‘TeeBot’: A High Throughput Robotic Fermentation and Sampling System
Abstract
:1. Introduction
2. Materials and Methods
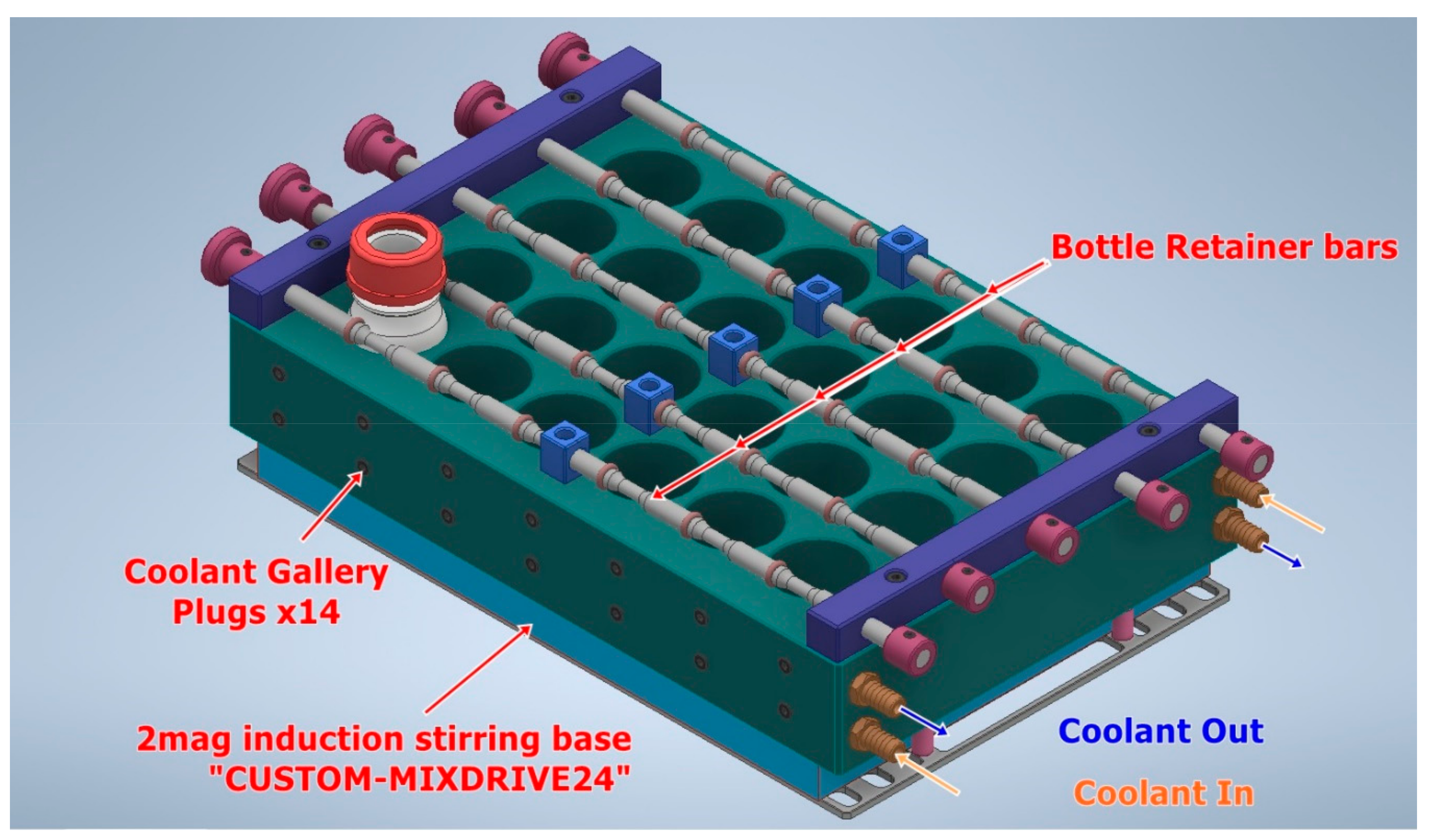
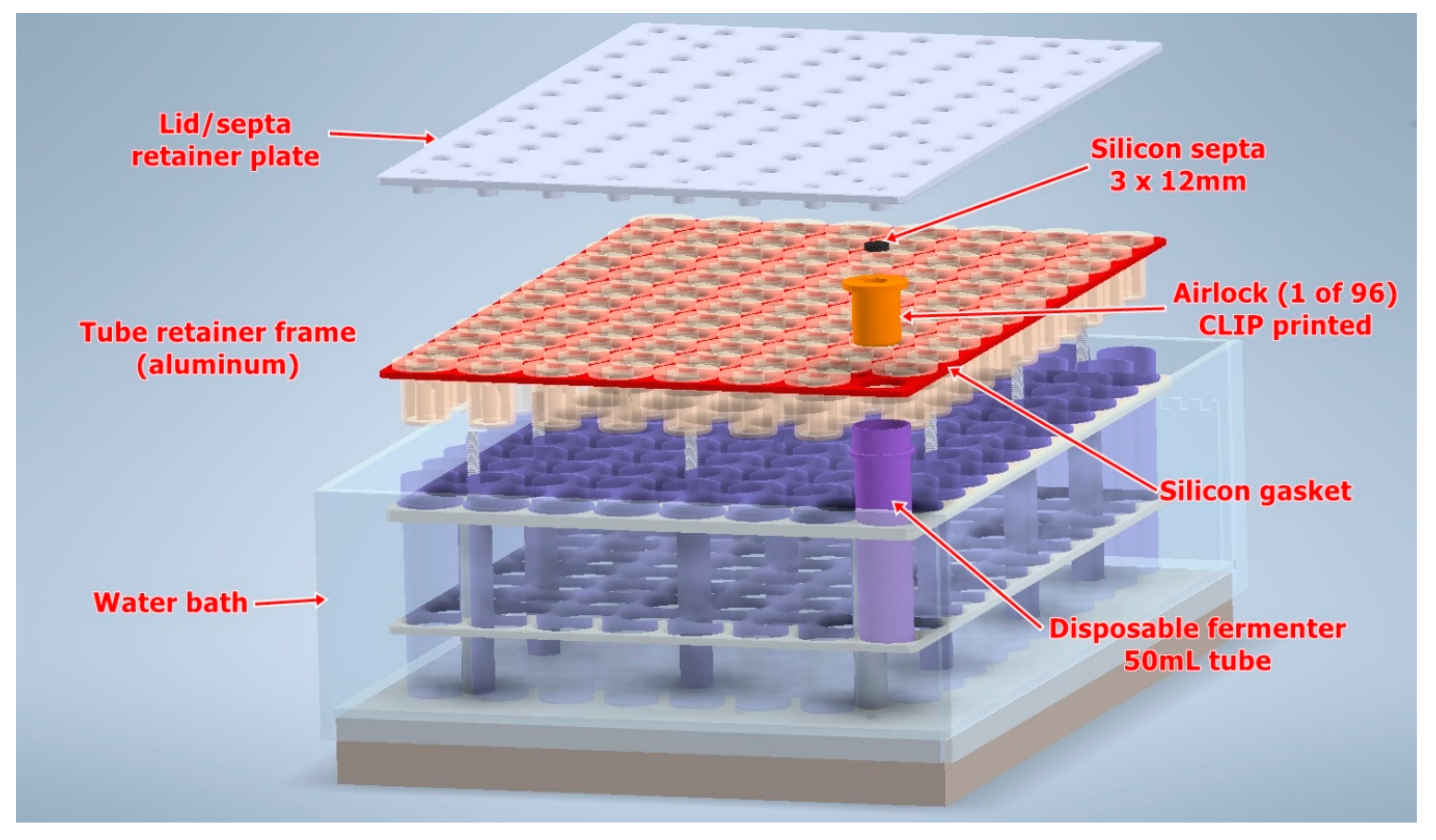
Design of Robotic Fermentation Platforms
- 96 ×100 mL ferments (Generation#1 instrument)
- II.
- 384 × 30 mL ferments (Generation#2)
- III.
- Airlock design
- IV.
- Temperature Control
- V.
- Software
- VI.
- Laboratory scale fermentations
3. Results
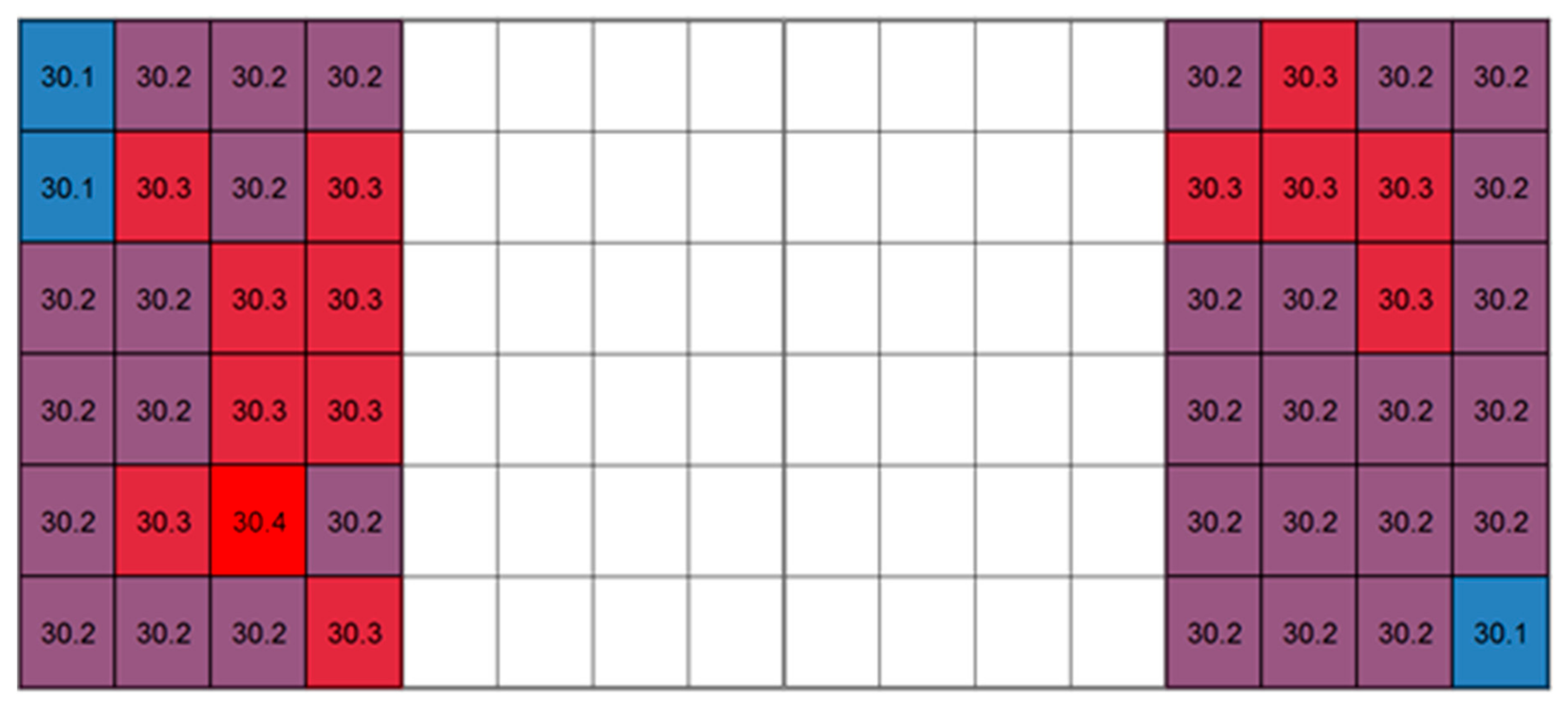
3.1. Maintenance of Temperature Uniformity
3.2. Establishment of an Anaerobic Environment
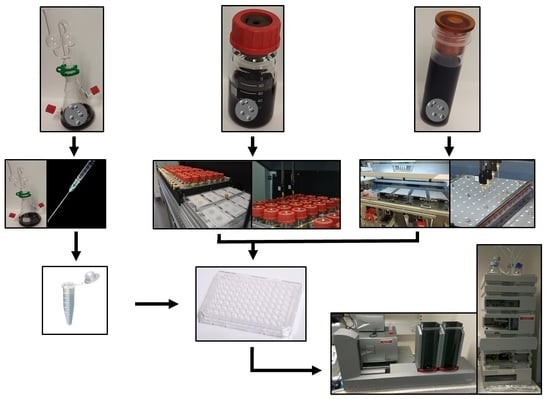
3.3. Preservation of Sterility of Individual Samples Per Se during Sampling
3.4. Reproducibility of Fermentations without Artefact, Comparable to or Better Than ‘Best Practice’
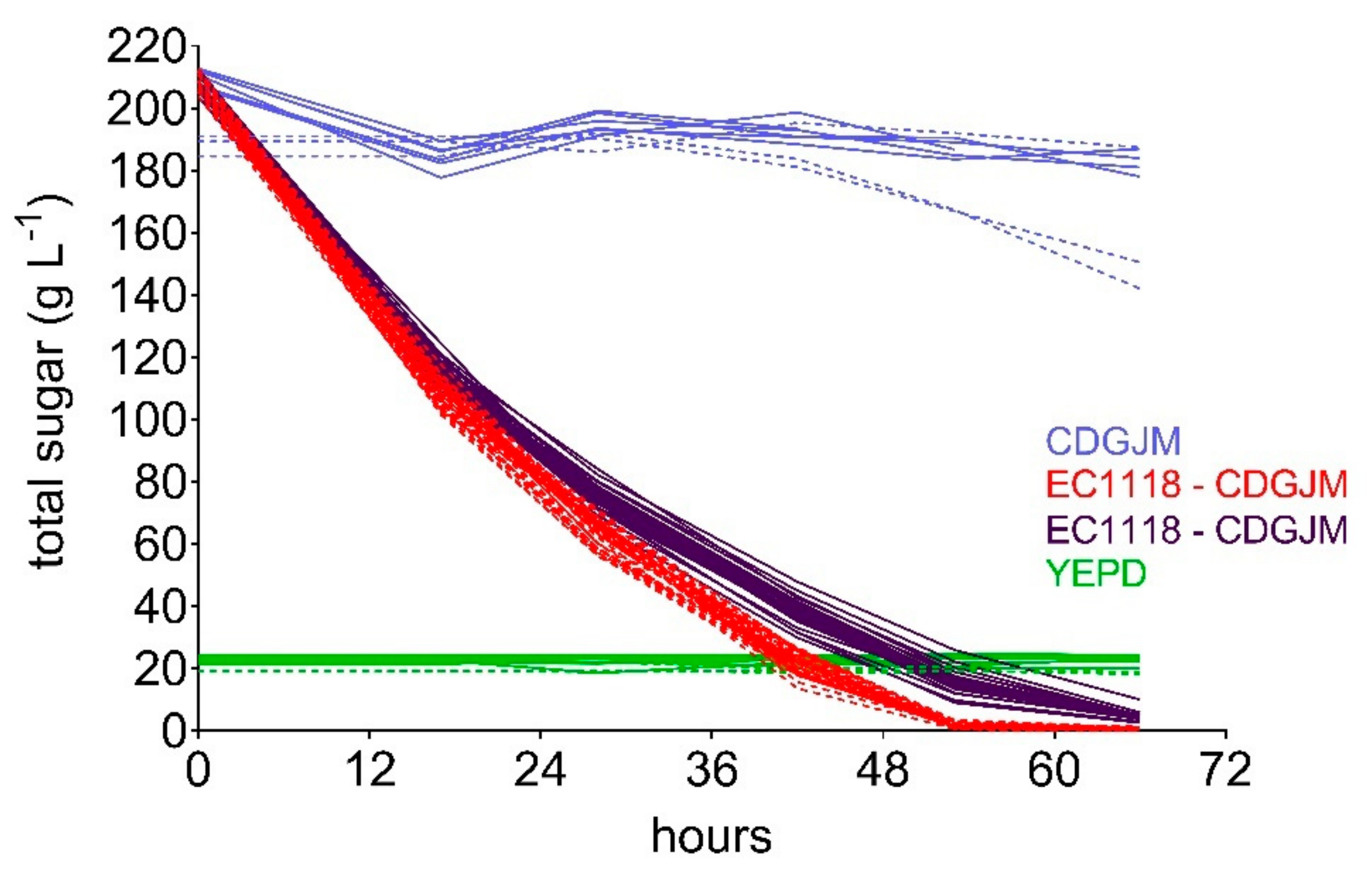
3.5. Provision of Sample Density Sufficient to Address Experimental Needs
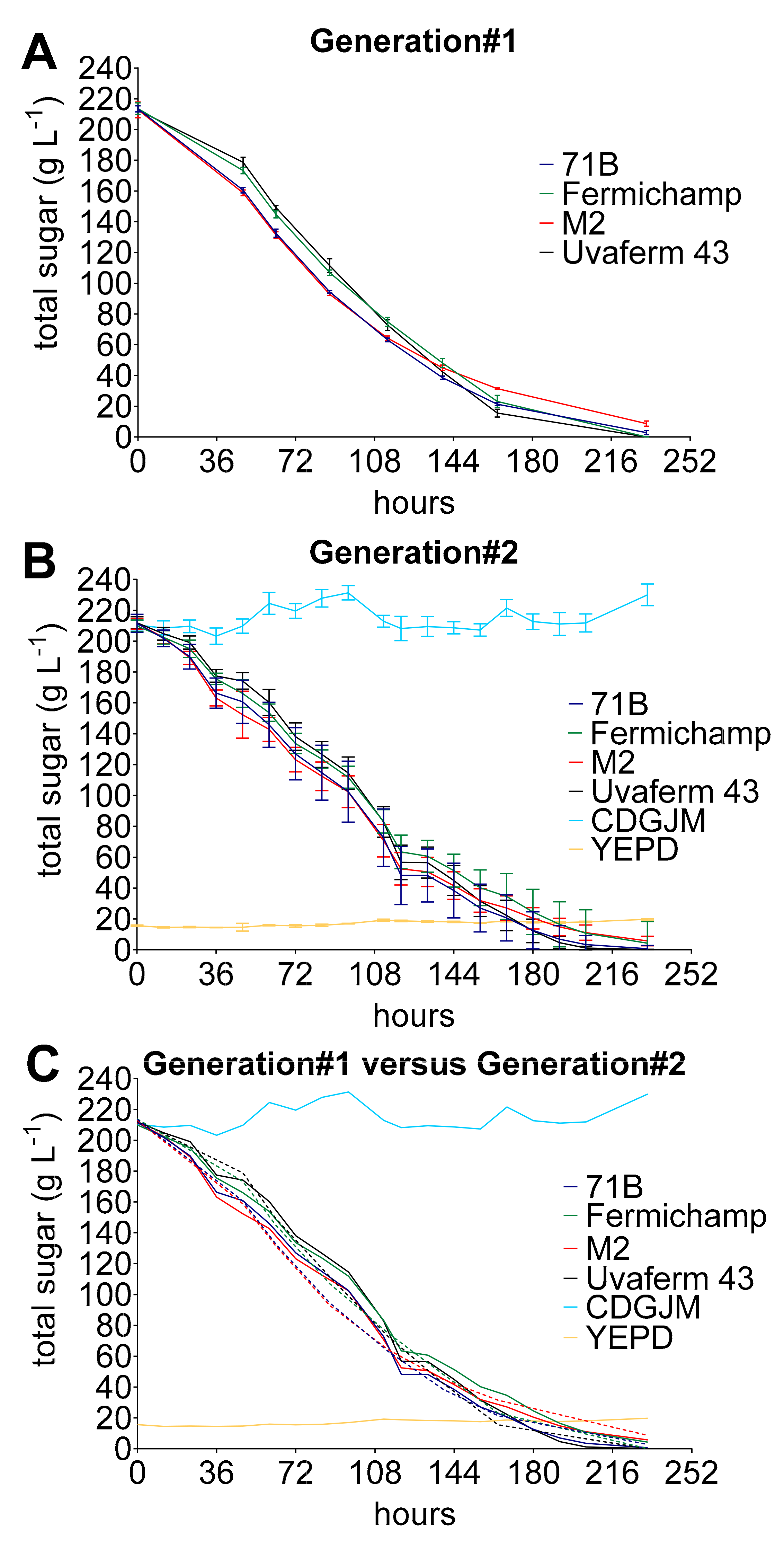
4. Discussion
5. Emerging Trends and Future Prospects
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Bartle, L.; Peltier, E.; Sundstrom, J.F.; Sumby, K.; Mitchell, J.G.; Jiranek, V.; Marullo, P. QTL mapping: An innovative method for investigating the genetic determinism of yeast-bacteria interactions in wine. Appl. Microbiol. Biotechnol. 2021, 105, 5053–5066. [Google Scholar] [CrossRef] [PubMed]
- McBryde, C.; Gardner, J.M.; de Barros Lopes, M.; Jiranek, V. Generation of novel wine yeast strains by adaptive evolution. Am. J. Enol. Vitic. 2006, 57, 423. [Google Scholar]
- Boehringer-Mannheim, Methods of Biochemical Analysis and Food Analysis Using Test-Combinations, D-Glucose/D-Fructose: UV Method; Boehringer-Mannheim GmbH Biochemicals: Mannheim, Germany, 1989; pp. 50–55.
- Gardner, J.M.; Walker, M.E.; Boss, P.K.; Jiranek, V. The effect of grape juice dilution on oenological fermentation. bioRxiv 2020. [Google Scholar] [CrossRef]
- Hranilovic, A.; Gambetta, J.M.; Schmidtke, L.; Boss, P.K.; Grbin, P.R.; Masneuf-Pomarede, I.; Bely, M.; Albertin, W.; Jiranek, V. Oenological traits of Lachancea thermotolerans show signs of domestication and allopatric differentiation. Sci. Rep. 2018, 8, 14812. [Google Scholar] [CrossRef] [PubMed]
- Liccioli, T.; Chambers, P.J.; Jiranek, V. A novel methodology independent of fermentation rate for assessment of the fructophilic character of wine yeast strains. J. Ind. Microbiol. Biotechnol. 2011, 38, 833–843. [Google Scholar] [CrossRef] [PubMed]
- Liccioli, T.; Tran, T.M.T.; Cozzolino, D.; Jiranek, V.; Chambers, P.J.; Schmidt, S.A. Microvinification—How small can we go? Appl. Microbiol. Biotechnol. 2010, 89, 1621–1628. [Google Scholar] [CrossRef] [PubMed]
- Morgan, S.C.; Haggerty, J.J.; Jiranek, V.; Durall, D.M. Competition between Saccharomyces cerevisiae and Saccharomyces uvarum in controlled Chardonnay wine fermentations. Am. J. Enol. Vitic. 2020, 71, 198–207. [Google Scholar] [CrossRef]
- Morgan, S.C.; Haggerty, J.J.; Johnston, B.; Jiranek, V.; Durall, D.M. Response to sulfur dioxide addition by two commercial Saccharomyces cerevisiae strains. Fermentation 2019, 5, 69. [Google Scholar] [CrossRef] [Green Version]
- Peter, J.J.; Watson, T.L.; E Walker, M.; Gardner, J.; A Lang, T.; Borneman, A.; Forgan, A.; Tran, T.; Jiranek, V. Use of a wine yeast deletion collection reveals genes that influence fermentation performance under low-nitrogen conditions. FEMS Yeast Res. 2018, 18, foy009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rankine, B.; Pocock, K. Alkalimetric determination of sulphur dioxide in wine. Aust. Wine Brew. Spirit Review 1970, 88, 40–44. [Google Scholar]
- Sumby, K.; Niimi, J.; Betteridge, A.; Jiranek, V. Ethanol-tolerant lactic acid bacteria strains as a basis for efficient malolactic fermentation in wine: Evaluation of experimentally evolved lactic acid bacteria and winery isolates. Aust. J. Grape Wine Res. 2019, 25, 404–413. [Google Scholar] [CrossRef]
- Tai, S.L.; Boer, V.M.; Daran-Lapujade, P.; Walsh, M.C.; de Winde, J.H.; Daran, J.M.; Pronk, J.T. Two-dimensional transcriptome analysis in chemostat cultures. Combinatorial effects of oxygen availability and macronutrient limitation in Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 437–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, M.E.; Gardner, J.M.; Vystavelova, A.; McBryde, C.; Lopes, M.D.B.; Jiranek, V. Application of the reuseable, KanMX selectable marker to industrial yeast: Construction and evaluation of heterothallic wine strains of Saccharomyces cerevisiae, possessing minimal foreign DNA sequences. FEMS Yeast Res. 2003, 4, 339–347. [Google Scholar] [CrossRef] [Green Version]
- Walker, M.E.; Nguyen, T.D.; Liccioli, T.; Schmid, F.; Kalatzis, N.; Sundstrom, J.F.; Gardner, J.M.; Jiranek, V. Genome-wide identification of the Fermentome; genes required for successful and timely completion of wine-like fermentation by Saccharomyces cerevisiae. BMC Genom. 2014, 15, 552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bitacura, J.G. The use of baker’s yeast in the Resazurin Reduction Test: A simple, low-cost method for determining cell viability in proliferation and cytotoxicity assays. J. Microbiol. Biol. Educ. 2018, 19, 19–87. [Google Scholar] [CrossRef] [PubMed]
- Wardrop, F.; Liti, G.; Cardinali, G.; Walker, G. Physiological responses of Crabtree positive and Crabtree negative yeasts to glucose upshifts in a chemostat. Ann. Microbiol. 2004, 54, 103–114. [Google Scholar]
- Aceituno, F.F.; Orellana, M.; Torres, J.; Mendoza, S.; Slater, A.W.; Melo, F.; Agosin, E. Oxygen response of the wine yeast Saccharomyces cerevisiae EC1118 grown under carbon-sufficient, nitrogen-limited enological conditions. Appl. Environ. Microbiol. 2012, 78, 8340–8352. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Strain | Generation#1 | Generation#2 |
|---|---|---|
| AUC ± Stdev (n = 3) | AUC ± Stdev (n = 19) | |
| 71B | 18,887 ± 101 | 19,261 ± 524 |
| Fermichamp | 20,383 ± 196 | 21,269 ± 448 |
| M2 | 19,608 ± 155 | 19,641 ± 311 |
| Uvaferm43 | 20,181 ± 206 | 20,693 ± 285 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Holst Pellekaan, N.; Walker, M.E.; Watson, T.L.; Jiranek, V. ‘TeeBot’: A High Throughput Robotic Fermentation and Sampling System. Fermentation 2021, 7, 205. https://doi.org/10.3390/fermentation7040205
van Holst Pellekaan N, Walker ME, Watson TL, Jiranek V. ‘TeeBot’: A High Throughput Robotic Fermentation and Sampling System. Fermentation. 2021; 7(4):205. https://doi.org/10.3390/fermentation7040205
Chicago/Turabian Stylevan Holst Pellekaan, Nicholas, Michelle E. Walker, Tommaso L. Watson, and Vladimir Jiranek. 2021. "‘TeeBot’: A High Throughput Robotic Fermentation and Sampling System" Fermentation 7, no. 4: 205. https://doi.org/10.3390/fermentation7040205
APA Stylevan Holst Pellekaan, N., Walker, M. E., Watson, T. L., & Jiranek, V. (2021). ‘TeeBot’: A High Throughput Robotic Fermentation and Sampling System. Fermentation, 7(4), 205. https://doi.org/10.3390/fermentation7040205