The effect of fermentation time and temperature (independent variables) on pH, TSS, TTA and consistency (dependent variables) of
amasi were investigated. Central composite design (CCD) was used to study the relationship between the independent and dependent parameters, and later used to quantify the optimum conditions.
Table 3 represents the physicochemical analysis (pH, TSS, TTA and consistency), and
Table 4 represents the analysis of variance (ANOVA) that was done based on the experimental runs (
Table 2). The data was later analyzed using multi-response numerical optimization (Minitab) and k-means to obtain the optimal sample (OP), and physicochemical analysis was also repeated on the optimal sample.
3.1. Optimization of Fermentation Conditions (Time and Temperature) of Amasi
The experimental results on the effect of fermentation time and temperature on the processing of
amasi from raw milk are shown in
Table 3. The ANOVA (
Table 4) was used as part of the search for a solution to examine the best fit of the produced model. The capability of the models was obtained by the coefficients of the determination values and lack-of-fit tests [
16,
34]. The optimized data is represented in
Table 5; the experimental data was analyzed to investigate the lack-of-fit and the importance of the quadratic model, as well as the effect of interaction between the input and output variables.
The lack-of-fit test was defined by Gan et al. [
35] as a dimension of the let-down of a model to display the trial results at the point not shown in the regression, and Lima et al. [
36] found that the goodness-of-fit of the model was obtained by the constant of determination (R
2) and can be approximately 80%. Accordingly, the
p-value can be used to determine the significance of each constant in a data set. When the F-value is high and the
p-value is low, it means that the conforming constant is more important.
The F-values and
p-values of ≤ 0.05 indicate that the quadratic model of
amasi development is statistically important at 95% confidence. Except for accuracy, which was negligible with a
p-value of 0.440, the lack of fit for each response factor is important. The R
2 values for all the responses were less than or equal to 80%, indicating that the model clearly explains a large proportion of the variability [
37]. The 3D response surface plots for pH, TTA, TSS and consistency are given in
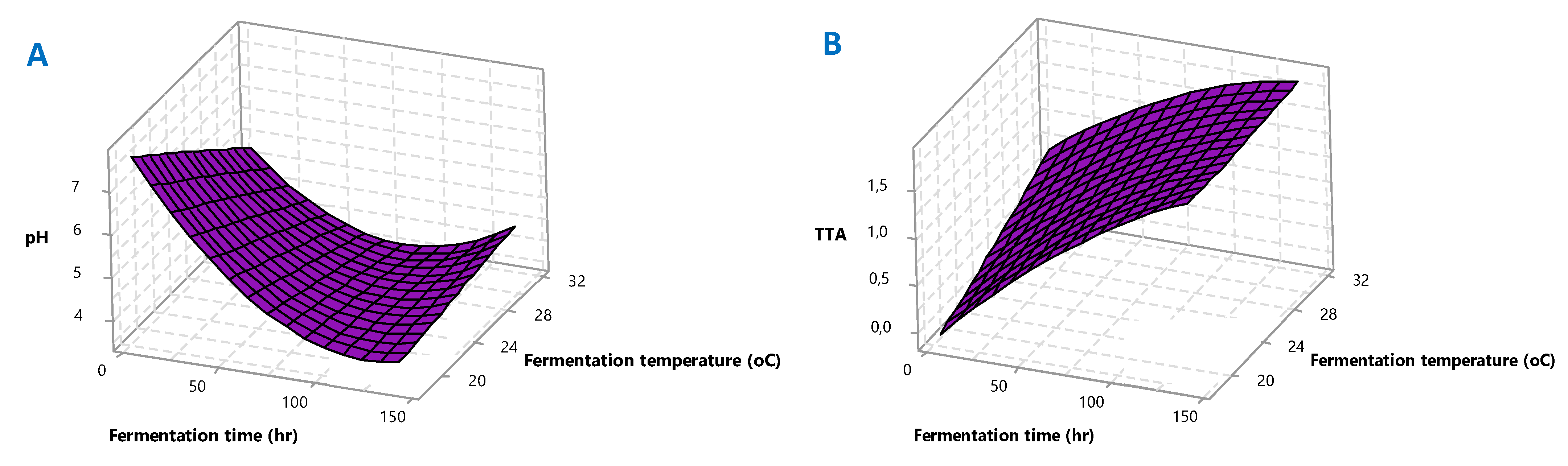
Figure 2A–D for better visualization. The polynomial equations and 3D plots were used to identify the mathematical solution of optimization.
3.2. The Effect of Fermentation Conditions on Physicochemical Properties of Amasi
The pH, TTA, TSS and consistency of
amasi were taken at the end of each fermentation, and is illustrated in
Table 3. The changes of physicochemical properties were affected by the fermentation conditions (time and temperature). As anticipated, the pH value of raw milk significantly (
p ≤ 0.05) decreased from the initial value of 6.64 (
Table 5) throughout the whole fermentation time. It was observed that the highest pH value of 6.76 was recorded at 4.11 h, which decreased to 3.83 (43.34% decrease) after 139 h. This was in correspondence with the TTA of 0.02% at 4.11 h, and this increased to 1.17% at the longest time. This was expected since the value of pH is inversely proportional to the lactic/organic acids content [
38]. A similar decrease in pH and simultaneous increase in TTA with the increasing fermentation time have been reported in other studies [
8,
16]. The acidic conditions within the environment allow most fermenting microorganisms such as LAB species (Lactococcus and Streptococcus) to be more active, thus causing a low pH and high TTA value. These microorganisms produce lactic acid and other organic acids; this might be due to the degradation of some carbohydrate and sugar-related components [
39,
40]. The inverse relationship of pH and TTA is good for the product because, in acidic conditions, most pathogenic microorganisms are inhibited, thereby improving the microbiological safety of the product [
41]. The use of low fermentation temperatures (20–25 °C) resulted in high pH and low TTA values (6.32–6.76; 0.02–0.02%), as compared to the high fermentation temperatures (25–32 °C) that resulted in low pH and high TTA values (3.83–3.96; 0.11–0.17%). The high pH values observed could be associated with the increased metabolic activities of the fermenting microorganisms [
19].
Central composite design was used to optimize the fermentation conditions using the mini tab; the polynomial model for responses (
Y1—pH,
Y2—titratable acidity,
Y3—total soluble solids,
Y4—consistency) was regressed by taking only the significant terms into consideration, and the equations for each response are represented below:
where
Y1 is the pH,
X1 is the fermentation time, and
X2 is the fermentation temperature. The R
2 value of the equation was 0.92 (92%); this indicated that the regression model could explain 92% of the data variability. The one-degree term
X1 and
X2 were significant at
p ≤ 0.05, suggesting that fermentation temperature and fermentation time have a significant linear effect on pH values. The effect is also reflected in the response surface plot (
Figure 2A). As observed in
Table 6, only one-degree term
X1 and
X2 were extremely significant (
p ≤ 0.05); this means that fermentation time and fermentation temperature have a significant effect on TTA values (
Y2). It can also be visually observed in
Figure 2B. On the other hand,
X22 had no significant effect on all the investigated parameters. For the TSS model (
Y3), only
X1,
X12, and
X1X2 were significant (
Table 6), and R
2 was found to be 0.92 (
Table 4), portraying 92% reproducibility. On the other hand, in consistency, it was
X1,
X2 and
X12 that had an R
2 value of 0.80. The singular effects
X1 and
X2 were significant at
p ≤ 0.05, suggesting that fermentation temperature and time have a significant linear effect on TSS and consistency. The effect is also reflected in the response surface plot (
Figure 2C,D).
The experimental values of TSS (
Y3) and consistency (
Y4) of the
amasi samples are represented in
Table 3. The F-value (16.21) and
p-value (0.001) indicated that the model for optimizing TSS in
amasi was significant (
Table 4). The TSS value of
amasi decreased with an increasing fermentation time. The lowest TSS value of
amasi observed was with a higher fermentation time (140 h) which was 5.80 °Brix and the highest was 10 °Brix and 9.60 °Brix at a lower fermentation time (24 and 4 h). This suggests that TSS of
amasi depends on the acid production and as the pH values were decreasing, the TSS values also decreased. This is also an indication that longer fermentation periods resulted in increased sugar metabolism and biotransformation of these components [
42]. A similar trend was reported in yoghurt samples, where the TSS values decreased with the decreasing pH values and increased fermentation temperature [
43].
The consistency (cm) of
amasi samples show a similar trend to TSS; a more viscous sample was observed at higher temperatures. For instance, at 32 °C, a consistency of 11 cm was observed, while a higher consistency of 23 cm was observed at lower temperatures (20 °C). A higher consistency indicates that the sample was less viscous compared to that of a higher consistency.
Amasi at 32 °C had more whey as it was fermented at higher temperatures, and this caused the product to be thicker because the whey was removed at the end of the fermentation. The result was also similar to that reported in
mabisi from Zambia [
8], as well as in a fermented milk product from Malaysia [
16]. There are limited studies that have reported the consistency of natural fermented dairy products using Bostwick or any equipment to measure consistency. Monyane and Jideani [
44] only analysed the viscosity of commercially produced
amasi using a viscometer.
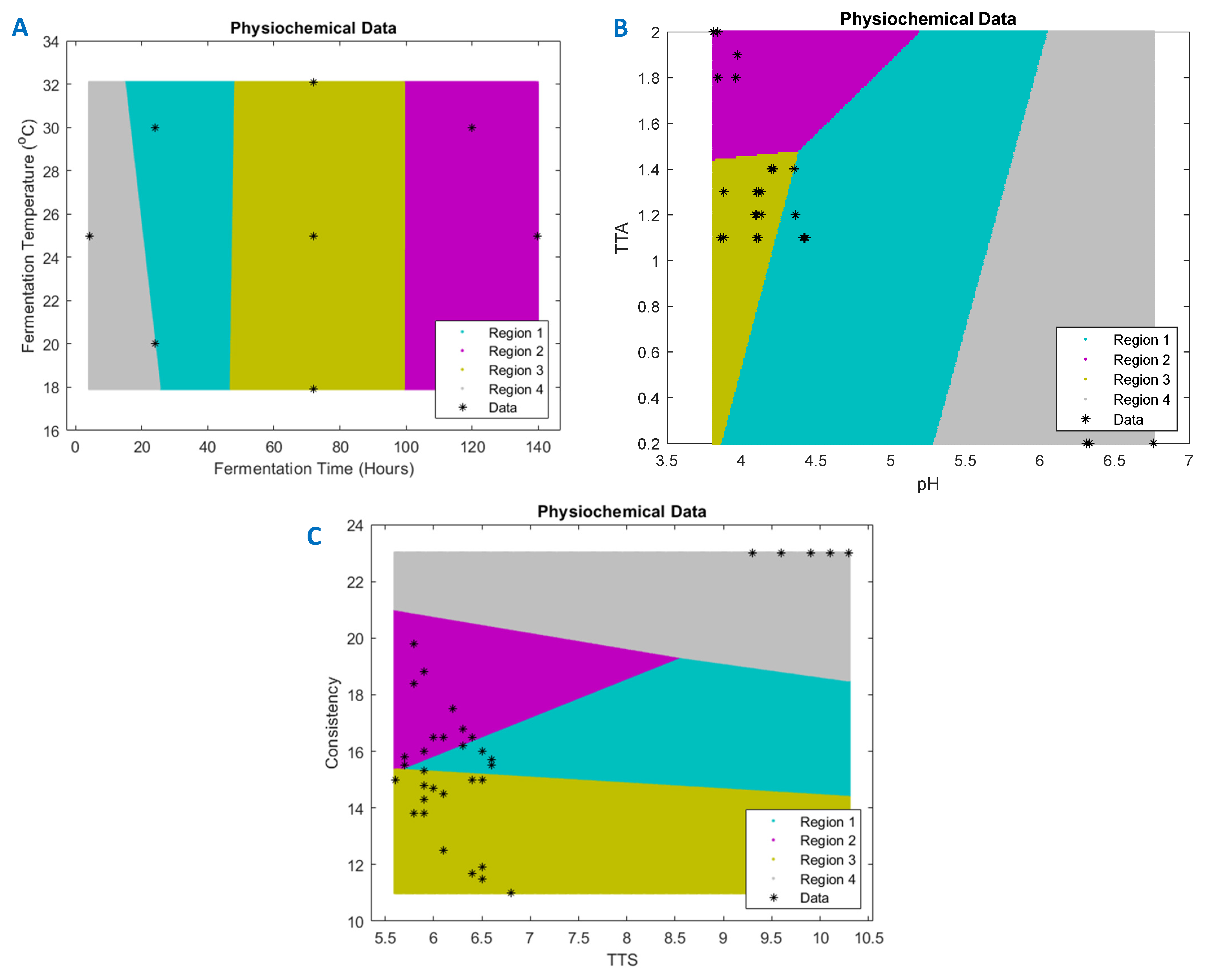
3.3. RSM Model Validation Using k-Means Clustering
The k-means clustering is a method used to obtain clusters and cluster centres in a data set. The data set obtained from the RSM experimental runs (
Table 2) was used to generate the k-means clustering on MATLAB R2020a (MathWorks, Natick, MA, USA). The input features analysed in this study were fermentation time and fermentation temperature, while the output features were pH, TTA, TSS, and consistency. The rows were labelled as observations and columns as variables. The model generated four regions per plot (
Figure 1A–C), and each cluster was represented in a different colour of a region. The desired number of clusters that was chosen for this study was four, and the k-means procedure iteratively moved the centres to reduce the total with the cluster variance.
The k-means clustering technique is a very simple technique that can be used in combination with the RSM technique to go beyond point estimates and group experimental instances into clusters and analyse regions of optimality. Additionally, it can competently deal with a large data set at once, although there are some weaknesses of using k-means. A drawback of this technique is that it is unsupervised and thus the number of clusters need to be specified. The resulting optimal fermentation conditions were 32 °C at 140 h and 25 °C at 120 h. According to the k-means clustered results, these optimal points are within two of the clustered ‘regions’ or solution subspaces. The response variables for these conditions, which were done in triplicate, are represented in
Table 5.
3.4. Amplicon Sequencing/Metabarcoding of the Naturally Fermented-Amasi
Most natural fermented dairy products are dominated by species of acetic acid and lactic acid bacteria. The bacterial composition between samples of the same product may differ due to variations in their source of raw material, processing, fermentation time, and temperature. Moonga et al. [
45] observed that Zambian fermented milk,
mabisi (a product similar to
amasi), depended on the production method, pH, and geographical location of the sample. This study investigated the bacterial composition of three different samples using next generation sequencing; the extracted DNA was sequenced for 16S rRNA on an Illumina MiSeq at Inqaba Biotech. The raw cow milk sample (RM) was compared with two
amasi samples fermented at different temperatures: OP25 and OP32. The results obtained showed that fermentation conditions have an influence on the microbial community of fermented dairy products. Previous studies on the microbiota of
amasi processed from raw milk included various species from LAB, Enterococcus, and yeast [
4,
13,
15,
45].
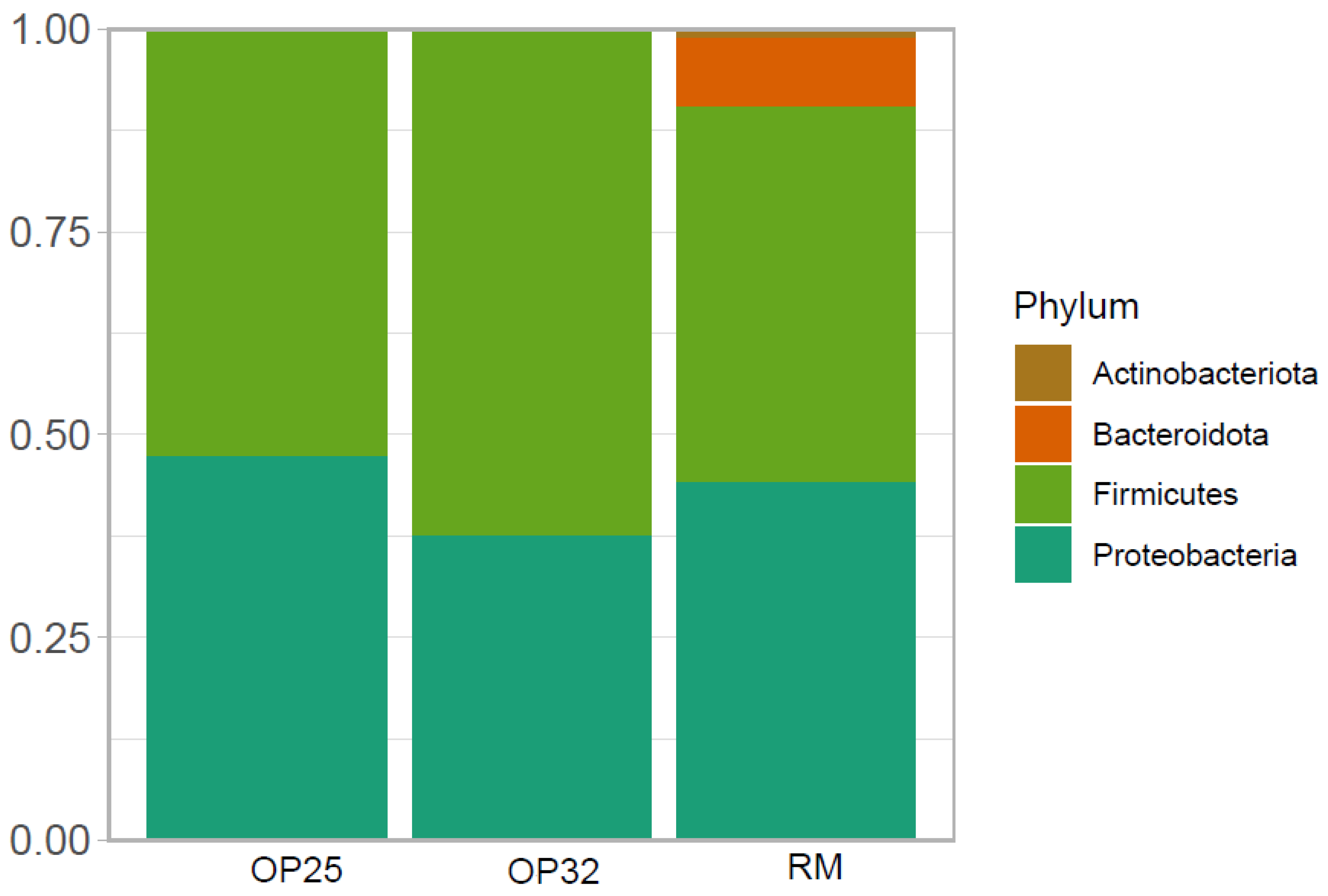
3.4.1. Microbiological Composition of Bacterial Communities at Phyla Level
In the present study, four phyla were detected in the three samples (RM, OP25 and OP32), and are summarized in
Figure 3. The bacterial reads of
amasi were clustered further into six classes, 20 families, and six species (discussed in subsequent sections). The bacterial composition of all three samples were dominated by Firmicutes and Proteobacteria. Bacteroidetes was only dominant in raw milk as compared to the two
amasi samples. The most dominant phylum in all three samples was Firmicutes. The members of the phyla Firmicutes and Proteobacteria were also reported by [
15] in
mabisi, a fermented dairy product from Zambia. Comparing the raw milk with the two
amasi samples, the bacterial composition at the phylum level seems to be decreasing excessively from the raw milk sample. As the fermentation temperature and time were increased for
amasi, the Firmicutes also increased in the M32 sample due to a decreased pH which could give an increased bacterial composition. A similar trend was also reported by [
43], where an elevation of fermentation temperature and low pH favoured the survival of
Lactobacillus, which belongs to the phyla Firmicutes.
3.4.2. Microbiological Composition of Bacterial Communities at Class Level
The bacterial community at class level was more abundant in raw milk than in fermented milk. In the three classes observed (
Figure 4),
Bacilli was the most abundant in all three samples. Within the phylum Firmicutes,
Bacilli was dominant in raw milk, followed by
Bacteroidetes and
Gammaproteobacteria from the phyla
Bacteroidetes and
Proteobacteria.
Alphaproteobacteria was the second most abundant class in both
amasi fermented at 25 and 32 °C. In
amasi fermented at 25 °C,
Gammaproteobacteria was the third abundant. At low pH levels between 3.83–4.10, the two classes (
Clostridia and
Actinobacteria) were observed to be less abundant in
amasi fermented at 25 and 32 °C.
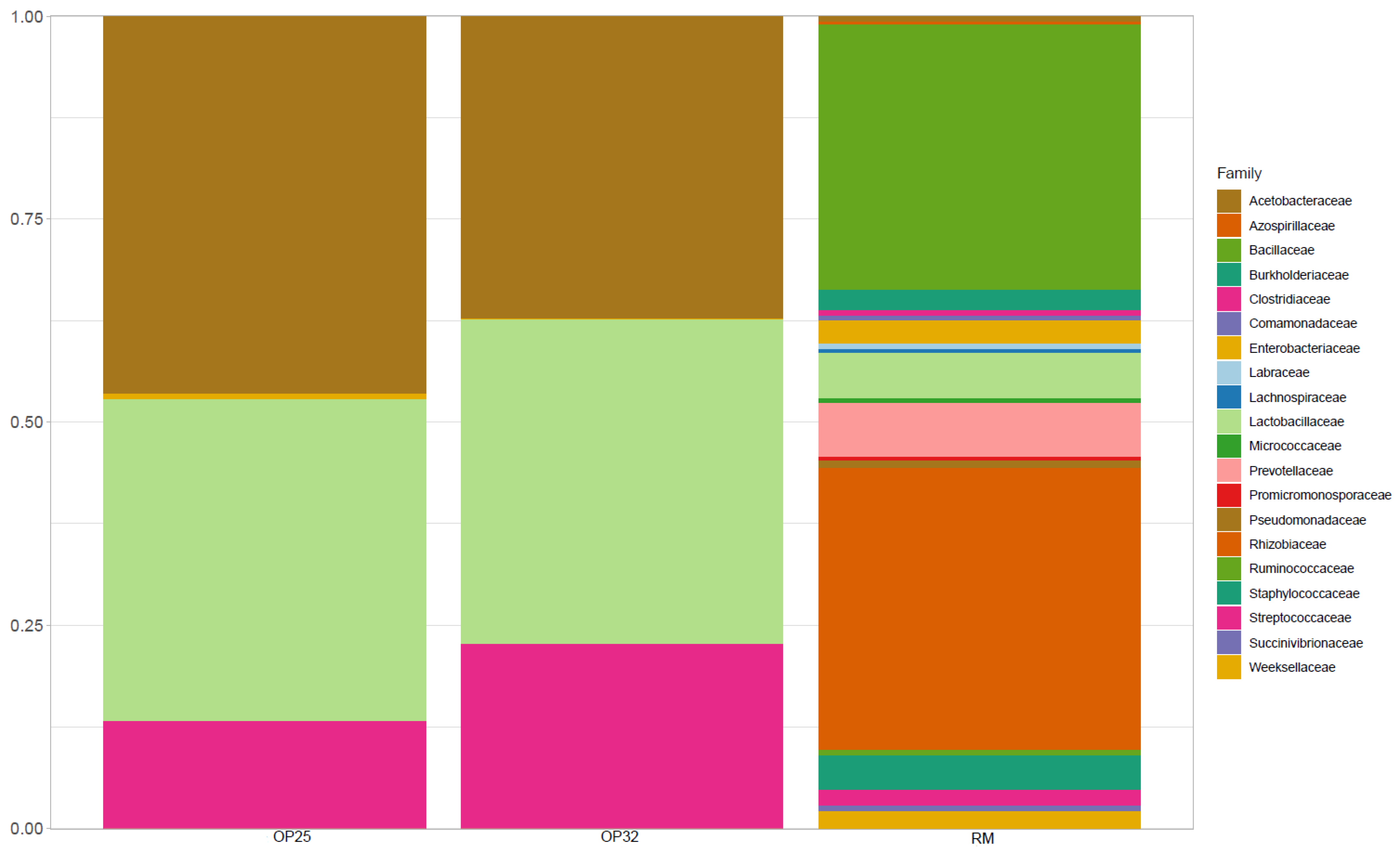
3.4.3. Microbiological Composition of Bacterial Communities at Family Level
A total of 20 bacterial communities at the family level were reported (
Figure 5).
Lactobacillaceae was found to be the most abundant in both amasi samples fermented at 25 and 32 °C, but was less abundant in the raw milk sample.
Prevotellaceae was the most dominant in raw milk, while it was found in less abundance for the
amasi samples.
Streptococcaceae is observed to be in high abundance, followed by
Acetobacteraceae in M32, but found to be decreasing in M25. They were low in abundance from RM, and started to increase in M25 as the pH decreased from 6.64 to 3.99 (25 °C), and it further increased in M32 as pH decreased to 3.44 (32 °C).
Prevotellaceae, which was more dominated in the raw milk, started to decrease when the fermentation was initiated. De Waal [
10], reported the presence of
Prevotell copri (AB064923) species within family
Prevotellaceae on fresh unfermented milk, while it was absent in fermented milk. The abundance of
Lactobacillaceae and
Streptococcaceae over other families suggested the dominance of LAB during the fermentation process, and this was equally reported in other studies [
8,
10,
46]. In Southern Africa, raw cow milk is rarely consumed due to the occurrence of lactose intolerance and other safety reasons. During the processing of
amasi, most of the lactose is converted into lactic acid and other compounds by microorganisms present during fermentation. This also facilitates better nutritional contents in
amasi as compared to raw milk. The dominance of
Lactobacillaceae and
Streptococcaceae in
amasi is acceptable due to LAB being considered as a healthy bacterium that may allow shifts in gut microbiota composition and healthier composition [
46].
According to Moonga [
15], fermentation temperature (25–30 °C) and pH (4.1–4.3) have an influence in determining the composition of bacteria in fermented milk. In this study, fermentation temperature and pH allowed the growth of some species from the families that were observed in
Figure 5. At species level, most species were not detected; this was also reported in previous studies [
10,
13,
46]. This could be due to the short fragment that was used for sequencing because only V3–V4 hypervariable regions were sequenced in this study.
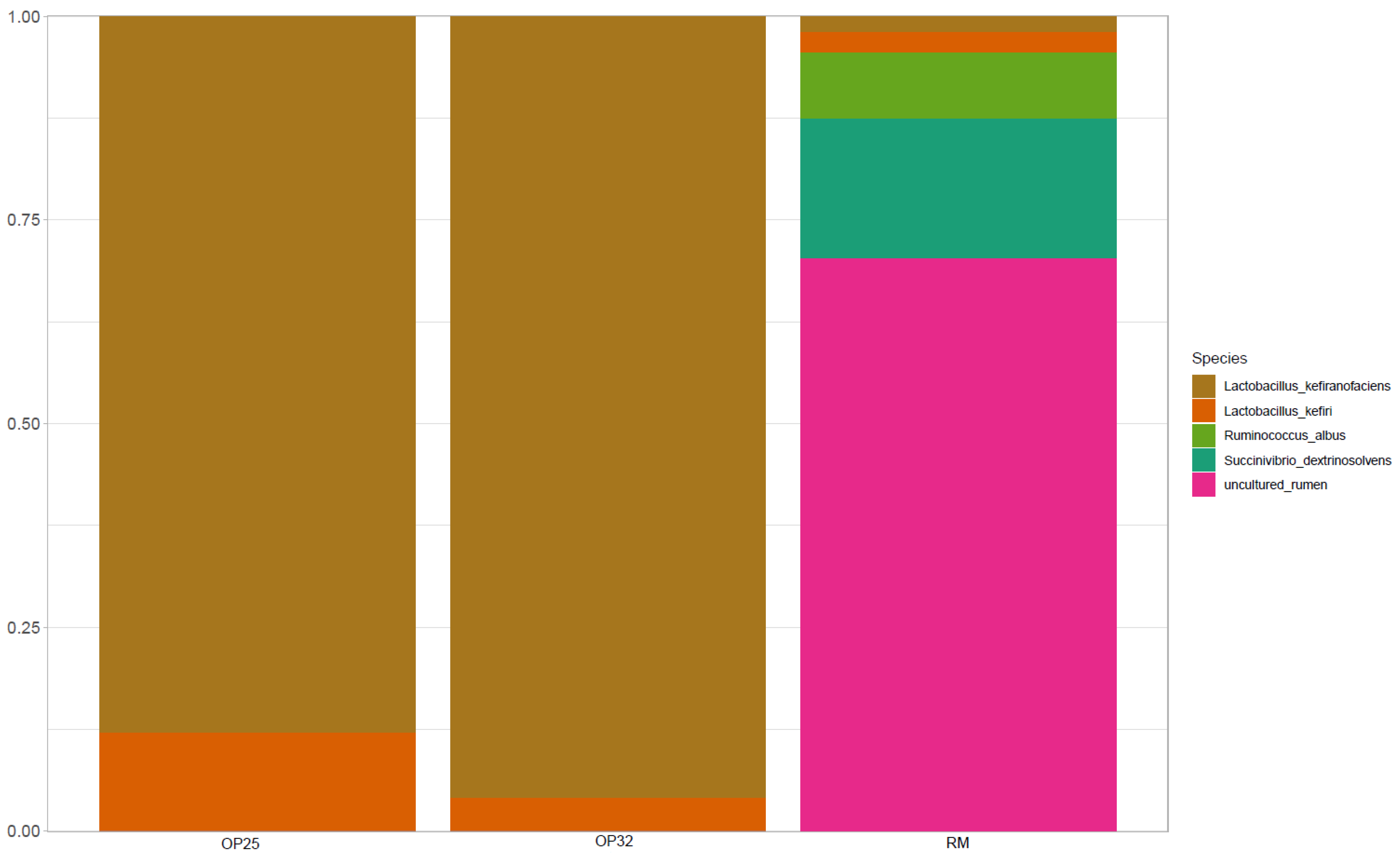
3.4.4. Microbiological Composition of Bacterial Communities at Species Level
At species level, the dominant species in all three samples (
Figure 6) were potentially classified as ‘N/A’, followed by uncultured rumen, which was only dominant in raw milk. While in OP32 and OP25 it was observed in low abundance,
Lactobacillus kefiranofaciens was found to be the second dominant species, but less abundant in OP25. The
Lactobacillus kefiranofaciens species was observed to be in low abundance in raw milk.
Lactobacillus kefiri slightly increased as fermentation was initiated.
Lactobacillus kefiranofaciens was reported to be in abundance in
mabisi in a study reported by Schoustra et al. [
4], while
Lactobacillus kefiri was reported in
mursik, a fermented milk product from Kenya [
6], and also in
omashikawa from Namibia [
3]. The results show that fermentation temperature has a significant influence on the bacterial community composition. The low pH also plays an important role, as an acidic environment is required by most microorganisms for the fermentation process.