Investigation of Microbial Hydrolysis of Hen Combs with Bacterial Concentrates
Abstract
:1. Introduction
2. Materials and Methods
2.1. Raw Materials and Ingredients
2.2. Biotechnological Treatment of the Combs
2.3. Optimization of Hydrolysis
2.4. Determination of Chemical Properties
2.5. Determination of Free Amino Acids in Comb Hydrolysates
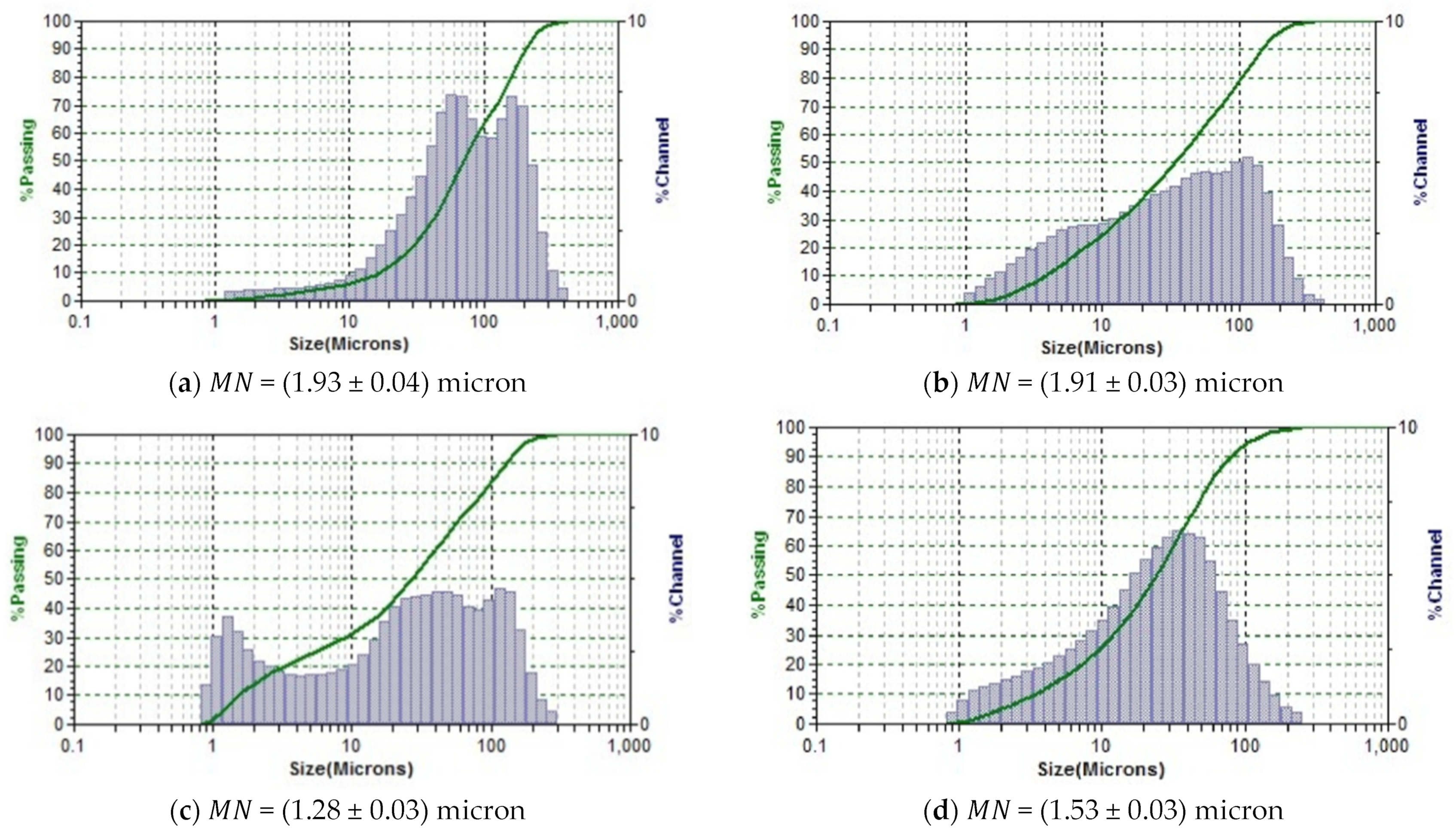
2.6. Determination of the Dispersed Composition
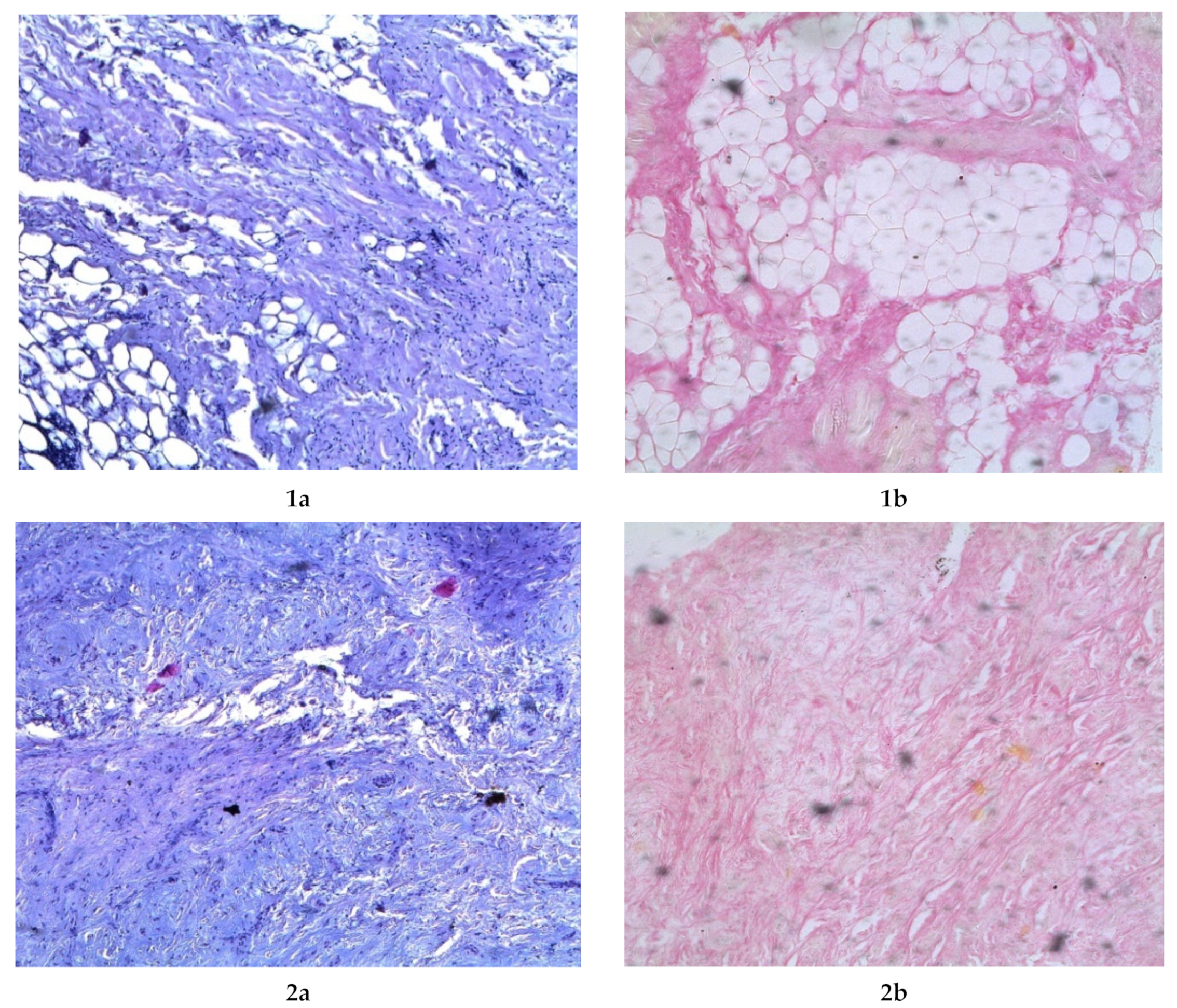
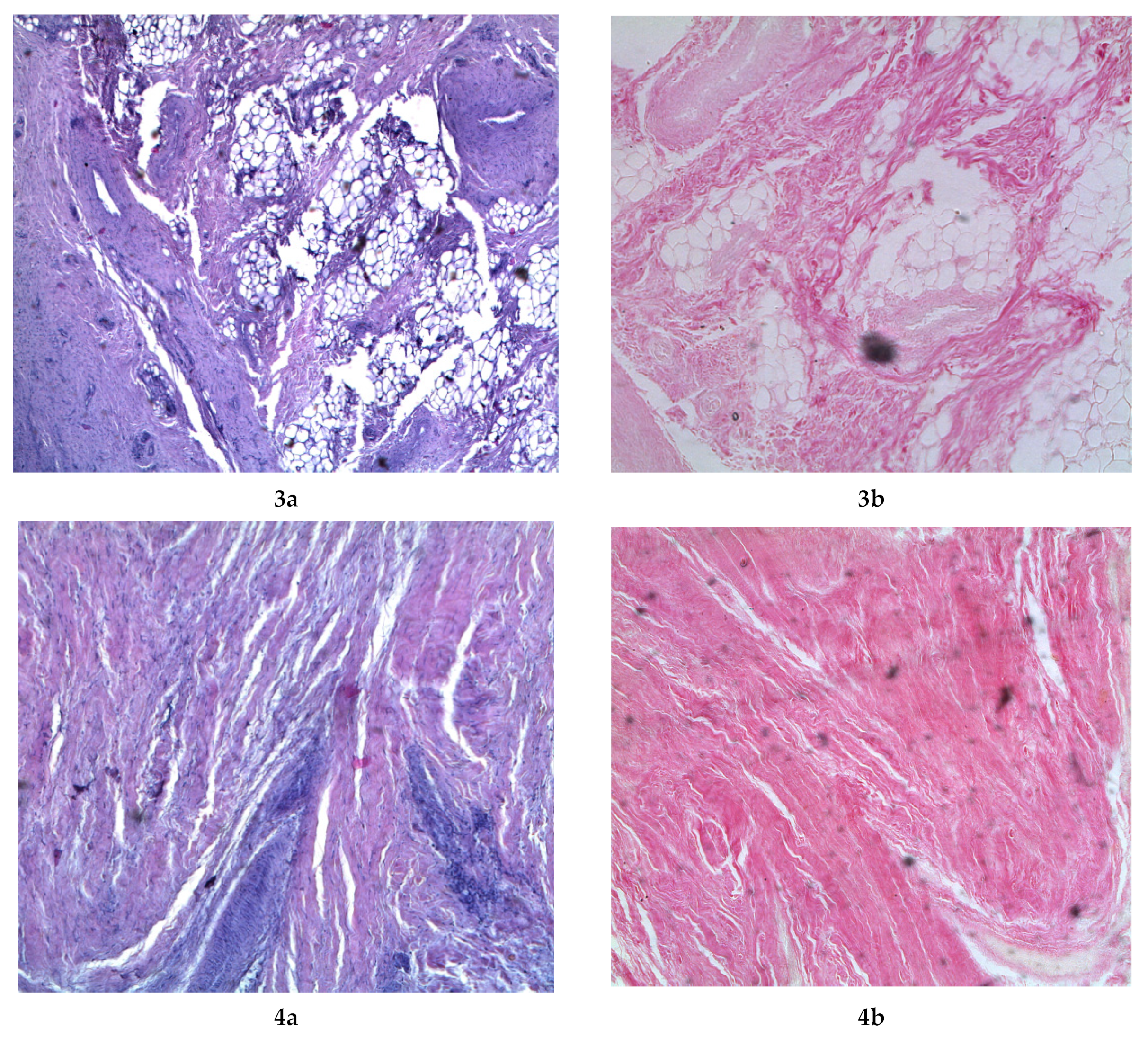
2.7. Investigation of Microstructure
2.8. Statistical Analysis
3. Results and Discussion
3.1. Optimization of Hydrolysis
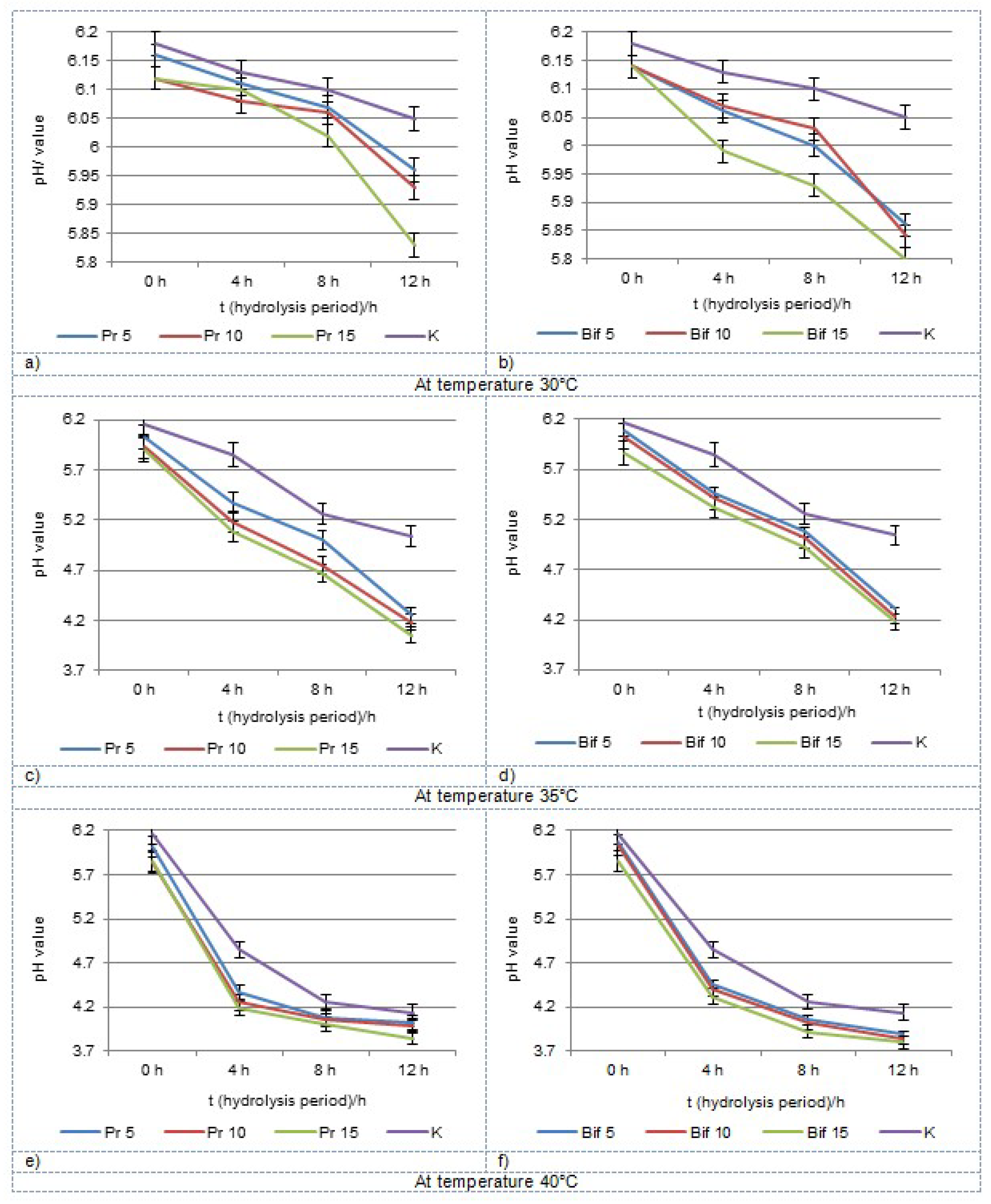
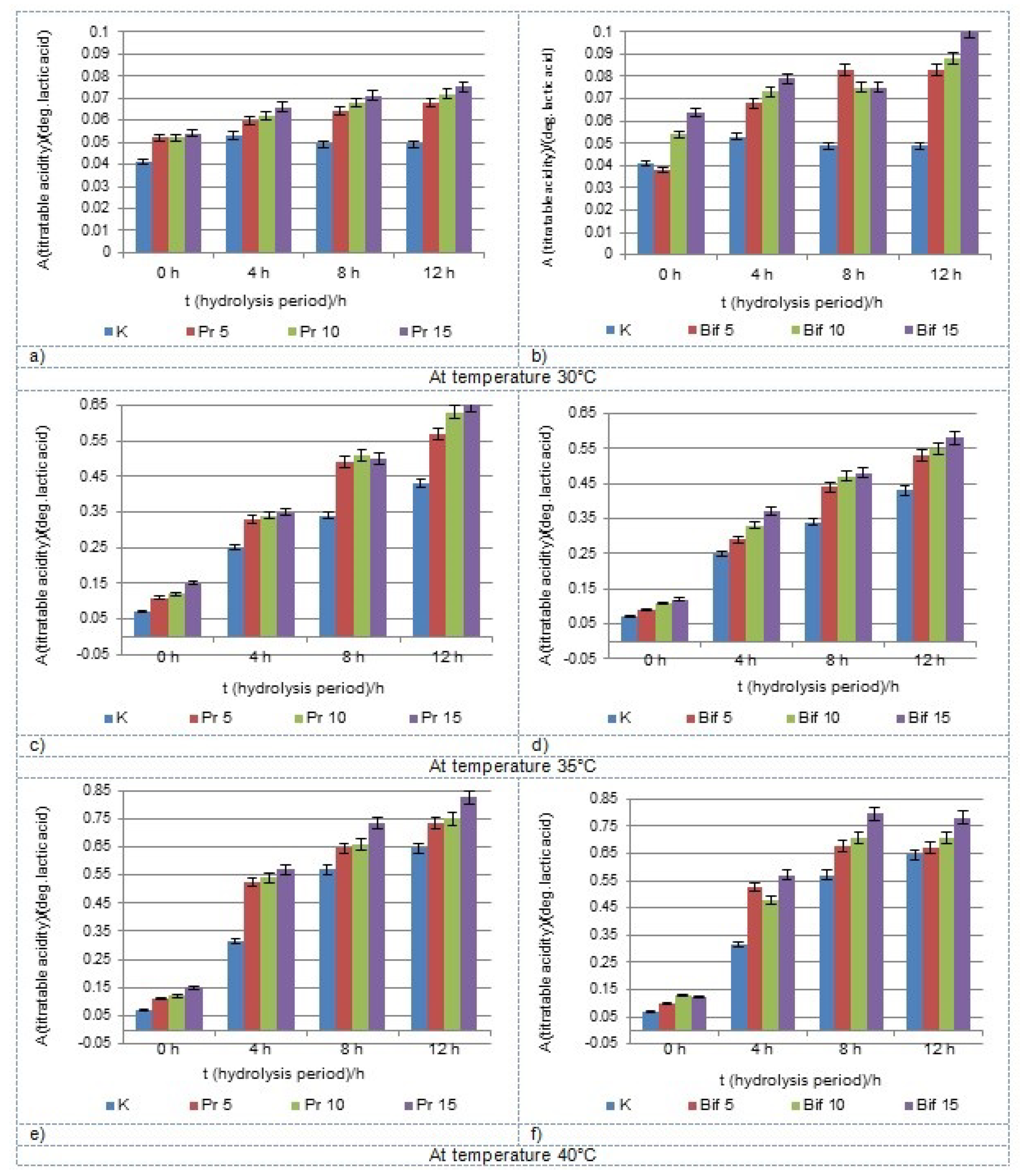
3.2. Chemical Properties
3.3. Dispersed Composition, Free Amino Acids and Microstructure of the Comb Hydrolysates
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mullen, A.M.; Álvarez, C.; Zeugolis, D.I.; Henchion, M.; O’Neill, E.; Drummond, L. Alternative uses for co-products: Harnessing the potential of valuable compounds from meat processing chains. Meat Sci. 2017, 132, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Brandelli, A.; Sala, L.; Kalil, S.J. Microbial enzymes for bioconversion of poultry waste into added-value products. Food Res. Int. 2015, 73, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, T.; Ismail, A.; Ahmad, S.A.; Khalil, K.A.; Kumar, Y.; Adeyemi, K.D.; Sazili, A.Q. Recent advances on the role of process variables affecting gelatin yield and characteristics with special reference to enzymatic extraction: A review. Food Hydrocoll. 2017, 63, 85–96. [Google Scholar] [CrossRef]
- Toldrá, F.; Reig, M.; Mora, L. Management of meat by- and co-products for an improved meat processing sustainability. Meat Sci. 2021, 181, 108608. [Google Scholar] [CrossRef] [PubMed]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M.; Hrynkiewicz, M. Food protein originating peptides as tastants-physiological, technological, sensory, and bioinformatic approaches. Food Res. Int. 2016, 89, 27–38. [Google Scholar] [CrossRef]
- Toldrá, F.; Reig, M.; Aristoy, M.C.; Mora, L. Generation of bioactive peptides during food processing. Food Chem. 2018, 267, 395–404. [Google Scholar] [CrossRef]
- Herregods, G.; Camp, J.V.; Morel, N.; Ghesquière, B.; Gevaert, K.; Vercruysse, L.; Dierckx, S.; Quanten, E.; Smagghe, G. Angiotensin I-converting enzyme inhibitory activity of gelatin hydrolysates and identification of bioactive peptides. J. Agric. Food Chem. 2011, 59, 552–558. [Google Scholar] [CrossRef]
- Verma, A.K.; Chatli, M.K.; Kumar, P.A.V.A.N.; Mehta, N. Antioxidant and antimicrobial activity of protein hydrolysate extracted from porcine liver. Indian J. Anim. Sci. 2017, 87, 711–717. [Google Scholar]
- Borrajo, P.; Pateiro, M.; Barba, F.J.; Mora, L.; Franco, D.; Toldrá, F.; Lorenzo, J.M. Antioxidant and Antimicrobial Activity of Peptides Extracted from Meat By-products: A Review. Food Anal. Meth. 2019, 12, 2401–2415. [Google Scholar] [CrossRef]
- Adhikari, B.B.; Chae, M.; Bressler, D.C. Utilization of slaughterhouse waste in value-added applications: Recent advances in the development of wood adhesives. Polymers 2018, 10, 176. [Google Scholar] [CrossRef] [Green Version]
- Bezerra, T.; Estévez, M.; Lacerda, J.T.; Dias, M.; Juliano, M.; Mendes, M.A.; Morgano, M.; Pacheco, M.T.; Madruga, M. Chicken Combs and Wattles as Sources of Bioactive Peptides: Optimization of Hydrolysis, Identification by LC-ESI-MS2 and Bioactivity Assessment. Molecules 2020, 25, 1698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryder, K.; Bekhit, A.E.D.; McConnell, M.; Carne, A. Towards generation of bioactive peptides from meat industry waste proteins: Generation of peptides using commercial microbial proteases. Food Chem. 2016, 208, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Zinina, O.; Merenkova, S.; Soloveva, A.; Savostina, T.; Sayfulmulyukov, E.; Lykasova, I.; Mizhevikina, A. The effect of starter cultures on the qualitative indicators of dry fermented sausages made from poultry meat. Agron. Res. 2018, 16, 2265–2281. [Google Scholar]
- Merenkova, S.; Zinina, O.; Loretz, O.; Neverova, O.; Sharaviev, P. Effect of transglutaminase and bacterial concentrates on the development of functional and technological properties of minced meat. Pol. J. Food Nutr. Sci. 2019, 69, 387–396. [Google Scholar] [CrossRef]
- Hong, H.; Fan, H.; Chalamaiah, M.; Wu, J. Preparation of low-molecular-weight, collagen hydrolysates (peptides): Current progress, challenges, and future perspectives. Food Chem. 2019, 301, 125222. [Google Scholar] [CrossRef] [PubMed]
- Teshnizi, Z.M.; Robatjazi, S.M.; Mosaabadi, J.M. Optimization of the Enzymatic Hydrolysis of Poultry Slaughterhouse Wastes Using Alcalase Enzyme for the Preparation of Protein Hydrolysates. Appl. Food Biotechnol. 2020, 7, 153–160. [Google Scholar]
- Assaad, H.; Zhou, L.; Carroll, R.J.; Wu, G. Rapid publication-ready MS-Word tables 597 for one-way ANOVA. SpringerPlus 2014, 3, 474. [Google Scholar] [CrossRef] [Green Version]
- Santos, B.A.S.; Azambuja, S.P.H.; Ávila, P.F.; Pacheco, M.T.B.; Goldbeck, R. n-Butanol production by Saccharomyces cerevisiae from protein-rich agro-industrial by-products. Braz. J. Microbiol. 2020, 51, 1655–1664. [Google Scholar] [CrossRef]
- Kurozawa, L.; Park, K.; Hubinger, M. Optimization of the enzymatic hydrolysis of chicken meat using response surface methodology. J. Food Sci. 2008, 73, 405–412. [Google Scholar] [CrossRef]
- Lindberg, D.; Kristoffersen, K.A.; Vogel-van den Bosch, H.; Wubshet, S.G.; Böcker, U.; Rieder, A.; Nils, E.F.; Afseth, K. Effects of poultry raw material variation and choice of protease on protein hydrolysate quality. Process. Biochem. 2021, 110, 85–93. [Google Scholar] [CrossRef]
- Lafarga, T.; Hayes, M. Bioactive peptides from meat muscle and by-products: Generation, functionality and application as functional ingredients. Meat Sci. 2014, 98, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Sun, Q.; Zhang, H.; Kong, B.; Xia, X. Purification and biochemical characteristics of the microbial extracellular protease from Lactobacillus curvatus isolated from Harbin dry sausages. Int. J. Biol. Macromol. 2019, 133, 987–997. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Tao, R.; Liu, Q.; Wang, H.; Kong, B. Effects of temperature and pH on the structure of a metalloprotease from Lactobacillus fermentum R6 isolated from Harbin dry sausages and molecular docking between protease and meat protein. J. Sci. Food Agric. 2021, 101, 5016–5027. [Google Scholar] [CrossRef] [PubMed]
- Thoresen, P.P.; Alvarez, R.G.; Vaka, M.R.; Rustad, T.; Sone, I.; Fernandez, E.N. Potential of innovative pre-treatment technologies for the revalorisation of residual materials from the chicken industry through enzymatic hydrolysis Innovative. Food Sci. Emerg. Technol. 2020, 64, 102377. [Google Scholar] [CrossRef]
- Wilkins, D.K.; Grimshaw, S.B.; Receveur, V.; Dobson, C.M.; Jones, J.A.; Smith, L.J. Hydrodynamic Radii of Native and Denatured Proteins Measured by Pulse Field Gradient NMR Techniques. Biochemistry 1999, 38, 16424–16431. [Google Scholar] [CrossRef]
- Hou, Y.; Wu, Z.; Dai, Z.; Wang, G.; Wu, G. Protein hydrolysates in animal nutrition: Industrial production, bioactive peptides, and functional significance. J. Anim. Sci. Biotechnol. 2017, 8, 24. [Google Scholar] [CrossRef] [Green Version]
- Glomm, W.R.; Wubshet, S.G.; Lindberg, D.; Dankel, K.R.; Afseth, N.K.; Stenstad, P.M.; Johnsen, H. Immobilized protease on magnetic particles for enzymatic protein hydrolysis of poultry by-products. LWT 2021, 152, 112327. [Google Scholar] [CrossRef]
- Biavati, B.; Vescovo, M.; Torriani, S.; Bottazzi, V. Bifidobacteria: History, ecology, physiology and applications. Ann. Microbiol. 2000, 50, 117–131. [Google Scholar]
- Aktas, N.; Kaya, M. The influence of marinating with weak organic acids and salts on the intramuscular connective tissue and sensory properties of beef. Eur. Food Res. Technol. 2001, 213, 88–94. [Google Scholar] [CrossRef]
- Rawdkuen, S.; Benjakul, S. Biochemical and microstructural characteristics of meat samples treated with different plant proteases. Afr. J. Biotechnol. 2012, 11, 14088–14095. [Google Scholar] [CrossRef]
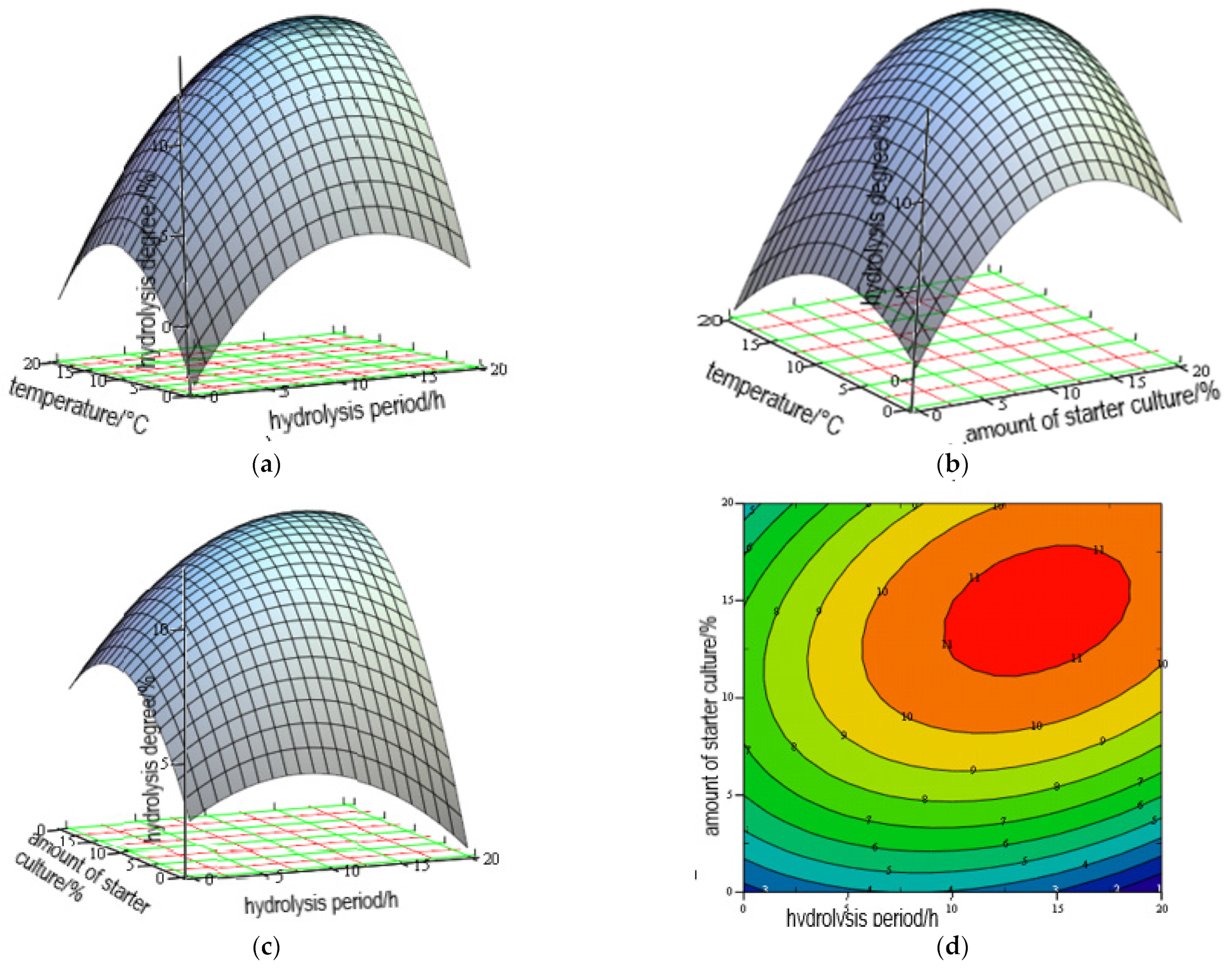
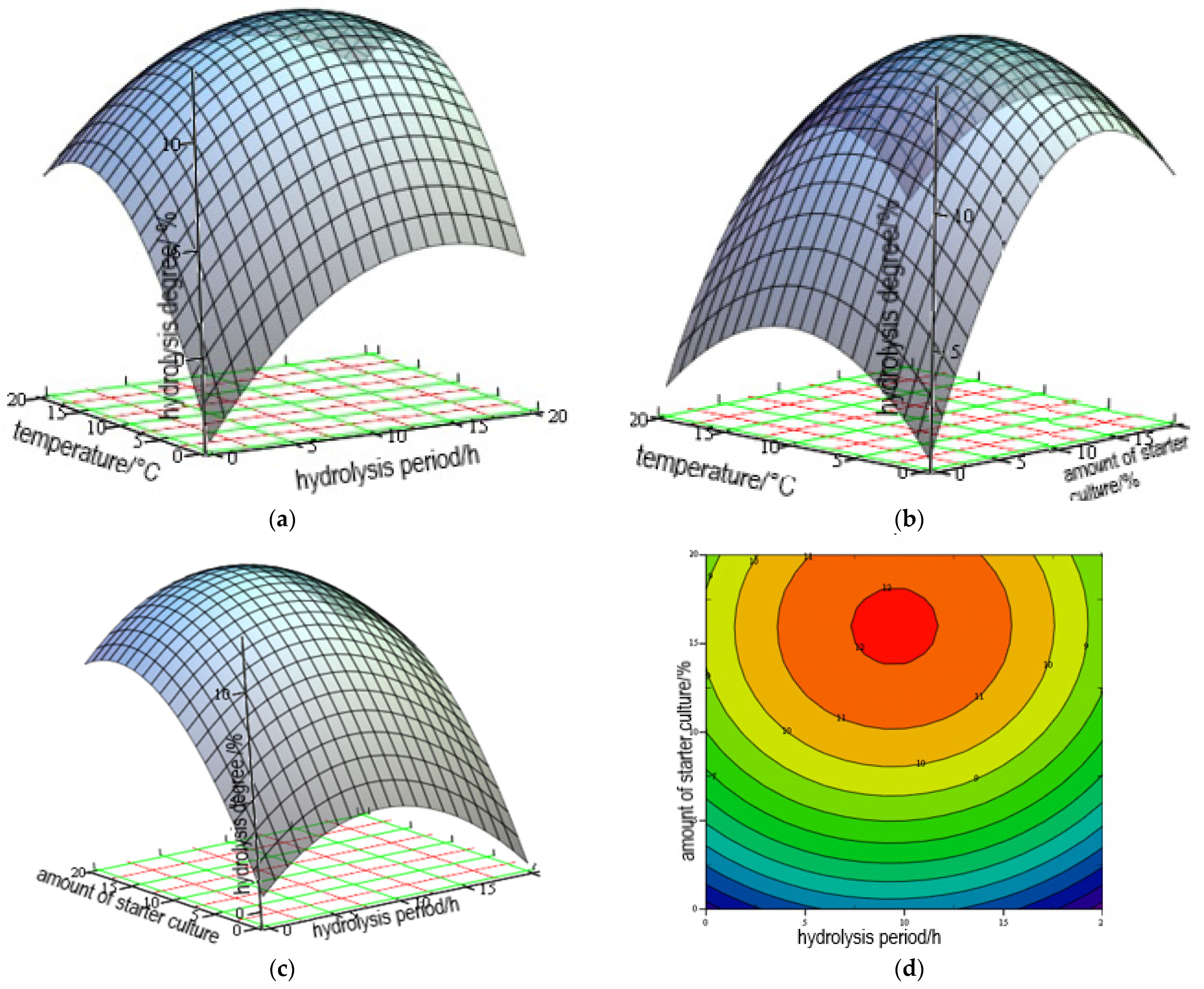
| Run | X1 | X2 | X3 | DH of Hen Combs | |
|---|---|---|---|---|---|
| Propionix LCSC | BLC | ||||
| 1 | 30 (−1) | 5 (−1) | 4 (−1) | 1.9 | 3.9 |
| 2 | 30 (−1) | 15 (+1) | 4 (−1) | 1.9 | 3.3 |
| 3 | 30 (−1) | 5 (−1) | 12 (+1) | 7.1 | 9.1 |
| 4 | 30 (−1) | 15 (+1) | 12 (+1) | 9.1 | 13.1 |
| 5 | 40 (+1) | 5 (−1) | 4 (−1) | 5.7 | 4.0 |
| 6 | 40 (+1) | 15 (+1) | 4 (−1) | 3.3 | 3.9 |
| 7 | 40 (+1) | 5 (−1) | 12 (+1) | 11.5 | 12.3 |
| 8 | 40 (+1) | 15 (+1) | 12 (+1) | 15.7 | 12.4 |
| 9 | 35 (0) | 10 (0) | 8 (0) | 4.5 | 9.1 |
| 10 | 35 (0) | 10 (0) | 8 (0) | 4.3 | 8.9 |
| 11 | 35 (0) | 10 (0) | 8 (0) | 4.8 | 9.3 |
| 12 | 48.6 (1, 2154) | 10 (0) | 8 (0) | 11.6 | 13.0 |
| 13 | 23.5 (−1, 2154) | 10 (0) | 8 (0) | 3.4 | 3.9 |
| 14 | 35 (0) | 18.2 (1, 2154) | 8 (0) | 5.9 | 5.9 |
| 15 | 35 (0) | 3.9 (−1, 2154) | 8 (0) | 4.5 | 5.2 |
| 16 | 35 (0) | 10 (0) | 14.6 (1, 2154) | 9.1 | 9.1 |
| 17 | 35 (0) | 10 (0) | 3.1 (−1, 2154) | 3.2 | 3.2 |
| Variables | Hydrolyzing by Propionix LCSC | Hydrolyzing by BLC | ||||
|---|---|---|---|---|---|---|
| Regression Coefficient (b) | t-Student’s Criterion | p-Value | Regression Coefficient (b) | t-Student’s Criterion | p-Value | |
| Y-intersection | 6.560 | 89.607 | p ≤ 0.001 | 7.427 | 125.009 | p ≤ 0.001 |
| Variable X1 | 1.744 | 19.127 | p ≤ 0.001 | 0.951 | 12.845 | p ≤ 0.01 |
| Variable X2 | 0.367 | 4.046 | p ≤ 0.05 | 0.283 | 3.829 | p ≤ 0.05 |
| Variable X3 | 2.518 | 27.780 | p ≤ 0.001 | 2.598 | 35.104 | p ≤ 0.001 |
| Variable X1·X2 | −0.013 | −0.125 | p ≥ 0.05 | −0.226 | −2.617 | p ≥ 0.05 |
| Variable X1·X3 | 0.387 | 3.645 | p ≤ 0.05 | 0.120 | 1.385 | p ≥ 0.05 |
| Variable X2·X3 | 0.573 | 5.405 | p ≤ 0.05 | 0.320 | 3.695 | p ≤ 0.05 |
| Variable X1·X2·X3 | 0.307 | 2.891 | p ≥ 0.05 | −0.293 | −3.387 | p ≥ 0.05 |
| Variable X12 | 0.433 | 3.015 | p ≥ 0.05 | 0.374 | 3.189 | p ≥ 0.05 |
| Variable X22 | −0.019 | −0.138 | p ≥ 0.05 | −0.197 | −1.682 | p ≥ 0.05 |
| Variable X32 | 0.167 | 1.164 | p ≥ 0.05 | −0.079 | −0.674 | p ≥ 0.05 |
| Amino Acid (AA) | Control Sample | Hydrolyzed Sample with Propionix LCSC | Hydrolyzed Sample with BLC |
|---|---|---|---|
| Aspa | 1.036 ± 0.0093 c | 2.238 ± 0.0102 a | 1.242 ± 0.0107 b |
| Glta | 3.464 ± 0.0093 b | 6.53 ± 0.0114 a | 3.212 ± 0.0086 c |
| Serine | not detected | 0.848 ± 0.0124 a | 0.18 ± 0.0063 b |
| Histidine | 3.58 ± 0.0089 c | 6.282 ± 0.0073 a | 4.034 ± 0.0108 b |
| Glycine | not detected | 2.03 ± 0.0070 a | 0.09 ± 0.0071 b |
| Threonine | 0.81 ± 0.0109 b | 0.852 ± 0.0097 a | 0.812 ± 0.0073 b |
| Arginine | 2.012 ± 0.0058 c | 7.35 ± 0.0118 a | 2.64 ± 0.0927 b |
| Alanin | 1.57 ± 0.0084 c | 3.574 ± 0.0081 a | 1.946 ± 0.006 b |
| Tyrosine | not detected | 1.564 ± 0.006 a | 1.442 ± 0.0097 b |
| Cystine | 3.046 ± 0.0068 b | 3.018 ± 0.0073 c | 3.162 ± 0.0073 a |
| Valine | 1.342 ± 0.0102 b | 1.614 ± 0.006 a | 1.61 ± 0.0055 a |
| Methionine | 1.654 ± 0.0087 b | 1.526 ± 0.0081 c | 1.774 ± 0.006 a |
| Phny | 1.876 ± 0.0112 b | 1.79 ± 0.0095 c | 2.142 ± 0.0107 a |
| Isoleucine | 1.654 ± 0.0081 c | 1.774 ± 0.006 b | 1.902 ± 0.0102 a |
| Leucine | 1.682 ± 0.0086 b | 1.464 ± 0.0087 c | 2.402 ± 0.0073 a |
| Proline | 1.858 ± 0.0058 c | 2.148 ± 0.0049 b | 2.904 ± 0.0108 a |
| Total AA | 25.45 ± 0.1205 c | 44.528 ± 0.1270 a | 31.51 ± 0.0911 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zinina, O.; Merenkova, S.; Rebezov, M.; Zhumanova, G.; Burkov, P.; Knyazeva, A. Investigation of Microbial Hydrolysis of Hen Combs with Bacterial Concentrates. Fermentation 2022, 8, 56. https://doi.org/10.3390/fermentation8020056
Zinina O, Merenkova S, Rebezov M, Zhumanova G, Burkov P, Knyazeva A. Investigation of Microbial Hydrolysis of Hen Combs with Bacterial Concentrates. Fermentation. 2022; 8(2):56. https://doi.org/10.3390/fermentation8020056
Chicago/Turabian StyleZinina, Oksana, Svetlana Merenkova, Maksim Rebezov, Gulnara Zhumanova, Pavel Burkov, and Alexandra Knyazeva. 2022. "Investigation of Microbial Hydrolysis of Hen Combs with Bacterial Concentrates" Fermentation 8, no. 2: 56. https://doi.org/10.3390/fermentation8020056
APA StyleZinina, O., Merenkova, S., Rebezov, M., Zhumanova, G., Burkov, P., & Knyazeva, A. (2022). Investigation of Microbial Hydrolysis of Hen Combs with Bacterial Concentrates. Fermentation, 8(2), 56. https://doi.org/10.3390/fermentation8020056






