactis A32 changed significantly (p < 0.005), of which 27 were significantly upregulated and 65 were significantly downregulated, as shown in Figure 4B. GO enrichment analysis was performed on the significant DEGs (Figure 4C) to understand their biological functions. DEG pathway enrichment analysis is helpful for further understanding the gene-related metabolic networks. The main metabolic pathways of significantly differentially expressed genes were metabolic pathways, ABC transporters, two-component systems, biosynthesis of secondary metabolites, quorum sensing, arginine biosynthesis and butanoate metabolism (Figure 4D).
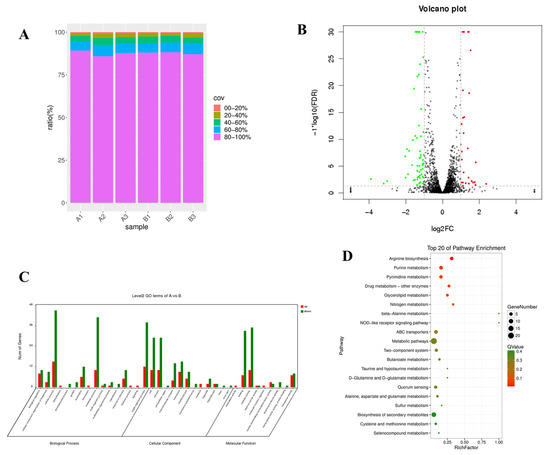
Figure 4.
Transcriptome sequencing results and analysis. (A) L. lactis and A32 gene coverage; (B) map of significant DEGs; (C) GO classification of DEGs; (D) statistical characteristics of DEG enrichment in KEGG pathways. Genes with no significant difference in expression are shown in black, upregulated genes are shown in red and downregulated genes are shown in green.
3.8. Comprehensive Analysis
The fermentation conditions and growth states of L. lactis strains A32 and lxl were observed. Comparative genomic analysis and transcriptomic analysis were then performed in combination to identify associated genes, and try to infer the regulatory pathway of nisin biosynthesis.
3.8.1. DEGs Associated with Nisin Immunity Genes
Nisin mainly targets Gram-positive bacteria, and Lactococcus itself is also a gram-positive bacterium. The nisin immunity genes nisEFG and nisI were found to be present in the nisin synthesis-related gene cluster [18,29]. Due to the existence of immunity genes, Lactococcus has immune activity against nisin. As also mentioned in the introduction of this paper, as early as 1998, Kim tried to increase the production of nisin by overexpressing the nisin immunity gene [30]. Therefore, the improvement of nisin immunological activity must depend on the upregulation of immunity-related gene expression. The results of transcriptomic analysis verified this conclusion.
3.8.2. DEGs Related to DNA Replication, Transcription and Translation
During the fermentation and growth of L. lactis strains A32 and lxl, the logarithmic growth period of A32 obviously lagged behind that of lxl, which made the cumulative nisin production time longer, and the nisin yield higher, in the former. When performing our comparative omics analysis, we focused on the DNA enzyme synthesis-related gene dnaG, which had a base mutation. Although the change in its expression was not obvious in transcriptomics analysis, the expression of a single-stranded DNA-binding (SSB) protein bound to dnaG decreased significantly. This SSB protein is very important for DNA metabolism. First, it can stabilize the intermediate products of single-stranded DNA (ssDNA) produced during DNA processing. In addition, it can interact with 14 proteins and deliver reserve enzymes to DNA when needed to repair DNA damage [31,32]. It is speculated that the oligonucleotide/oligosaccharide-binding fold (OB-fold) of the dnaG protein was changed due to the gene mutation in dnaG [33], resulting in a decrease in its ability to bind to the SSB protein.
3.8.3. DEGs Related to ABC ATPase
ABC proteins refer only to proteins containing an ATP-binding box or an ABC-ATPase domain, and these proteins are involved in coupling ATP hydrolysis energy with many physiological processes that are not always, but usually, related to transport. On the other hand, when cytoplasmic ABC-ATPase binds to a hydrophobic membrane domain (MD), it forms an ABC transporter, which is synonymous with a trafficking ATPase (or permeability of the input system). Most (but not all) ABC transporters form six hypothetical helical transmembrane segments [34]. Romano showed that the ABC transporter mcjd has high specificity for its substrate, and the experiment also proved that the nisin transporter nisT cannot recognize MccJ25. Similarly, nisT is also an ABC transporter, and these transporters depend on the energy generated by ATP hydrolysis as an energy source [35,36]. In this study, many ABC-ATPase-related genes, including patM, tcyc, tcyj, adcA, yfmr and rexb, were mutated. According to the transcriptomic analysis, the expression of some phosphoamino acid transporter-related genes, such as phnE, phnC and rbsA, was significantly downregulated. It is speculated that changes occur in the distribution of ATP energy, and a portion of ATP energy flows into the pathway related to ABC transport, which promotes the excretion of nisin outside the cell.
3.8.4. DEGs Related to Cysteine Thiometabolism Translation
Nisin is a small-molecular polypeptide composed of various amino acid molecules. Amino acid metabolism in all cells is closely related to nisin synthesis. Nisin, as a lanthanide compound, has an intramolecular sulfide bridge similar to those in other lanthanide compounds. During the modification process from the nisin precursor to mature nisin, serine and threonine in the core peptide must be hydrated by the cysteine protease NisB, followed by the coupling of these dehydrated residues with cysteine by cyclase (NisC), which is homologous to asparagine synthase, to form a sulfide ring [37]. Studies have shown that the automatic regulation of signal transduction in nisin biosynthesis [38] and the antibacterial mechanism of nisin, including binding with lipid II and the inhibition of bacterial cell wall synthesis [39,40,41], are related to the presence of sulfide bonds.
In this study, the mutated genes also included some genes involved in cysteine sulfur metabolism, such as cysteine tRNA synthase (CysS) and iron sulfur reductase (YtqA). According to our transcriptomic analysis, the expression of cysteine synthase (CysK), which is a gene related to cysS, was significantly upregulated, and the expression of the whole cysteine metabolic pathway was upregulated and enriched. With respect to the biosynthesis of nisin, the cysteine thiometabolism pathway is related to the formation of sulfide bonds. CysS catalyses the CO conversion of cysteine polysulfide [42], which improves the efficiency of nisin modification.
3.8.5. DEGs Related to Purine Metabolism
The de novo biosynthesis of purine plays an important role in various metabolic pathways. It is involved in not only nucleotide synthesis, but also other biosynthetic processes. Pur-type purine synthesis-related genes are related to different physiological activities of microorganisms. Yanfei Xia proved that purL, whose product catalyses the transformation of 5’-phosphoribosyl-N-formylglycinamide (FGAR) into 5’-phosphoribosyl-N-formyl-glycinamidine (FGAM) in the purine synthesis pathway, is a gene that affects Bacillus subtilis nematocidal activity [43]. Buendia-Claver analysed Sinorhizobium fredii with a mutation in the purL gene, and found that its specific lipopolysaccharide (LPS) changed [44]. Studies have shown that mutation of the pur gene usually leads to significant weakening of the virulence of human and animal pathogens. Mutation of the purE gene in Brucella melitensis reduces its ability to infect mice [45], and mutations of purL and purF in Francisella tularensis were also shown to significantly reduce the toxicity of the strain [46]. Maegawa analysed the relationship between the production of Clostridium difficile toxin and purine synthesis, and found that the production of C. difficile toxin was negatively correlated with the expression of the purL gene [47].
In this study, comparative genomic analysis revealed base mutations in the purL gene. According to the transcriptomic analysis, the expression levels of purL, purC, purD and purE, which are involved in the purine metabolism pathway, were significantly downregulated. Quanli Liu tested the characteristics of a purL mutant (Bacillus amyloliquefaciens L4) and proved, for the first time, that purl inactivation can significantly enhance the production of subtilosin A by B. amyloliquefaciens L4, resulting in a production level three times higher than that in the wild-type strain [36]. It can be inferred that mutations in the purL gene reduce the activity of its protein and improve the ability of L. lactis A32 to produce nisin.
4. Conclusions
In this study, the mutant L. lactis A32 was screened from among lxl strain bacteria subjected to ARTP and UV irradiation. Mutant A32 presented improved nisin production ability and nisin resistance. The results of comprehensive analysis show that strain A32 obtained in this study had a delayed growth rate compared with that of the original lxl strain. The comparative genomics results show that 107 genes in the A32 genome had mutations, and most base mutations were not located in the four well-researched nisin-related operons. The overall transcriptome sequencing and analysis showed that the output of nisin increased in L. lactis strain A32, which was accompanied by changes in the DNA replication-related gene dnaG, the ABC-ATPase transport-related genes patM and tcyC, the cysteine thiometabolism-related gene cysS, and the purine metabolism-related gene purL. This research provides novel insights into the traditional genetic mechanisms involved nisin production in L. lactis, which was far beyond the four traditional well-known nisin-related operons. This study revealed a series of DEGs related to high nisin yields, which provides important genomic information and highlights potential influencing factors for further improving nisin yields.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation8060255/s1. Table S1: Results of the nisin production stability of strains L. lactis A32 and A225; Table S2: Statistics of the genome sequencing data of L. lactis A32 and lxl; Table S3: GO functional categories of L. lactis A32 and lxl; Table S4: Numbers and lengths of reads and numbers of expressed genes detected by transcriptome sequencing in control and acid-treated samples; Figure S1: Nisin titre standard curve; Figure S2: Biomass curve of L. lactis A32 and lxl with increasing fermentation times.
Author Contributions
The research was conceived and designed by L.H., X.L. (Xiaomeng Liu), D.Z. and X.L. (Xinli Liu); compost experiments and microbial communities were analysed by L.H., X.L. (Xiaomeng Liu), J.L., C.W., Q.W., S.P. and D.Z.; and the manuscript was written and revised by L.H., X.R., D.Z. and X.L. (Xinli Liu). All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (No. 31901821), as well as a grant from the State Key Laboratory of Biobased Material and Green Papermaking, Qilu University of Technology, Shandong Academy of Sciences (No. ZZ20200137).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that they have no competing financial interests.
References
- Yi, Y.; Zhang, Q.; Wilfred, A.V. Insights into the evolution of lanthipeptide biosynthesis. Protein Sci. 2013, 22, 1478–1489. [Google Scholar] [CrossRef]
- Joint FAO/WHO Expert Committee on Food Additives. Specifications for the Identity and Purity of Food Additives and Their Toxicological Evaluation: Some Antibiotics, Twelfth Report of the Joint FAO/WHO Expert Committee on Food Additives, Geneva, 1–8 July 1968; World Health Organization: Geneva, Switzerland, 1969; Available online: https://extranet.who.int/iris/restricted/handle/10665/40752 (accessed on 16 June 2012).
- Wu, H.; Teng, C.; Liu, B.; Tian, H.; Wang, J.G. Characterization and long-term antimicrobial activity of the nisin anchored cellulose films. Int. J. Biol. Macromol. 2018, 113, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Wenting, L.; Rong, Z.; Tengteng, J.; Dur, E.; Sameena; Saeed, A.; Wen, Q. Improving nisin production by encapsulated Lactococcus lactis with starch/carboxymethyl cellulose edible films. Carbohydr. Polym. 2021, 251, 117062. [Google Scholar] [CrossRef]
- Jung, Y.; Alayande, A.B.; Chae, S.; Kim, I.S. Applications of nisin for biofouling mitigation of reverse osmosis membranes. Desalination 2018, 429, 52–59. [Google Scholar] [CrossRef]
- Andre, C.; de Jesus Pimentel-Filho, N.; de Almeida Costa, P.M.; Vanetti, M.C.D. Changes in the composition and architecture of Staphylococcal biofilm by nisin. Braz. J. Microbiol. 2019, 50, 1083–1090. [Google Scholar] [CrossRef] [PubMed]
- Eming, S.A.; Martin, P. Wound repair and regeneration: Mechanisms, signaling, and translation. Sci. Transl. Med. 2014, 6, 265sr6. [Google Scholar] [CrossRef]
- Mouritzen, M.V.; Andrea, A.; Qvist, K.; Poulsen, S.S.; Jenssen, H. Immunomodulatory potential of Nisin A with application in wound healing. Wound Repair Regen. 2019, 27, 650–660. [Google Scholar] [CrossRef]
- Wei, Q.; Kun, Y.; Jun, L.; Ke, L.; Fuqiang, L.; Junhui, J.; Wei, Z. Precise management of chronic wound by nisin with antibacterial selectivity. Biomed. Mater. 2019, 14, 045008. [Google Scholar] [CrossRef]
- Malvano, F.; Pilloton, R.; Albanese, D. A novel impedimetric biosensor based on the antimicrobial activity of the peptide nisin for the detection of Salmonella spp. Food Chem. 2020, 325, 126868. [Google Scholar] [CrossRef]
- Malaczewska, J.; Kaczorek-Łukowska, E.; Wójcik, R.; Rękawek, W.; Andrzej, K.S. In vitro immunomodulatory effect of nisin on porcine leucocytes. J. Anim. Physiol. Anim. Nutr. 2019, 103, 882–893. [Google Scholar] [CrossRef]
- Bagde, P.; Nadanathangam, V. Improving the stability of bacteriocin extracted from Enterococcus faecium by immobilization onto cellulose nanocrystals. Carbohydr. Polym. 2019, 209, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Rana, K.; Sharma, R.; Preet, S. Augmented therapeutic efficacy of 5-fluorouracil in conjunction with lantibiotic nisin against skin cancer. Biochem. Biophys. Res. Commun. 2019, 520, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Joo, N.E.; Ritchie, K.; Kamarajan, P.; Miao, D.; Kapila, Y.L. Nisin, an apoptogenic bacteriocin and food preservative, attenuates HNSCC tumorigenesis via CHAC1. Cancer Med. 2012, 1, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Siegers, K.; Entian, K.D. Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl. Environ. Microbiol. 1995, 61, 1082–1089. [Google Scholar] [CrossRef]
- Ra, S.R.; Qiao, M.; Immonen, T.; Pujana, I. Genes responsible for nisin synthesis, regulation and immunity form a regulon of two operons and are induced by nisin in Lactoccocus lactis N8. Microbiology 1996, 142, 1281–1288. [Google Scholar] [CrossRef]
- Qiao, M.; Ye, S.; Koponen, O.; Ra, R.; Usabiaga, M.; Immonen, T.; Saris, P.E.J. Regulation of the nisin operons in Lactococcus lactis N8. J. Appl. Bacteriol. 1996, 80, 626–634. [Google Scholar] [CrossRef]
- Li, H.; O’Sullivan, D.J. Identification of a nisI promoter within the nisABCTIP operon that may enable establishment of nisin immunity prior to induction of the operon via signal transduction. J. Bacteriol. 2006, 188, 8496–8503. [Google Scholar] [CrossRef]
- Patnaik, R. Engineering complex phenotypes in industrial strains. Biotechnol. Prog. 2008, 24, 38–47. [Google Scholar] [CrossRef]
- Jian, Z.; Qinggele, C.; Wenjing, F.; Xiuli, Z.; Bin, Q.; Guangrong, Z.; Jianjun, Q. Enhance nisin yield via improving acid-tolerant capability of Lactococcus lactis F44. Sci. Rep. 2016, 6, 27973. [Google Scholar] [CrossRef]
- Shifu, C.; Yanqing, Z.; Yaru, C.; Jia, G. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Yu, L.; Pevzner, P.A. Assembly of long error-prone reads using de Bruijn graphs. Proc. Natl. Acad. Sci. USA 2016, 113, 8396–8405. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Tjaden, B. De novo assembly of bacterial transcriptomes from RNA-seq data. Genome Biol. 2015, 16, 1. [Google Scholar] [CrossRef]
- Hacker, C.; Christ, N.A.; Duchardt-Ferner, E.; Korn, S.; Berninger, L. The Solution Structure of the Lantibiotic Immunity Protein NisI and Its Interactions with Nisin. J. Biol. Chem. 2015, 290, 28869–28886. [Google Scholar] [CrossRef]
- Kim, W.S.; Hall, R.J.; Dunn, N.W. Improving nisin production by increasing nisin immunity/resistance genes in the producer organism Lactococcus lactis. Appl. Microbiol. Biotechnol. 1998, 50, 429–433. [Google Scholar] [CrossRef]
- Huang, Y.H.; Huang, C.Y. Comparing SSB-PriA Functional and Physical Interactions in Gram-Positive and -Negative Bacteria. Methods Mol. Biol. 2021, 2281, 67–80. [Google Scholar] [CrossRef]
- Bianco, P.R. The mechanism of action of the SSB interactome reveals it is the first OB-fold family of genome guardians in prokaryotes. Protein Sci. 2021, 30, 1757–1775. [Google Scholar] [CrossRef] [PubMed]
- Bianco, P.R.; Pottinger, S.; Tan, H.Y.; Nguyenduc, T.; Rex, K.; Varshney, U. The IDL of E. coli SSB links ssDNA and protein binding by mediating protein-protein interactions. Protein Sci. 2017, 26, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Young, J.; Holland, I.B. ABC transporters: Bacterial exporters-revisited five years on. Biochim. Biophys. Acta (BBA) Biomembr. 1999, 1461, 177–200. [Google Scholar] [CrossRef]
- Romano, M.; Fusco, G.; Choudhury, H.G.; Mehmood, S.; Robinson, C.V.; Zirah, S.; Hegemann, J.D. Structural basis for natural product selection and export by bacterial ABC Transporters. ACS Chem. Biol. 2018, 13, 1598–1609. [Google Scholar] [CrossRef]
- Beis, K.; Rebuffat, S. Multifaceted ABC transporters associated to microcin and bacteriocin export. Res. Microbiol. 2019, 170, 399–406. [Google Scholar] [CrossRef]
- Hegemann, J.D.; Fouque, K.J.D.; Santos-Fernandez, M.; Fernandez-Lima, F. A bifunctional leader peptidase/ABC transporter protein is involved in the maturation of the lasso peptide cochonodin I from Streptococcus suis. J. Nat. Prod. 2021, 84, 2683–2691. [Google Scholar] [CrossRef]
- Kuipers, O.P.; Beerthuyzen, M.M.; Pascalle, G.; Luesink, E.J.; Willem, M.V. Autoregulation of Nisin Biosynthesis in Lactococcus lactis by Signal Transduction. J. Biol. Chem. 1995, 270, 27299–27304. [Google Scholar] [CrossRef]
- Sahl, H.G.; Jack, R.W.; Bierbaum, G. Biosynthesis and biological activities of lantibiotics with unique post-translational modifications. Eur. J. Biochem. 1995, 230, 827–853. [Google Scholar] [CrossRef]
- Driessen, A.J.M.; Hooven, H.W.; Kuiper, W.; Kamp, M.; Sahl, H.G.; Konings, R.N.H.; Konings, W.N. Mechanistic studies of lantibiotic-induced permeabilization of phospholipid vesicles. Biochemistry 1995, 34, 1606–1614. [Google Scholar] [CrossRef]
- Moll, G.N.; Konings, W.N.; Driessen, A.J. The lantibiotic nisin induces transmembrane movement of a fluorescent phospholipid. J. Bacteriol. 1998, 180, 6565–6570. [Google Scholar] [CrossRef]
- Akaike, T.; Ida, T.; Wei, F.Y.; Nishida, M.; Kumagai, Y.; Alam, M.M. Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat. Commun. 2017, 8, 177. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.F.; Xie, S.S.; Ma, X.; Wu, H.J.; Wang, X.; Gao, X.W. The purL gene of Bacillus subtilis is associated with nematicidal activity. FEMS Microbiol. Lett. 2011, 322, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Buendía-Clavería, A.M.; Moussaid, A.; Ollero, F.J.; Vinardell, J.M.; Torres, A. A purL mutant of Sinorhizobium fredii HH103 is symbiotically defective and altered in its lipopolysaccharide. Microbiology 2003, 149, 1807–1818. [Google Scholar] [CrossRef]
- Crawford, R.M.; Verg, L.V.D.; Yuan, L.; Hadfield, T.L.; Warren, R.L.; Drazek, E.S.; Houng, H.H.; Hammack, C. Deletion of purE attenuates Brucella melitensis infection in mice. Infect. Immun. 1996, 64, 2188–2192. [Google Scholar] [CrossRef]
- Kadzhaev, K.; Zingmark, C.; Golovliov, I.; Bolanowski, M.; Shen, H. Identification of genes contributing to the virulence of Francisella tularensis SCHU S4 in a mouse intradermal infection model. PLoS ONE 2009, 4, e5463. [Google Scholar] [CrossRef] [PubMed]
- Maegawa, T.; Karasawa, T.; Ohta, T.; Wang, X.; Kato, H.; Hayashi, H.; Nakamura, S. Linkage between toxin production and purine biosynthesis in Clostridium difficile. J. Med. Microbiol. 2002, 51, 34–41. [Google Scholar] [CrossRef] [PubMed][Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).