Potentially Probiotic Fermented Glutinous Rice (Oryza sativa L.) with Lactiplantibacillus plantarum Improved Immune System Response in a Small Sample of BALB/cByJ Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Lactic acid Bacteria (LAB)
2.2. Acid Tolerance, Bile Tolerance, Enzyme Resistance, and High-Temperature Resistance
2.3. Antibiotic Susceptibility
2.4. Phenotypic and Genotypic Identifications
2.5. Growth Condition for the Probiotic Bacteria
2.6. Preparation of Tapai Pulut
2.7. Chemical and Microbial Analyses
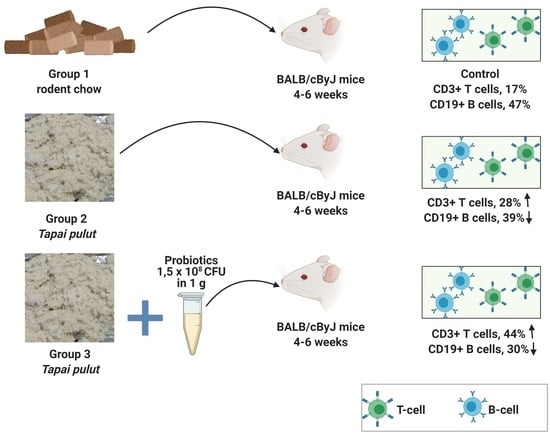
2.8. Animals and Diets
2.9. Biochemical Analysis for Blood Samples
2.10. Flow Cytometry Analysis
2.11. Statistical Analysis
3. Results
3.1. Probiotic Properties
3.2. Antibiotic Susceptibility
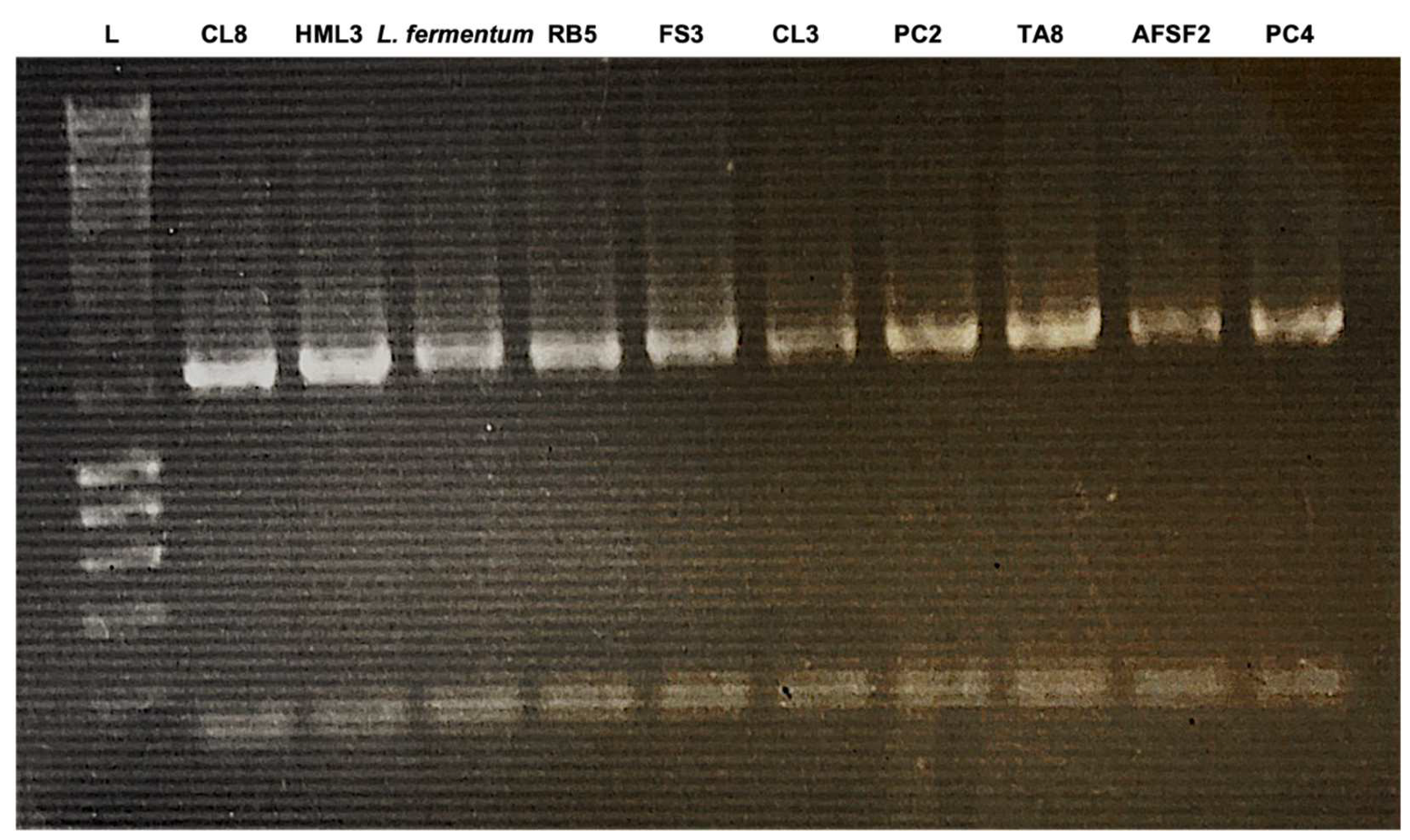
3.3. Phenotypic and Genotypic Identification
3.4. Feeding Samples and Growth Performances
3.5. Haematology Analysis
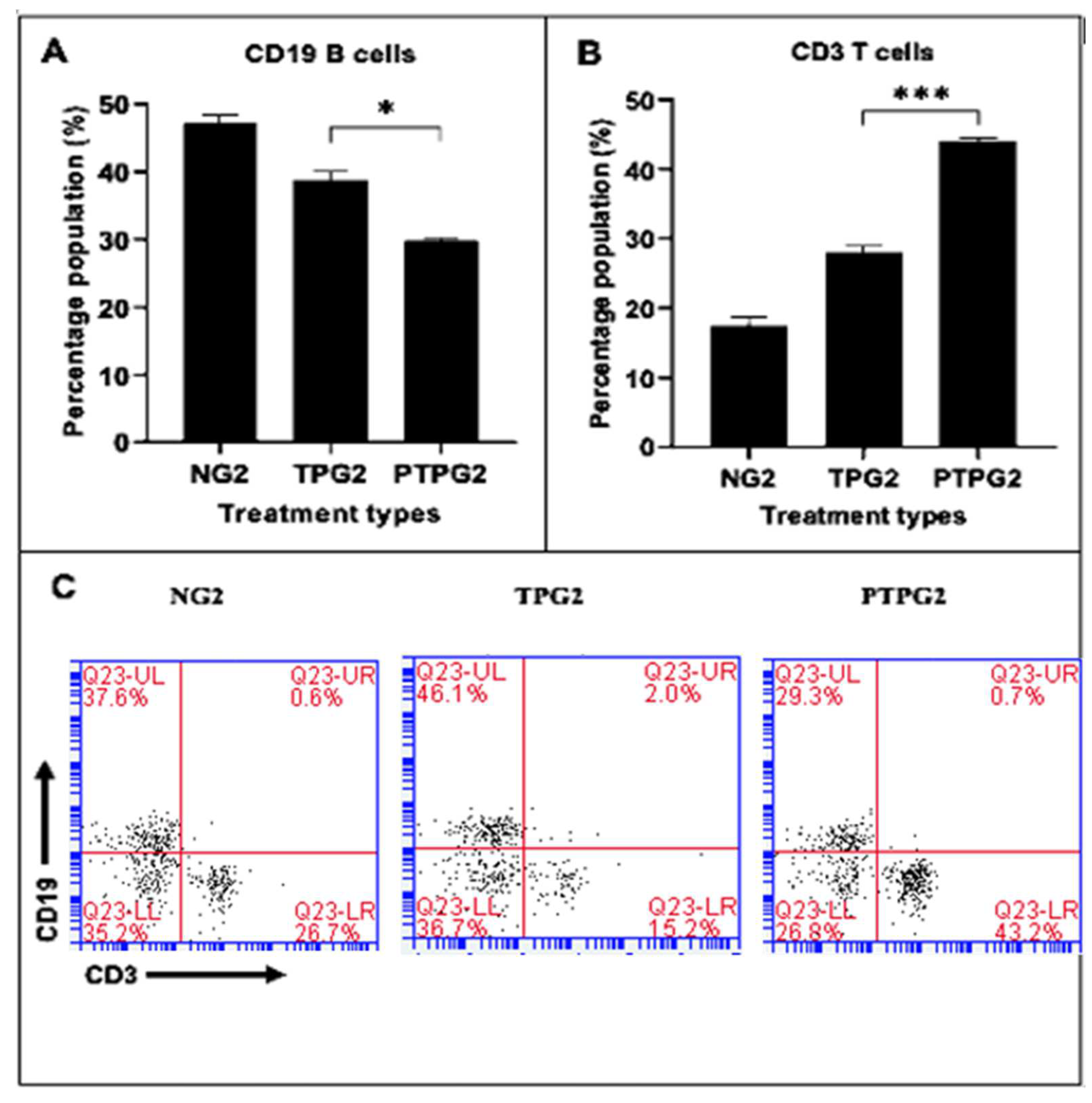
3.6. T-Cell and B-Cell Analysis
4. Discussion
4.1. Probiotic Properties
4.2. Phenotypic and Genotypic Identification
4.3. Feeding Samples and Growth Performance
4.4. Haematology Analysis
4.5. T-Cell and B-Cell Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Gerez, C.L.; Torres, M.J.; De Valdez, G.F.; Rollán., G. Control of spoilage fungi by lactic acid bacteria. Biol. Control 2013, 64, 231–237. [Google Scholar] [CrossRef]
- Samtiya, M.; Aluko, R.E.; Puniya, A.K.; Dhewa, T. Enhancing micronutrients bioavailability through fermentation of plant-based foods: A concise review. Fermentation 2021, 7, 63. [Google Scholar] [CrossRef]
- Azad, M.; Kalam, A.; Sarker, M.; Wan, D. Immunomodulatory effects of probiotics on cytokine profiles. BioMed Res. Int. 2018, 2018, 8063647. [Google Scholar] [CrossRef]
- Lee, Y.J.; Lee, A.; Yoo, H.J.; Kim, M.; Noh, G.M.; Lee, J.H. Supplementation with the probiotic strain Weissella cibaria JW15 enhances natural killer cell activity in nondiabetic subjects. J. Funct. Foods 2018, 48, 153–158. [Google Scholar] [CrossRef]
- Markov, A.V.; Sen’kova, A.V.; Babich, V.O.; Odarenko, K.V.; Talyshev, V.A.; Salomatina, O.V.; Salakhutdinov, N.F.; Zenkova, M.A.; Logashenko, E.B. Dual effect of soloxolone methyl on LPS-induced inflammation in vitro and in vivo. Int. J. Mol. Sci. 2020, 21, 7876. [Google Scholar] [CrossRef]
- Fukushima, Y.; Kawata, Y.; Mizumachi, K.; Kurisaki, J.I.; Mitsuoka, T. Effect of bifidobacteria feeding on fecal flora and production of immunoglobulins in lactating mouse. Int. J. Food Microbiol. 1999, 46, 193–197. [Google Scholar] [CrossRef]
- Merican, Z.; Quee-Lan, Y. Tapai processing in Malaysia: A technology in transition. In Industrialization of Indigenous Fermented Foods, Revised and Expanded, 2nd ed.; Steinkraus, K., Ed.; Marcel Dekker, Inc.: New York, NY, USA, 2004; pp. 247–269. [Google Scholar]
- Kofli, N.T.; Dayaon, S.H.M. Identification of Microorganism from Ragi for Bioethanol Production by API Kit. J. Appl. Sci. 2010, 10, 2751–2753. [Google Scholar] [CrossRef]
- Hermanto, F.E.; Warsito, W.; Rifa’I, M.; Widodo, N.; Jatmiko, Y.D. Metabarcoding dataset on the elicitation of Soybean and Mungbean using Ragi Tape as elicitors for enhancing secondary metabolites production. Data Brief 2022, 42, 108209. [Google Scholar] [CrossRef]
- Hamid, T.H.T.A.; Amsya, N.F. Lactic acid bacterium with antimicrobial properties from selected malay traditional fermented foods. Int. J. Life Sci. Biotechnol. 2020, 4, 13–24. [Google Scholar] [CrossRef]
- Sujaya, I.; Amachi, S.; Yokota, A.; Asano, K.; Tomita, F. Identification and characterization of lactic acid bacteria in ragi tape. World J. Microbiol. Biotechnol. 2001, 17, 349–357. [Google Scholar] [CrossRef]
- Martinez, R.C.; Aynaou, A.E.; Albrecht, S.; Schols, H.A.; De Martinis, E.C.; Zoetendal, E.G., Venema; Zoetendal, E.G.; Venema, K.; Smidt, H. In vitro evaluation of gastrointestinal survival of Lactobacillus amylovorus DSM 16698 alone and combined with galactooligosaccharides, milk and/or Bifidobacterium animalis subsp. lactis Bb-12. Int. J. Food Microbio. 2011, 149, 152–158. [Google Scholar] [CrossRef]
- Liasi, S.A.; Azmi, T.I.; Hassan, M.D.; Shuhaimi, M.; Rosfarizan, M.; Ariff, A.B. Antimicrobial activity and antibiotic sensitivity of three isolates of lactic acid bacteria from fermented fish product, Budu. Malays. J. Microb. 2009, 5, 33–37. [Google Scholar]
- D’Aimmo, M.R.; Modesto, M.; Biavati, B. Antibiotic resistance of lactic acid bacteria and Bifidobacterium spp. isolated from dairy and pharmaceutical products. Int. J. Food. Microbiol. 2007, 115, 35–42. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematic; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Association of Official Analytical Chemists (AOAC). Official Methods of Analysis of the Association Analytical Chemists, 18th ed.; AOAC: Gaithersburg, MD, USA, 2005. [Google Scholar]
- Zhao, J.; Fleet, G. Yeasts are essential for cocoa bean fermentation. Int. J. Food Microbiol. 2014, 174, 72–87. [Google Scholar] [CrossRef]
- Chuah, L.O.; Shamila-Syuhada, A.K.; Liong, M.T.; Rosma, A.; Thong, K.L.; Rusul, G. Physio-chemical, microbiological properties of tempoyak and molecular characterisation of lactic acid bacteria isolated from tempoyak. Food Microbiol. 2016, 58, 95–104. [Google Scholar] [CrossRef]
- Adnan, A.F.M.; Tan, I.K. Isolation of lactic acid bacteria from Malaysian foods and assessment of the isolates for industrial potential. Bioresour. Technol. 2007, 98, 1380–1385. [Google Scholar] [CrossRef]
- Swain, M.R.; Anandharaj, M.; Ray, R.C.; Rani, R.P. Fermented fruits and vegetables of Asia: A potential source of probiotics. Biotechnol. Res. Int. 2014, 2014, 250424. [Google Scholar] [CrossRef]
- Ruas-Madiedo, P.; Moreno, J.A.; Salazar, N.; Delgado, S.; Mayo, B.; Margolles, A.; de Los Reyes-Gavilán, C.G. Screening of exopolysaccharide-producing Lactobacillus and Bifidobacterium strains isolated from the human intestinal microbiota. Appl. Environ. Microbiol. 2007, 73, 4385–4388. [Google Scholar] [CrossRef]
- Ren, D.; Li, C.; Qin, Y.; Yin, R.; Du, S.; Ye, F.; Liu, C.; Liu, H.; Wang, M.; Li, Y.; et al. In vitro evaluation of the probiotic and functional potential of Lactobacillus strains isolated from fermented food and human intestine. Anaerobe 2014, 30, 1–10. [Google Scholar] [CrossRef]
- Liu, W.J.; Chen, Y.F.; Kwok, L.Y.; Li, M.H.; Sun, T.; Sun, C.L.; Wang, X.N.; Dan, T.; Zhang, H.P.; Sun, T.S. Preliminary selection for potential probiotic Bifidobacterium isolated from subjects of different Chinese ethnic groups and evaluation of their fermentation and storage characteristics in bovine milk. J. Dairy Sci. 2013, 96, 6807–6817. [Google Scholar] [CrossRef]
- Anandharaj, M.; Sivasankari, B.; Santhanakaruppu, R.; Manimaran, M.; Rani, R.P.; Sivakumar, S. Determining the probiotic potential of cholesterol-reducing Lactobacillus and Weissella strains isolated from gherkins (fermented cucumber) and south Indian fermented koozh. Res. Microbiol. 2015, 166, 428–439. [Google Scholar] [CrossRef]
- García-Ruiz, A.; de Llano, D.G.; Esteban-Fernández, A.; Requena, T.; Bartolomé, B.; Moreno-Arribas, M.V. Assessment of probiotic properties in lactic acid bacteria isolated from wine. Food Microbiol. 2014, 44, 220–225. [Google Scholar] [CrossRef]
- Noriega, L.; Gueimonde, M.; Sánchez, B.; Margolles, A.; de los Reyes-Gavilán, C.G. Effect of the adaptation to high bile salts concentrations on glycosidic activity, survival at low pH and cross-resistance to bile salts in Bifidobacterium. Int. J. Food Microbiol. 2004, 94, 79–86. [Google Scholar] [CrossRef]
- Shokryazdan, P.; Faseleh Jahromi, M.; Liang, J.B.; Ramasamy, K.; Sieo, C.C.; Ho, Y.W. Effects of a Lactobacillus salivarius mixture on performance, intestinal health and serum lipids of broiler chickens. PLoS ONE 2017, 12, e0175959. [Google Scholar] [CrossRef]
- Meybodi, N.M.; Mortazavian, A.M.; Arab, M.; Nematollahi, A. Probiotic viability in yoghurt: A review of influential factors. Int. Dairy J. 2020, 109, 104793. [Google Scholar] [CrossRef]
- Mortazavian, A.M.; Ehsani, M.R.; Mousavi, S.M.; Reinheimer, J.A.; Emamdjomeh, Z.; Sohrabvandi, S.; Rezaei, K. Preliminary investigation of the combined effect of heat treatment and incubation temperature on the viability of the probiotic micro-organisms in freshly made yogurt. Int. J. Dairy Technol. 2006, 59, 8–11. [Google Scholar] [CrossRef]
- Corona-Hernandez, R.I.; Álvarez-Parrilla, E.; Lizardi-Mendoza, J.; Islas-Rubio, A.R.; de la Rosa, L.A.; Wall-Medrano, A. Structural stability and viability of microencapsulated probiotic bacteria: A review. Compr. Rev. Food Sci. Food Saf. 2013, 12, 614–628. [Google Scholar] [CrossRef]
- Cruz, A.G.D.; Castro, W.F.; Faria, J.; Bolini, H.M.A.; Celeghini, R.M.D.S.; Raices, R.S.L.; Oliveira, C.A.F.; Freitas, M.Q.; Júnior, C.C.; Mársico, E.T. Stability of probiotic yogurt added with glucose oxidase in plastic materials with different permeability oxygen rates during the refrigerated storage. Food Res. Int. 2013, 51, 723–728. [Google Scholar] [CrossRef]
- Zoumpopoulou, G.; Tzouvanou, A.; Mavrogonatou, E.; Alexandraki, V.; Georgalaki, M.; Anastasiou, R.; Papadelli, M.; Manolopoulou, E.; Kazou, M.; Kletsas, D.; et al. Probiotic features of lactic acid bacteria isolated from a diverse pool of traditional Greek dairy products regarding specific strain-host interactions. Probiotics Antimicrob. Proteins 2018, 10, 313–322. [Google Scholar] [CrossRef]
- Rahayu, H.M.; Qurbaniah, M. Selection of Tempoyak Lactic Acid Bacteria as Candidate Strain for Yoghurt Starter Culture. Biosaintifika: J. Biol. Educ. 2019, 11, 39–46. [Google Scholar] [CrossRef]
- Cao, M.; Feng, Y.; Zhang, Y.; Kang, W.; Lian, K.; Ai, L. Studies on the metabolism and degradation of vancomycin in simulated in vitro and aquatic environment by UHPLC-Triple-TOF-MS/MS. Sci. Rep. 2018, 8, 15471. [Google Scholar] [CrossRef]
- Jafari-Nasab, T.; Khaleghi, M.; Farsinejad, A.; Khorrami, S. Probiotic potential and anticancer properties of Pediococcus sp. isolated from traditional dairy products. Biotechnol. Rep. 2021, 29, e00593. [Google Scholar] [CrossRef]
- Gueimonde, M.; Sánchez, B.G.; de los Reyes-Gavilán, C.; Margolles, A. Antibiotic resistance in probiotic bacteria. Front. Microbiol. 2013, 4, 202. [Google Scholar] [CrossRef]
- Adesulu-Dahunsi, A.T.; Sanni, A.I.; Jeyaram, K. Rapid differentiation among Lactobacillus, Pediococcus and Weissella species from some Nigerian indigenous fermented foods. LWT 2017, 77, 39–44. [Google Scholar] [CrossRef][Green Version]
- Zheng, J.; Ruan, L.; Sun, M.; Gänzle, M. A genomic view of lactobacilli and pediococci demonstrates that phylogeny matches ecology and physiology. Appl. Environ. Microbiol. 2015, 81, 7233–7243. [Google Scholar] [CrossRef]
- Xu, X.; Zhou, S.; McClements, D.J.; Huang, L.; Meng, L.; Xia, X.; Dong, M. Multistarter fermentation of glutinous rice with Fu brick tea: Effects on microbial, chemical, and volatile compositions. Food Chem. 2020, 309, 125790. [Google Scholar] [CrossRef]
- Champagne, C.P.; da Cruz, A.G.; Daga, M. Strategies to improve the functionality of probiotics in supplements and foods. Curr. Opin. Food Sci. 2018, 22, 160–166. [Google Scholar] [CrossRef]
- Shiby, V.K.; Mishra, H.N. Fermented milks and milk products as functional foods—A review. Crit. Rev. Food Sci. Nutr. 2013, 53, 482–496. [Google Scholar] [CrossRef]
- Huang, Z.R.; Chen, M.; Guo, W.L.; Li, T.T.; Liu, B.; Bai, W.D.; Ai, L.Z.; Rao, P.F.; Ni, L.; Lv, X.C. Monascus purpureus-fermented common buckwheat protects against dyslipidemia and non-alcoholic fatty liver disease through the regulation of liver metabolome and intestinal microbiome. Food Res. Int. 2020, 136, 109511. [Google Scholar] [CrossRef] [PubMed]
- Ayyat, M.S.; Al-Sagheer, A.A.; El-Latif, A.; Khaled, M.; Khalil, B.A. Organic selenium, probiotics, and prebiotics effects on growth, blood biochemistry, and carcass traits of growing rabbits during summer and winter seasons. Biol. Trace Elem. Res. 2018, 186, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Kong, C.; Gao, R.; Yan, X.; Huang, L.; Qin, H. Probiotics improve gut microbiota dysbiosis in obese mice fed a high-fat or high-sucrose diet. Nutrition 2019, 60, 175–184. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho Marchesin, J.; Celiberto, L.S.; Orlando, A.B.; de Medeiros, A.I.; Pinto, R.A.; Zuanon, J.A.S.; Spolidorio, L.C.; dos Santos, A.; Taranto, M.P.; Cavallini, D.C.U. A soy-based probiotic drink modulates the microbiota and reduces body weight gain in diet-induced obese mice. J. Funct. Foods 2018, 48, 302–313. [Google Scholar] [CrossRef]
- Lehtoranta, L.; Latvala, S.; Lehtinen, M.J. Role of probiotics in stimulating the immune system in viral respiratory tract infections: A narrative review. Nutrients 2020, 12, 3163. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, S.; Sabikhi, L.; Singh, A.K. Effect of whey-pearl millet-barley based probiotic beverage on Shigella-induced pathogenicity in murine model. J. Funct. Foods 2019, 54, 498–505. [Google Scholar] [CrossRef]
- Tang, C.; Zhaoxin, L. Health promoting activities of probiotics. J. Food Biochem. 2019, 43, e12944. [Google Scholar] [CrossRef]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef]
- Wan, M.L.Y.; Forsythe, S.J.; El-Nezami, H. Probiotics interaction with foodborne pathogens: A potential alternative to antibiotics and future challenges. Crit. Rev. Food Sci. Nutr. 2019, 59, 3320–3333. [Google Scholar] [CrossRef]
- Niamah, A.K.; Sahi, A.A.; Al-Sharifi, A.S. Effect of feeding soy milk fermented by probiotic bacteria on some blood criteria and weight of experimental animals. Probiotics Antimicrob. Proteins 2017, 9, 284–291. [Google Scholar] [CrossRef]
- Sharma, R.; Kumari, M.; Kumari, A.; Sharma, A.; Gulati, A.; Gupta, M.; Padwad, Y. Diet supplemented with phytochemical epigallocatechin gallate and probiotic Lactobacillus fermentum confers second generation synbiotic effects by modulating cellular immune responses and antioxidant capacity in aging mice. Eur. J. Nutr. 2019, 58, 2943–2957. [Google Scholar] [CrossRef] [PubMed]
- Mortaz, E.; Alipoor, S.D.; Adcock, I.M.; Mumby, S.; Koenderman, L. Update on neutrophil function in severe inflammation. Front. Immunol. 2018, 9, 2171. [Google Scholar] [CrossRef] [PubMed]
- Frayn, K.N.; Kingman, S.M. Dietary sugars and lipid metabolism in humans. Am. J. Clin. Nutr. 1995, 62, 250S–261S. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Ahn, J.; Kim, N.S.; Lee, B.K. High carbohydrate diets are positively associated with the risk of metabolic syndrome irrespective to fatty acid composition in women: The KNHANES 2007–2014. Int. J. Food Sci. Nutr. 2017, 68, 479–487. [Google Scholar] [CrossRef]
- Aminlari, L.; Shekarforoush, S.S.; Hosseinzadeh, S.; Nazifi, S.; Sajedianfard, J.; Eskandari, M.H. Effect of probiotics Bacillus coagulans and Lactobacillus plantarum on lipid profile and feces bacteria of rats fed cholesterol-enriched diet. Probiotics Antimicrob. Proteins 2019, 11, 1163–1171. [Google Scholar] [CrossRef]
- van Beek, A.A.; Sovran, B.; Hugenholtz, F.; Meijer, B.; Hoogerland, J.A.; Mihailova, V.; van der Ploeg, C.; Belzer, C.; Boekschoten, M.V.; Hoeijmakers, J.H.; et al. Supplementation with Lactobacillus plantarum WCFS1 prevents decline of mucus barrier in colon of accelerated aging Ercc1−/Δ7 mice. Front. Immunol. 2016, 7, 408. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Ding, Y.H.; Qian, L.Y.; Pang, J.; Lin, J.Y.; Xu, Q.; Wang, L.H.; Huang, D.S.; Zou, H. The regulation of immune cells by Lactobacilli: A potential therapeutic target for anti-atherosclerosis therapy. Oncotarget 2017, 8, 59915. [Google Scholar] [CrossRef]
- Zhu, Y.H.; Li, X.Q.; Zhang, W.; Zhou, D.; Liu, H.Y.; Wang, J.F. Dose-dependent effects of Lactobacillus rhamnosus on serum interleukin-17 production and intestinal T-cell responses in pigs challenged with Escherichia coli. Appl. Environ. Microbiol. 2014, 80, 1787–1798. [Google Scholar] [CrossRef]
- Smelt, M.J.; de Haan, B.J.; Bron, P.A.; van Swam, I.; Meijerink, M.; Wells, J.M.; Faas, M.M.; de Vos, P. Probiotics can generate FoxP3 T-cell responses in the small intestine and simultaneously inducing CD4 and CD8 T cell activation in the large intestine. PLoS ONE 2013, 8, e68952. [Google Scholar] [CrossRef]
- Xie, J.; Nie, S.; Yu, Q.; Yin, J.; Xiong, T.; Gong, D.; Xie, M. Lactobacillus plantarum NCU116 attenuates cyclophosphamide-induced immunosuppression and regulates Th17/Treg cell immune responses in mice. J. Agric. Food Chem. 2016, 64, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Santiago-López, L.; Hernández-Mendoza, A.; Garcia, H.S.; Mata-Haro, V.; Vallejo-Cordoba, B.; González-Córdova, A.F. The effects of consuming probiotic-fermented milk on the immune system: A review of scientific evidence. Int. J. Dairy Technol. 2015, 68, 153–165. [Google Scholar] [CrossRef]
| Strains | Control | Acid (pH 3.0) | Temperature (40 °C) | Bile Salts (3%) | Enzyme (Pepsin) | Overall SR (%) 1 |
|---|---|---|---|---|---|---|
| CL3 | 9.35 ± 0.01 | 8.35 ± 0.01 | 6.57 ± 0.02 | 7.12 ± 0.01 | 6.24 ± 0.05 | 67.64 a |
| PC2 | 8.34 ± 0.10 | 7.34 ± 0.10 | 6.38 ± 0.10 | 7.78 ± 0.03 | 6.03 ± 0.02 | 77.21 a |
| TA8 | 8.44 ± 0.01 | 7.44 ± 0.01 | 6.52 ± 0.05 | 7.00 ± 0.01 | 6.03 ± 0.01 | 66.14 a,b |
| PC4 | 7.97 ± 0.10 | 7.07 ± 0.10 | 6.38 ± 0.01 | 7.13 ± 0.02 | 6.39 ± 0.04 | 89.62 b |
| AFSF2 | 8.78 ± 0.01 | 7.48 ± 0.01 | 6.59 ± 0.06 | 8.13 ± 0.10 | 6.13 ± 0.01 | 77.21 c |
| L. fermentum ATCC:14931 | 8.99 ± 0.03 | 7.39 ± 0.03 | 6.37 ± 0.05 | 7.13 ± 0.03 | 6.06 ± 0.01 | 84.99 a,b |
| FS3 | 9.20 ± 0.02 | 7.20 ± 0.02 | 6.26 ± 0.01 | 8.13 ± 0.07 | 6.21 ± 0.02 | 76.38 a |
| RB5 | 8.35 ± 0.15 | 8.35 ± 0.15 | 6.78 ± 0.00 | 7.04 ± 0.07 | 6.91 ± 0.01 | 98.94 a |
| CL8 | 8.49 ± 0.02 | 8.49 ± 0.02 | 6.52 ± 0.02 | 7.00 ± 0.02 | 6.23 ± 0.06 | 88.74 b |
| HML3 | 7.07 ± 0.08 | 8.07 ± 0.08 | 6.24 ± 0.04 | 7.41± 0.09 | 6.37 ± 0.06 | 65.96 a,b,c |
| Strain | B | AMP | DA | C | E | IPM | TE | VA | AMC | GN |
|---|---|---|---|---|---|---|---|---|---|---|
| CL3 | R | R | R | S | R | R | S | R | R | S |
| PC2 | R | S | R | S | R | R | S | R | S | S |
| TA8 | R | S | R | S | R | R | R | R | S | R |
| PC4 | R | R | R | S | R | R | R | R | R | R |
| AFSF2 | R | S | R | S | R | R | S | R | R | R |
| L. fermentum ATCC:14931 | R | R | R | S | R | R | S | R | S | R |
| FS3 | R | R | R | S | R | R | S | R | R | R |
| RB5 | R | R | R | S | R | R | S | R | S | R |
| CL8 | R | R | R | S | R | R | R | R | R | R |
| HML3 | R | R | R | S | R | R | S | R | S | R |
| Sugars Fermented | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Strains Name | Cell Shape | Gas from Glucose | NH3 from Arginine | Lactate Isomer | Growth at 45 °C | Arabinose | Cellobiose | Esculin | Galactose | Lactose | Maltose |
| CL3 * | rod | − | − | DL | − | − | + | − | + | − | + |
| PC2 * | rod | − | − | DL | − | − | + | − | + | − | + |
| TA8 * | rod | − | − | DL | − | − | + | − | + | − | + |
| PC4 * | rod | − | − | DL | − | − | + | − | + | − | + |
| AFSF2 * | rod | − | − | DL | − | − | + | − | + | − | + |
| L. fermentum ATCC:14931 * | rod | − | − | DL | − | − | + | − | + | − | + |
| FS3 * | rod | − | − | DL | − | − | + | − | + | − | + |
| RB5 * | rod | − | − | DL | − | − | + | − | + | − | + |
| CL8 * | coccoid | − | + | DL | + | + | 4/1 | 4/1 | + | − | + |
| HML3 * | rod | − | − | DL | − | − | + | − | + | − | + |
| Strain | Phenotypic Identification | Genotypic Identification | Gene Accession No. |
|---|---|---|---|
| TA8 | Lactobacillus plantarum 1 | Lactiplantibacillus plantarum strain 10334 | MW090367.1 |
| PC4 | Lactobacillus plantarum 1 | Lactiplantibacillus plantarum strain 11112 | MW092969.1 |
| AFSF2 | Lactobacillus plantarum 1 | Lactiplantibacillus plantarum strain 11905 sequence | MW116637.1 |
| RB5 | Lactobacillus plantarum 1 | Lactiplantibacillus plantarum strain NHD2 16S ribosomal RNA gene, partial sequence | MW345831.1 |
| CL8 | Pediococcus spp. | Pediococcus acidilactici strain FCP-1 | MN367973.1 |
| HML3 | Lactobacillus plantarum 1 | Lactiplantibacillus plantarum strain XYY2 | MW301118.1 |
| Sample | pH Value | Acidity (% w/v) | Moisture (%) | °Brix | Cell Count (log10 CFU g−1) |
|---|---|---|---|---|---|
| NG | 5.96 ± 0.03 a | 0.17 ± 0.04 c | 38.69 ± 0.51 a | 37.6 ± 1.43 c | ND |
| PTG | 4.54 ± 0.12 b | 0.26 ± 0.06 b | 33.29 ± 0.41 b | 53.7 ± 0.98 a | ND |
| PTPG | 3.77 ± 0.06 c | 0.531 ± 0.04 a | 34.15 ± 0.48 b | 42.8 ± 1.62 b | 8.11 ± 0.45 |
| Groups | Average Weekly Weight Gain (g) | Average Feed Intake | Feed Efficiency |
|---|---|---|---|
| NG | Week 1: 0.52 ± 0.82 c | Week 1: 17.89 ± 1.35 | Week 1: 0.02 |
| Week 2: 0.52 ± 0.74 c | Week 2: 14.25 ± 0.91 | Week 2: 0.03 | |
| Week 3: 0.54 ± 0.76 b | Week 3: 13.59 ± 0.87 | Week 3: 0.03 | |
| Week 4: 1.32 ± 0.21 a | Week 4: 16.67 ± 0.54 | Week 4: 0.07 | |
| Overall: 2.90 ± 0.39 c | Overall: 15.62 ± 2.03 | Overall: 0.18 | |
| TPG | Week 1: 1.93 ± 0.49 a | Week 1: 15.96 ± 2.10 | Week 1: 0.12 |
| Week 2: 0.66 ± 0.69 b | Week 2: 15.98 ± 0.86 | Week 2: 0.04 | |
| Week 3: 0.87 ± 0.70 a | Week 3: 11.15 ± 0.54 | Week 3: 0.07 | |
| Week 4: 1.17 ± 0.23 b | Week 4: 16.84 ± 0.63 | Week 4: 0.06 | |
| Overall: 4.63 ± 0.55 a | Overall: 14.98 ± 2.58 | Overall: 0.30 | |
| PTPG | Week 1: 1.10 ± 0.92 b | Week 1: 17.21 ± 1.90 | Week 1: 0.06 |
| Week 2: 0.90 ± 0.55 a | Week 2: 18.46 ± 1.03 | Week 2: 0.04 | |
| Week 3: 0.78 ± 0.20 a | Week 3: 14.70 ± 1.42 | Week 3: 0.05 | |
| Week 4: 0.91 ± 0.18 b | Week 4: 15.09 ± 0.15 | Week 4: 0.06 | |
| Overall: 3.69 ± 0.13 b | Overall: 16.36 ± 1.77 | Overall: 0.22 |
| Test | NG | TPG | PTPG |
|---|---|---|---|
| WBC × 109/L | 3.70 ± 0.35 c | 4.30 ± 1.62 b | 4.60 ± 1.20 a |
| RBC × 1012/L | 9.69 ± 2.80 a | 9.35 ± 2.43 b | 9.66 ± 0.79 a,b |
| Neutrophil × 109/L | 0.59 ± 0.07 c | 0.69 ± 0.37 b | 0.74 ± 0.23 a |
| Lymp × 109/L | 2.81 ± 0.38 c | 3.27 ± 0.19 b | 3.54 ± 0.26 a |
| Mono × 109/L | 0.22 ± 0.04 b | 0.26 ± 0.06 a | 0.23 ± 0.01 a,b |
| Eosin × 109/L | 0.04 ± 0.00 a | 0.04 ± 0.00 a | 0.05 ± 0.01 a |
| AST (U/L) | 102.00 ± 17.00 c | 509.00 ± 24.00 a | 398.00 ± 22.00 b |
| ALT (U/L) | 36.00 ± 8.00 c | 184.00 ± 19.00 a | 54.00 ± 17.00 b |
| Gluc (mmol/L) | 11.90 ± 4.00 c | 18.30 ± 5.00 a | 14.10 ± 1.00 b |
| Tgl mmol/L | 1.51 ± 0.20 b | 2.66 ± 0.05 a | 1.56 ± 0.01 b |
| LDL mmol/L | 0.35 ± 0.33 b | 0.50 ± 0.21 a | 0.33 ± 0.01 b |
| HDL mmol/L | 2.67 ± 0.45 b | 3.04 ± 0.83 a | 3.03 ± 0.27 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussin, M.; Anzian, A.; Liew, C.X.-Q.; Muhialdin, B.J.; Mohsin, A.Z.; Fang, C.-M.; Saad, M.Z.; Ahmad, N.H.; Hassan, M.; Adnan, H.; et al. Potentially Probiotic Fermented Glutinous Rice (Oryza sativa L.) with Lactiplantibacillus plantarum Improved Immune System Response in a Small Sample of BALB/cByJ Mice. Fermentation 2022, 8, 612. https://doi.org/10.3390/fermentation8110612
Hussin M, Anzian A, Liew CX-Q, Muhialdin BJ, Mohsin AZ, Fang C-M, Saad MZ, Ahmad NH, Hassan M, Adnan H, et al. Potentially Probiotic Fermented Glutinous Rice (Oryza sativa L.) with Lactiplantibacillus plantarum Improved Immune System Response in a Small Sample of BALB/cByJ Mice. Fermentation. 2022; 8(11):612. https://doi.org/10.3390/fermentation8110612
Chicago/Turabian StyleHussin, Muhaini, Aliaa Anzian, Crystal Xiao-Qi Liew, Belal J. Muhialdin, Aliah Zannierah Mohsin, Chee-Mun Fang, Mohd Zamri Saad, Nurul Hawa Ahmad, Masriana Hassan, Hazniza Adnan, and et al. 2022. "Potentially Probiotic Fermented Glutinous Rice (Oryza sativa L.) with Lactiplantibacillus plantarum Improved Immune System Response in a Small Sample of BALB/cByJ Mice" Fermentation 8, no. 11: 612. https://doi.org/10.3390/fermentation8110612
APA StyleHussin, M., Anzian, A., Liew, C. X.-Q., Muhialdin, B. J., Mohsin, A. Z., Fang, C.-M., Saad, M. Z., Ahmad, N. H., Hassan, M., Adnan, H., & Meor Hussin, A. S. (2022). Potentially Probiotic Fermented Glutinous Rice (Oryza sativa L.) with Lactiplantibacillus plantarum Improved Immune System Response in a Small Sample of BALB/cByJ Mice. Fermentation, 8(11), 612. https://doi.org/10.3390/fermentation8110612