Ensiled Mixed Vegetables Enriched Carbohydrate Metabolism in Heterofermentative Lactic Acid Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Silage Production and Sampling
2.2. Chemical Composition
2.3. pH, Volatile Fatty Acids, and Organic Acids
2.4. Profiling of the 16S rRNA and ITS1 Microbial Communities
2.5. Aerobic Stability
2.6. In Vitro Rumen Fermentation, Gas, and Methane Production
2.7. Statistical Analysis
3. Results
3.1. Chemical Composition
3.2. Silage Organic Acids and Volatile Fatty Acids
3.2.1. Silage after 101 d Ensiling
3.2.2. Fermentation Acids during the Ensiling Period
3.3. Bacterial, Archaeal, and Fungal Alpha Diversity Indices
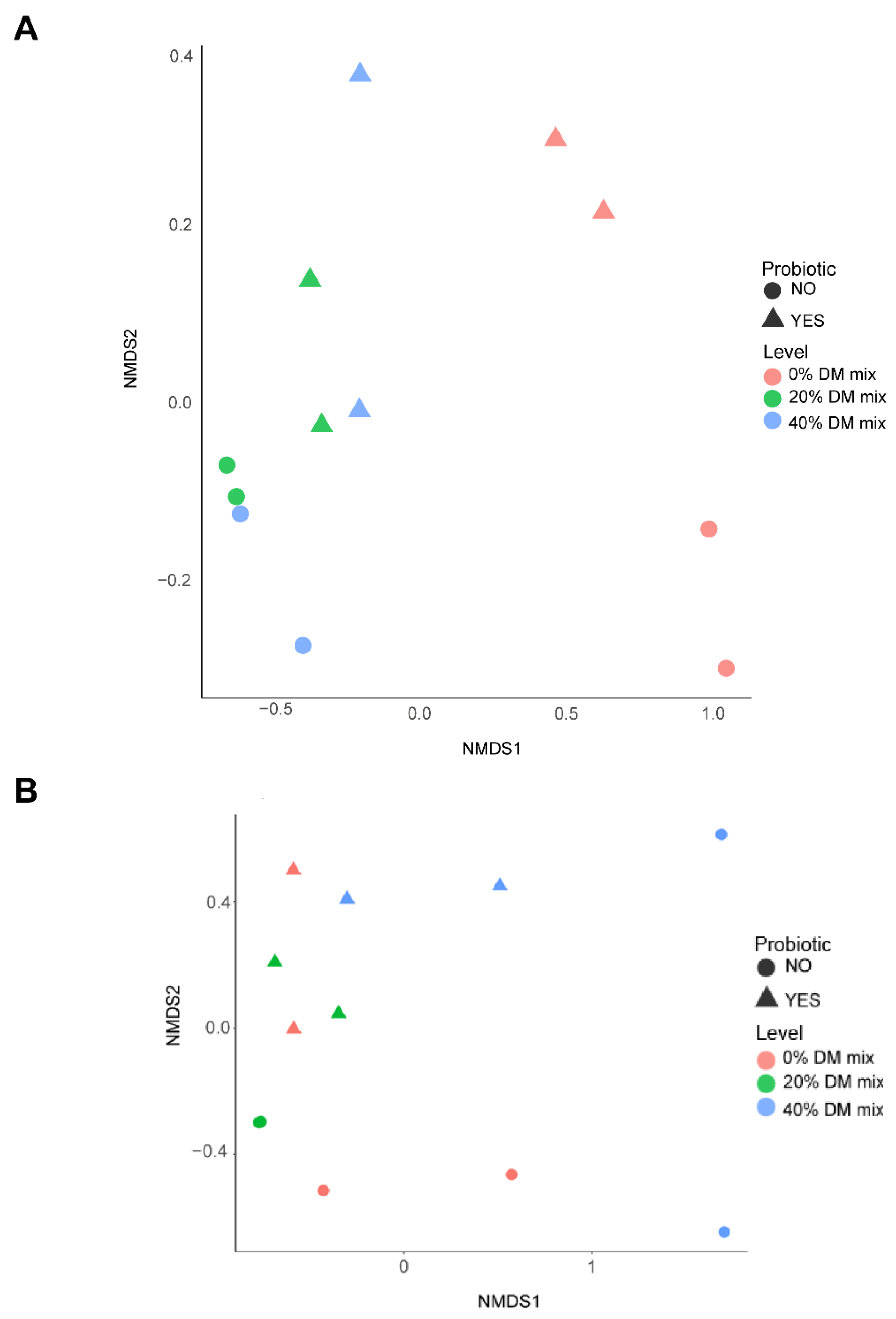
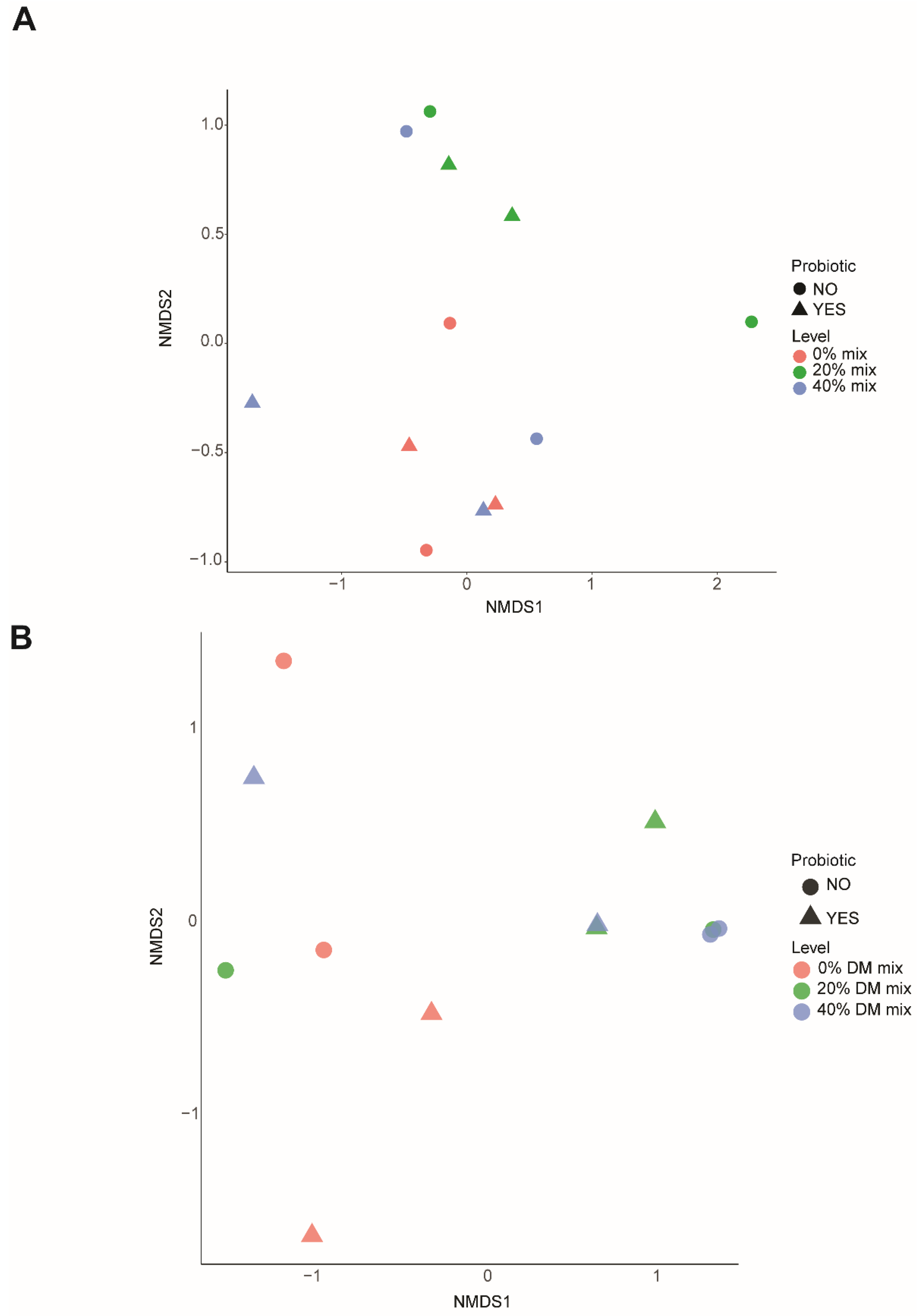
3.4. Bacterial and Archaeal Community Structure and Profile Abundance
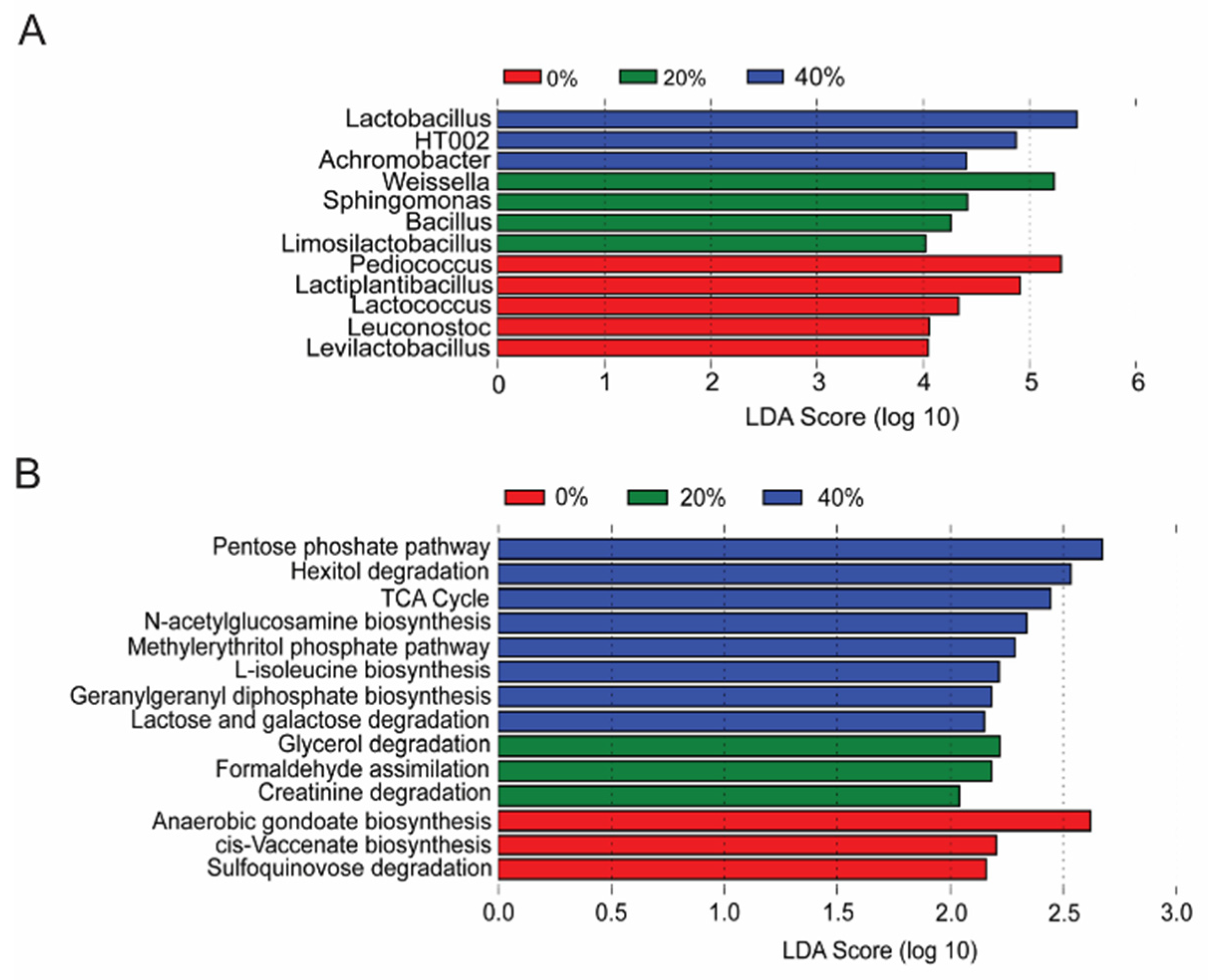
3.5. Differential Expression of Functional Pathways
3.6. Correlation Coefficients of Bacterial Communities, Fermentation Parameters, and Predicted Functional Pathways
3.7. Fungal Community Structure and Profile Abundance
3.8. Aerobic Stability
3.9. In Vitro Rumen Fermentation, Gas, and Methane Production
3.10. In Vitro Rumen Volatile Fatty Acids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, J.; Knox, K.; Rundle-Thiele, S. Waste Not Want Not: A Co-Created Food Waste Pilot, in Broadening Cultural Horizons in Social Marketing; Springer: Berlin/Heidelberg, Germany, 2021; pp. 47–65. [Google Scholar]
- Pablo, J.; Sanguansri, P.; Krause, D.; Villaddara Gamage, M.; Garcia-Flores, R. Mapping of Australian Fruit and Vegetable Losses Pre-Retail; CSIRO: Melbourne, Australia, 2019. [Google Scholar] [CrossRef]
- Keegan, E.; Breadsell, J.K. Food waste and social practices in Australian households. Sustainability 2021, 13, 3377. [Google Scholar] [CrossRef]
- Lou, X.F.; Nair, J.; Ho, G. Potential for energy generation from anaerobic digestion of food waste in Australia. Waste Manag. Res. 2013, 31, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Sabertanha, E.; Rouzbehan, Y.; Fazaeli, H.; Rezaei, J. Nutritive value of sorghum silage for sheep. J. Anim. Physiol. Anim. Nutr. 2021, 105, 1034–1045. [Google Scholar] [CrossRef] [PubMed]
- Hooker, K.; Forwood, D.L.; Caro, E.; Huo, Y.; Holman, D.B.; Chaves, A.V.; Meale, S.J. Microbial characterization and fermentative characteristics of crop maize ensiled with unsalable vegetables. Sci. Rep. 2019, 9, 13183. [Google Scholar] [CrossRef] [PubMed]
- Forwood, D.L.; Hooker, K.; Caro, E.; Huo, Y.; Holman, D.B.; Meale, S.J.; Chaves, A.V.J. Crop sorghum ensiled with unsalable vegetables increases silage microbial diversity. Front. Microbiol. 2019, 10, 2599. [Google Scholar] [CrossRef] [PubMed]
- Moon, N.J. Effect of inoculation of vegetable processing wastes with Lactobacillus plantarum on silage fermentation. J. Sci. Food Agric. 1981, 32, 675–683. [Google Scholar] [CrossRef]
- Gardner, N.J.; Savard, T.; Obermeier, P.; Caldwell, G.; Champagne, C.P. Selection and characterization of mixed starter cultures for lactic acid fermentation of carrot, cabbage, beet and onion vegetable mixtures. Int. J. Food Microbiol. 2001, 64, 261–275. [Google Scholar] [CrossRef]
- Carvalho, B.; Sales, G.; Schwan, R.; Ávila, C. Criteria for lactic acid bacteria screening to enhance silage quality. J. Appl. Microbiol. 2021, 130, 341–355. [Google Scholar] [CrossRef]
- Kung, L.; Shaver, R.D.; Grant, R.J.; Schmidt, R.J. Silage review: Interpretation of chemical, microbial, and organoleptic components of silages. J. Dairy Sci. 2018, 101, 4020–4033. [Google Scholar] [CrossRef]
- Nurk, L.; Graβ, R.; Pekrun, C.; Wachendorf, M. Methane yield and feed quality parameters of mixed silages from maize (Zea mays L.) and common bean (Phaseolus vulgaris L.). BioEnergy Res. 2017, 10, 64–73. [Google Scholar] [CrossRef]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Mertens, D.R.; Allen, M.; Carmany, J.; Clegg, J.; Davidowicz, A.; Drouches, M.; Frank, K.; Gambin, D.; Garkie, M.; Gildemeister, B.; et al. Gravimetric determination of amylase-treated neutral detergent fiber in feeds with refluxing in beakers or crucibles: Collaborative study. J. AOAC Int. 2002, 85, 1217–1240. [Google Scholar]
- Playne, M. Determination of ethanol, volatile fatty acids, lactic and succinic acids in fermentation liquids by gas chromatography. J. Sci. Food Agric. 1985, 36, 638–644. [Google Scholar] [CrossRef]
- Yu, Z.; Morrison, M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. BioTechniques 2004, 36, 808–812. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. 2021. Available online: https://www.R-project.org/ (accessed on 19 August 2021).
- R Core Team. RStudio: Integrated Development Environment for R; PBC: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 19 August 2021).
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. IEEE Signal Process. Lett. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson–Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2018, 47, D259–D264. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2:4-1. 2019. Available online: https://CRAN.R-project.org/package=vergan (accessed on 19 August 2021).
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018, 46, D633–D639. [Google Scholar] [CrossRef]
- Borreani, G.; Tabacco, E. The relationship of silage temperature with the microbiological status of the face of corn silage bunkers. J. Dairy Sci. 2010, 93, 2620–2629. [Google Scholar] [CrossRef] [PubMed]
- Meale, S.; Chaves, A.; Baah, J.; McAllister, T. Methane production of different forages in in vitro ruminal fermentation. Asian-Australas. J. Anim. Sci. 2012, 25, 86. [Google Scholar]
- Menke, K.; Raab, L.; Salewski, A.; Steingass, H.; Fritz, D.; Schneider, W. The estimation of the digestibility and metabolizable energy content of ruminant feeding stuffs from the gas production when they are incubated with rumen liquor in vitro. J. Agric. Sci. 1979, 93, 217–222. [Google Scholar] [CrossRef]
- Fedorah, P.M.; Hrudey, S.E. A simple apparatus for measuring gas production by methanogenic cultures in serum bottles. Environ. Technol. Lett. 1983, 4, 425–432. [Google Scholar] [CrossRef]
- SAS Institute Inc. SAS OnlineDoc® 9.4, Edn; SAS Institute Inc.: Cary, NC, USA, 2022. [Google Scholar]
- Jalili, V.; Afgan, E.; Gu, Q.; Clements, D.; Blankenberg, D.; Goecks, J.; Taylor, J.; Nekrutenko, A. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 2020, 48, W395–W402. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Li, R.; Jiang, D.; Zheng, M.; Tian, P.; Zheng, M.; Xu, C. Microbial community dynamics during alfalfa silage with or without clostridial fermentation. Sci. Rep. 2020, 10, 17782. [Google Scholar] [CrossRef]
- Forwood, D.L.; Bryce, E.K.; Caro, E.; Holman, D.B.; Meale, S.J.; Chaves, A.V. Influence of probiotics on biofilm formation and diversity of bacteria colonising crop sorghum ensiled with unsalable vegetables. Appl. Microbiol. Biotechnol. 2020, 104, 8825–8836. [Google Scholar] [CrossRef]
- Forwood, D.L.; Caro, E.; Holman, D.B.; Meale, S.J.; Chaves, A.V. Ensiling sorghum with unsalable pumpkin improves feed digestibility with minimal influence on the rumen microbial population using the rumen simulation technique. Appl. Microbiol. Biotechnol. 2021, 105, 3289–3300. [Google Scholar] [CrossRef]
- Malik, R.; Sharma, D.D. In vitro evaluation of different probiotics as feed supplement. Ind. J. Dairy Sci. 1998, 51, 357–362. [Google Scholar]
- Weinberg, Z.G.; Chen, Y.; Gamburg, M. The passage of lactic acid bacteria from silage into rumen fluid, in vitro studies. J. Dairy Sci. 2004, 87, 3386–3397. [Google Scholar] [CrossRef]
- Lahtinen, S.; Ouwehand, A.C.; Salminen, S.; von Wright, A. Lactic Acid Bacteria: Microbiological and Functional Aspects; CRC Press: Boca Raton, FL, USA, 2011. [Google Scholar]
- Carvalho, B.F.; Ávila, C.L.S.; Pinto, J.C.; Neri, J.; Schwan, R.F. Microbiological and chemical profile of sugar cane silage fermentation inoculated with wild strains of lactic acid bacteria. Anim. Feed Sci. Technol. 2014, 195, 1–13. [Google Scholar] [CrossRef][Green Version]
- Oude Elferink, S.J.; Krooneman, J.; Gottschal, J.C.; Spoelstra, S.F.; Faber, F.; Driehuis, F. Anaerobic conversion of lactic acid to acetic acid and 1, 2-propanediol by Lactobacillus buchneri. Appl. Environ. Microbiol. 2001, 67, 125–132. [Google Scholar] [CrossRef]
- Plengvidhya, V.; Breidt, F., Jr.; Lu, Z.; Fleming, H.P. DNA fingerprinting of lactic acid bacteria in sauerkraut fermentations. Appl. Environ. Microbiol. 2007, 73, 7697–7702. [Google Scholar] [CrossRef]
- Cao, Y.; Cai, Y.; Takahashi, T.; Yoshida, N.; Tohno, M.; Uegaki, R.; Nonaka, K.; Terada, F. Effect of lactic acid bacteria inoculant and beet pulp addition on fermentation characteristics and in vitro ruminal digestion of vegetable residue silage. J. Dairy Sci. 2011, 94, 3902–3912. [Google Scholar] [CrossRef]
- Burrichter, A.; Denger, K.; Franchini, P.; Huhn, T.; Müller, N.; Spiteller, D.; Schleheck, D. Anaerobic degradation of the plant sugar sulfoquinovose concomitant with H2S production: Escherichia coli K-12 and Desulfovibrio sp. strain DF1 as co-culture model. Front. Microbiol. 2018, 9, 2792. [Google Scholar] [CrossRef]
- Hanson, B.T.; Dimitri Kits, K.; Löffler, J.; Burrichter, A.G.; Fiedler, A.; Denger, K.; Frommeyer, B.; Herbold, C.W.; Rattei, T.; Karcher, N.; et al. Sulfoquinovose is a select nutrient of prominent bacteria and a source of hydrogen sulfide in the human gut. ISME J. 2021, 15, 2779–2791. [Google Scholar] [CrossRef]
- Kuriyama, I.; Musumi, K.; Yonezawa, Y.; Takemura, M.; Maeda, N.; Iijima, H.; Hada, T.; Yoshida, H.; Mizushina, Y. Inhibitory effects of glycolipids fraction from spinach on mammalian DNA polymerase activity and human cancer cell proliferation. J. Nutr. Biochem. 2005, 16, 594–601. [Google Scholar] [CrossRef]
- Haange, S.-B.; Groeger, N.; Froment, J.; Rausch, T.; Burkhardt, W.; Gonnermann, S.; Braune, A.; Blaut, M.; von Bergen, M.; Rolle-Kampczyk, U. Multiplexed quantitative assessment of the fate of taurine and sulfoquinovose in the intestinal microbiome. Metabolites 2020, 10, 430. [Google Scholar] [CrossRef]
- Speciale, G.; Jin, Y.; Davies, G.J.; Williams, S.J.; Goddard-Borger, E.D. YihQ is a sulfoquinovosidase that cleaves sulfoquinovosyl diacylglyceride sulfolipids. Nat. Chem. Biol. 2016, 12, 215–217. [Google Scholar] [CrossRef]
- Sharma, M.; Lingford, J.P.; Petricevic, M.; Snow, A.J.; Zhang, Y.; Järvå, M.A.; Mui, J.W.-Y.; Scott, N.E.; Saunders, E.C.; Mao, R. Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria. Proc. Natl. Acad. Sci. USA 2022, 119, e2116022119. [Google Scholar] [CrossRef] [PubMed]
- Filya, I.; Sucu, E.K.İ.N. The effects of lactic acid bacteria on the fermentation, aerobic stability and nutritive value of maize silage. Grass Forage Sci. 2010, 65, 446–455. [Google Scholar] [CrossRef]
- Blajman, J.E.; Paez, R.B.; Vinderola, C.G.; Lingua, M.S.; Signorini, M.L. A meta-analysis on the effectiveness of homofermentative and heterofermentative lactic acid bacteria for corn silage. J. Appl. Microbiol. 2018, 125, 1655–1669. [Google Scholar] [CrossRef] [PubMed]
- Yebra, M.J.; Zúñiga, M.; Beaufils, S.; Pérez-Martínez, G.; Deutscher, J.; Monedero, V. Identification of a gene cluster enabling Lactobacillus casei BL23 to utilize myo-inositol. Appl. Environ. Microbiol. 2007, 73, 3850–3858. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Q.; Davis, C.R.; Brooks, J.D. Growth and metabolism of selected lactic acid bacteria in synthetic wine. Am. J Enol. Vitic. 1995, 46, 166–174. [Google Scholar]
- Liu, S.Q. Practical implications of lactate and pyruvate metabolism by lactic acid bacteria in food and beverage fermentations. Int. J. Food Microbiol. 2003, 83, 115–131. [Google Scholar] [CrossRef]
- Tian, H.; Wang, Y.; Liu, Z.; Hu, Z.; Guo, Y.; Deng, M.; Liu, G.; Sun, B. Effects of Malic Acid and Sucrose on the Fermentation Parameters, CNCPS Nitrogen Fractions, and Bacterial Community of Moringa oleifera Leaves Silage. Microorganisms 2021, 9, 2102. [Google Scholar] [CrossRef]
- Tian, H.; Zhu, Y.; Dai, M.; Li, T.; Guo, Y.; Deng, M.; Sun, B. Additives Altered Bacterial Communities and Metabolic Profiles in Silage Hybrid Pennisetum. Front. Microbiol. 2021, 12, 770728. [Google Scholar] [CrossRef]
- Leyrolle, Q.; Cserjesi, R.; Mulders, M.D.; Zamariola, G.; Hiel, S.; Gianfrancesco, M.A.; Portheault, D.; Amadieu, C.; Bindels, L.B.; Leclercq, S. Prebiotic effect on mood in obese patients is determined by the initial gut microbiota composition: A randomized, controlled trial. Brain Behav. Immun. 2021, 94, 289–298. [Google Scholar] [CrossRef]
- Testa, C.; Brown, M. The methylerythritol phosphate pathway and its significance as a novel drug target. Curr. Pharm. Biotechnol. 2003, 4, 248–259. [Google Scholar] [CrossRef]
- Lauret, R.; Morel-Deville, F.; Berthier, F.; Champomier-Verges, M.; Postma, P.; Ehrlich, S.D.; Zagorec, M. Carbohydrate utilization in Lactobacillus sake. Appl. Environ. Microbiol. 1996, 62, 1922–1927. [Google Scholar] [CrossRef]
- Sánchez, B.; González-Tejedo, C.; Ruas-Madiedo, P.; Urdaci, M.C.; Margolles, A. Lactobacillus plantarum extracellular chitin-binding protein and its role in the interaction between chitin, Caco-2 cells, and mucin. Appl. Environ. Microbiol. 2011, 77, 1123–1126. [Google Scholar] [CrossRef]
- Chen, J.-K.; Shen, C.-R.; Liu, C.-L. N-acetylglucosamine: Production and applications. Mar. Drugs 2010, 8, 2493–2516. [Google Scholar] [CrossRef]
- Soundharrajan, I.; Kim, D.; Kuppusamy, P.; Muthusamy, K.; Lee, H.J.; Choi, K.C. Probiotic and Triticale silage fermentation potential of Pediococcus pentosaceus and Lactobacillus brevis and their impacts on pathogenic bacteria. Microorganisms 2019, 7, 318. [Google Scholar] [CrossRef]
- Ni, K.; Wang, Y.; Cai, Y.; Pang, H. Natural lactic acid bacteria population and silage fermentation of whole-crop wheat. Asian-Australas. J. Anim. Sci. 2015, 28, 1123. [Google Scholar]
- Lim, Y.; Joung, Y.; Nam, G.G.; Jahng, K.-Y.; Kim, S.-B.; Joh, K.-S.; Cha, C.-J.; Seong, C.-N.; Bae, J.-W.; Im, W.-T. A report on 33 unrecorded bacterial species of Korea isolated in 2014, belonging to the class Gammaproteobacteria. J. Species Res. 2016, 5, 241–253. [Google Scholar] [CrossRef][Green Version]
- Kalniņš, M.; Bērziņš, A.; Gudrā, D.; Megnis, K.; Fridmanis, D.; Danilko, P.; Muter, O. Selective enrichment of heterotrophic nitrifiers Alcaligenaceae and Alcanivorax spp. from industrial wastewaters. AIMS Microbiol. 2020, 6, 32. [Google Scholar] [CrossRef]
- Gallo, A.; Ghilardelli, F.; Atzori, A.S.; Zara, S.; Novak, B.; Faas, J.; Fancello, F. Co-occurrence of regulated and emerging mycotoxins in corn silage: Relationships with fermentation quality and bacterial communities. Toxins 2021, 13, 232. [Google Scholar] [CrossRef]
- Dutkiewicz, J.; Olenchock, S.; Sorenson, W.; Gerencser, V.; May, J.; Pratt, D.; Robinson, V. Levels of bacteria, fungi, and endotoxin in bulk and aerosolized corn silage. Appl. Environ. Microbiol. 1989, 55, 1093–1099. [Google Scholar] [CrossRef]
- Peles, F.; Sipos, P.; Kovács, S.; Győri, Z.; Pócsi, I.; Pusztahelyi, T. Biological control and mitigation of aflatoxin contamination in commodities. Toxins 2021, 13, 104. [Google Scholar] [CrossRef]
- Veiga da Cunha, M.; Foster, M.A. Sugar-glycerol cofermentations in lactobacilli: The fate of lactate. J. Bacteriol. 1992, 174, 1013–1019. [Google Scholar] [CrossRef] [PubMed]
- Williams, N.C.; O’Neill, L.A. A role for the Krebs cycle intermediate citrate in metabolic reprogramming in innate immunity and inflammation. Front. Immunol. 2018, 9, 141. [Google Scholar] [CrossRef] [PubMed]
- Driehuis, F.; Oude Elferink, S.; Van Wikselaar, P. Fermentation characteristics and aerobic stability of grass silage inoculated with Lactobacillus buchneri, with or without homofermentative lactic acid bacteria. Grass Forage Sci. 2001, 56, 330–343. [Google Scholar] [CrossRef]
- Kleinschmit, D.H.; Kung, L. Meta-Analysis of the Effects of Lactobacillus buchneri on the Fermentation and Aerobic Stability of Corn and Grass and Small-Grain Silages. J. Dairy Sci. 2006, 89, 4005–4013. [Google Scholar] [CrossRef] [PubMed]
- Lindgren, S.E.; Axelsson, L.T.; McFeeters, R.F. Anaerobic L-lactate degradation by Lactobacillus plantarum. FEMS Microbiol. Lett. 1990, 66, 209–213. [Google Scholar] [CrossRef]
- Klem, T.J. Alcaligenes. In Encyclopedia of Food Microbiology; Robinson, R.K., Ed.; Elsevier: Oxford, UK, 1999; pp. 38–42. [Google Scholar]
- Shoda, M. Chapter 2—Alcaligenes. In Beneficial Microbes in Agro-Ecology; Amaresan, N., Kumar, M.S., Annapurna, K., Kumar, K., Sankaranarayanan, A., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 13–26. [Google Scholar]
- Velázquez-Guerrero, J.J.; Cerna Chávez, E.; Delgado Ortiz, J.C.; Hernández Perez, A.; Quezada Tristán, T.; Ochoa Fuentes, Y.M. First report of Monascus purpureus in corn silage, oats, triticale and lucerne. Rev. Mexicana Cienc. 2019, 10, 1911–1917. [Google Scholar]
- Bridge, P.; Hawksworth, D. Biochemical tests as an aid to the identification of Monascus species. Lett. Appl. Microbiol. 1985, 1, 25–29. [Google Scholar] [CrossRef]
- Rogalski, E.; Ehrmann, M.A.; Vogel, R.F. Strain-specific interaction of Fructilactobacillus sanfranciscensis with yeasts in the sourdough fermentation. Eur. Food Res. Technol. 2021, 247, 1437–1447. [Google Scholar] [CrossRef]
- Rollin-Pinheiro, R.; de Meirelles, J.V.; Vila, T.V.; Fonseca, B.B.; Alves, V.; Frases, S.; Rozental, S.; Barreto-Bergter, E. Biofilm formation by Pseudallescheria/Scedosporium species: A comparative study. Front. Microbiol. 2017, 8, 1568. [Google Scholar] [CrossRef]
- Sperschneider, J.; Dodds, P.N.; Gardiner, D.M.; Manners, J.M.; Singh, K.B.; Taylor, J.M. Advances and challenges in computational prediction of effectors from plant pathogenic fungi. PLoS Path. 2015, 11, e1004806. [Google Scholar] [CrossRef]

| Probiotic (Prob) | NO | YES | p-Value | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Level, % DM | 0 | 20 | 40 | 0 | 20 | 40 | SEM | Level | Prob | Level × Prob | L | Q |
| DM content, % | 38.2 | 25.6 | 17.4 | 35.9 | 26.7 | 18.4 | 0.69 | <0.01 | 0.95 | 0.08 | <0.01 | 0.07 |
| CP, % DM | 9.16 a | 12.3 a | 8.47 a | 14.8 a | 10.7 b | 8.63 b | 1.17 | 0.05 | 0.20 | 0.05 | 0.03 | 0.26 |
| NFC, % DM | 20.7 | 19.8 | 26.4 | 12.8 | 21.4 | 23.8 | 2.79 | 0.06 | 0.24 | 0.30 | 0.02 | 0.91 |
| EE, % DM | 7.07 | 5.62 | 5.99 | 6.78 | 6.51 | 6.68 | 0.35 | 0.11 | 0.18 | 0.27 | 0.14 | 0.11 |
| NDF, % DM | 51.2 | 49.6 | 47.3 | 52.8 | 48.6 | 46.9 | 2.43 | 0.21 | 0.98 | 0.86 | 0.09 | 0.84 |
| Ash, % DM | 11.9 | 12.6 | 11.8 | 12.9 | 12.8 | 13.9 | 0.64 | 0.77 | 0.08 | 0.38 | 0.49 | 0.95 |
| Level, % DM | p-Value | ||||||
|---|---|---|---|---|---|---|---|
| 0 | 20 | 40 | SEM | Level | L | Q | |
| pH | 3.9 c | 4.3 b | 4.4 a | 0.17 | <0.01 | <0.01 | <0.01 |
| Organic acids, mM | |||||||
| Lactate | 5.93 a | 3.43 b | 2.24 c | 0.23 | <0.01 | <0.01 | 0.07 |
| Succinate | 0.39 | 0.42 | 0.40 | 0.02 | 0.39 | 0.70 | 0.19 |
| Ethanol | 8.42 b | 12.7 b | 20.4 a | 1.32 | <0.01 | <0.01 | 0.34 |
| Volatile fatty acids | |||||||
| Total VFA, mM | 15.2 | 17.2 | 18.0 | 1.78 | 0.54 | 0.29 | 0.79 |
| Volatile fatty acids as a percentage of total VFA | |||||||
| Acetate, % | 77.6 | 85.7 | 87.5 | 3.47 | 0.17 | 0.08 | 0.50 |
| Propionate, % | 3.61 | 5.07 | 4.72 | 0.37 | 0.07 | 0.07 | 0.09 |
| Butyrate, % | 8.41 | 4.60 | 5.43 | 1.27 | 0.16 | 0.14 | 0.19 |
| Minor VFA, % | 10.3 | 4.67 | 2.32 | 2.34 | 0.11 | 0.04 | 0.59 |
| Level, % DM | p-Value | ||||||
|---|---|---|---|---|---|---|---|
| 0 | 20 | 40 | SEM | Level | L | Q | |
| Bacteria and archaea | |||||||
| Pre-ensiled | |||||||
| Chao1 | 47.0 | 49.8 | 40.3 | 2.86 | 0.13 | 0.15 | 0.13 |
| Inverse Simpson | 3.61 b | 5.74 a | 4.10 b | 0.34 | 0.01 | 0.34 | <0.01 |
| Shannon | 1.82 b | 2.28 a | 2.02 b | 0.06 | 0.01 | 0.06 | <0.01 |
| 101 d Ensiled | |||||||
| Chao1 | 102.8 | 88.3 | 69.0 | 12.7 | 0.25 | 0.11 | 0.88 |
| Inverse Simpson | 6.01 | 4.99 | 5.01 | 0.63 | 0.47 | 0.30 | 0.53 |
| Shannon | 2.41 | 2.22 | 2.26 | 0.15 | 0.69 | 0.53 | 0.58 |
| 14 d aerobic exposure | |||||||
| Chao1 | 113 | 96.8 | 80.8 | 23.9 | 0.66 | 0.38 | 1.00 |
| Inverse Simpson | 10.3 | 8.29 | 7.93 | 4.37 | 0.92 | 0.71 | 0.88 |
| Shannon | 1.82 | 2.28 | 2.02 | 0.06 | 0.91 | 0.80 | 0.75 |
| Fungi | |||||||
| Pre-ensiled | |||||||
| Chao1 | 38.8 | 35.8 | 41.8 | 1.44 | 0.07 | 0.19 | 0.04 |
| Inverse Simpson | 4.26 | 3.95 | 4.63 | 0.27 | 0.28 | 0.37 | 0.18 |
| Shannon | 2.05 | 1.91 | 2.11 | 0.07 | 0.18 | 0.59 | 0.08 |
| 101 d Ensiled | |||||||
| Chao1 | 17.5 a | 8.75 b | 9.50 b | 1.59 | 0.01 | 0.01 | 0.05 |
| Inverse Simpson | 4.01 a | 1.47 b | 2.13 b | 0.52 | 0.03 | 0.04 | 0.05 |
| Shannon | 1.59 a | 0.52 b | 0.93 ab | 0.21 | 0.03 | 0.07 | 0.03 |
| 14 d aerobic exposure | |||||||
| Chao1 | 7.50 | 8.50 | 6.00 | 1.47 | 0.52 | 0.50 | 0.37 |
| Inverse Simpson | 2.66 | 2.02 | 2.19 | 0.33 | 0.43 | 0.35 | 0.36 |
| Shannon | 1.02 | 0.95 | 0.91 | 0.14 | 0.83 | 0.56 | 0.94 |
| Level, % DM | p-Value | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Day | 0 | 20 | 40 | SEM | Level | Prob | Level × Prob | L | Q |
| 0 | −2.74 | −2.59 | −2.79 | 0.09 | 0.28 | 0.19 | 0.62 | 0.70 | 0.13 |
| 1 | −2.18 a | −2.38 b | −2.69 c | 0.05 | <0.01 | 0.10 | 0.36 | <0.01 | 0.40 |
| 2 | −1.76 a | −2.14 b | −2.48 c | 0.05 | <0.01 | 0.57 | 0.67 | <0.01 | 0.76 |
| 3 | −1.56 a | −1.94 b | −2.18 c | 0.06 | <0.01 | 0.47 | 0.56 | <0.01 | 0.35 |
| 4 | −1.34 a | −1.66 b | −2.04 c | 0.07 | <0.01 | 0.62 | 0.27 | <0.01 | 0.78 |
| 5 | −1.00 a | −1.30 b | −1.61 c | 0.05 | <0.01 | 0.65 | 0.10 | <0.01 | 0.91 |
| 6 | −0.71 a | −1.04 b | −1.38 c | 0.06 | <0.01 | 0.31 | 0.85 | <0.01 | 0.94 |
| 7 | −0.44 a | −0.66 b | −0.99 c | 0.04 | <0.01 | 0.61 | 1.00 | <0.01 | 0.35 |
| 8 | −1.51 a | −1.83 b | −2.25c | 0.06 | <0.01 | 0.70 | 0.07 | <0.01 | 0.42 |
| 9 | −1.33 a | −1.46 a | −2.00 b | 0.06 | <0.01 | 0.91 | 0.12 | <0.01 | 0.04 |
| 10 | −0.55 a | −0.61 a | −1.41 b | 0.12 | <0.01 | 0.80 | 0.48 | <0.01 | 0.02 |
| 11 | −0.34 a | −0.50 a | −1.24 b | 0.10 | <0.01 | 0.68 | 0.32 | <0.01 | 0.06 |
| 12 | −0.20 a | −0.29 a | −1.13 b | 0.10 | <0.01 | 0.55 | 0.32 | <0.01 | 0.02 |
| 13 | −0.01 a | −0.14 a | −0.84 b | 0.11 | <0.01 | 0.95 | 0.50 | <0.01 | 0.09 |
| 14 | 0.05 a | −0.01 a | −0.66 b | 0.10 | <0.01 | 0.56 | 0.79 | <0.01 | 0.02 |
| Level, % DM | p-Value | ||||||
|---|---|---|---|---|---|---|---|
| 0 | 20 | 40 | SEM | Level | L | Q | |
| Total gas, mL | 39.7 | 40.4 | 39.2 | 1.64 | 0.66 | 0.73 | 0.40 |
| Total gas, mL/g DM | 82.4 | 83.8 | 81.6 | 3.38 | 0.68 | 0.75 | 0.42 |
| Total gas, mL/g digestible DM | 182.3 a | 157.7 b | 155.9 b | 6.69 | <0.01 | <0.01 | 0.01 |
| CH4, % | 16.3 | 17.1 | 16.8 | 0.49 | 0.13 | 0.17 | 0.14 |
| CH4, mL/g digestible DM | 29.2 a | 27.1 b | 26.4 b | 0.64 | 0.01 | <0.01 | 0.40 |
| CH4, mL/g incubated DM | 13.1 | 14.0 | 13.4 | 0.33 | 0.16 | 0.53 | 0.07 |
| IVDMD, % | 44.9 b | 51.6 a | 50.7 a | 0.53 | <0.01 | <0.01 | <0.01 |
| Level, % DM | p-Value | ||||||
|---|---|---|---|---|---|---|---|
| 0 | 20 | 40 | SEM | Level | L | Q | |
| pH | 6.7 | 6.7 | 6.7 | 0.01 | 0.08 | 0.06 | 0.22 |
| Total VFA, mM | 97.1 b | 103.6 a | 105.6 a | 4.71 | 0.01 | <0.01 | 0.38 |
| Volatile fatty acids as a percentage of total VFA | |||||||
| Acetate (A), % | 56.9 | 57.6 | 59.0 | 0.89 | 0.17 | 0.06 | 0.75 |
| Propionate (P), % | 19.7 | 21.2 | 20.5 | 0.86 | 0.04 | 0.15 | 0.03 |
| Butyrate, % | 18.1 a | 16.0 b | 15.3 b | 1.63 | 0.01 | <0.01 | 0.38 |
| BCVFA, % | 3.75 | 3.88 | 3.71 | 0.36 | 0.38 | 0.76 | 0.18 |
| Caproate, % | 0.50 | 0.48 | 0.47 | 0.05 | 0.89 | 0.67 | 0.84 |
| Valerate, % | 1.04 | 0.80 | 0.95 | 0.49 | 0.68 | 0.74 | 0.42 |
| Ratio A:P | 2.89 | 2.72 | 2.88 | 0.10 | 0.17 | 0.57 | 0.08 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Forwood, D.L.; Holman, D.B.; Meale, S.J.; Chaves, A.V. Ensiled Mixed Vegetables Enriched Carbohydrate Metabolism in Heterofermentative Lactic Acid Bacteria. Fermentation 2022, 8, 699. https://doi.org/10.3390/fermentation8120699
Forwood DL, Holman DB, Meale SJ, Chaves AV. Ensiled Mixed Vegetables Enriched Carbohydrate Metabolism in Heterofermentative Lactic Acid Bacteria. Fermentation. 2022; 8(12):699. https://doi.org/10.3390/fermentation8120699
Chicago/Turabian StyleForwood, Daniel L., Devin B. Holman, Sarah J. Meale, and Alex V. Chaves. 2022. "Ensiled Mixed Vegetables Enriched Carbohydrate Metabolism in Heterofermentative Lactic Acid Bacteria" Fermentation 8, no. 12: 699. https://doi.org/10.3390/fermentation8120699
APA StyleForwood, D. L., Holman, D. B., Meale, S. J., & Chaves, A. V. (2022). Ensiled Mixed Vegetables Enriched Carbohydrate Metabolism in Heterofermentative Lactic Acid Bacteria. Fermentation, 8(12), 699. https://doi.org/10.3390/fermentation8120699






