Abstract
Nattokinase is a serine protease in the subtilisin family which is produced by Bacillus subtilis subsp. natto and exhibits vigorous fibrinolytic activity that has been suggested to be able to prevent and treat thromboembolic diseases. In this study, WTC016, a spore-forming and rod-shaped bacterium with fibrinolytic activity was successfully isolated from soil, which was identified as Bacillus subtilis subsp. natto based on morphological and physiological tests, and phylogenetic analysis of 16S rRNA and gyrA. According to the growth curve of WTC016, the nattokinase production reached the highest amount in the stationary phase. To optimize the liquid fermentation condition for nattokinase yield of WTC016, further optimal tests of four factors, including the temperature, pH, inoculum size, and loading volume, followed by orthogonal test of all these factors, was performed. The optimal fermentation conditions were determined as 30 °C, 7.0 pH, 2% inoculum size, and 60 mL of loading volume in 250 mL conical flask, which indicates the highest nattokinase production of 3284 ± 58 IU/mL while fermented for 26 h. This work laid the foundation for producing nattokinase using Bacillus subtilis subsp. natto WTC016.
1. Introduction
Nattokinase (NK), introduced by Sumi from Natto in 1987 [1], was produced by Bacillus subtilis subsp. natto and composed of 275 amino acid residues (M.W. = 27,728 Dalton) [2]. NK is one of the most considerable extracellular serine proteinase and exhibits vigorous direct fibrinolytic activity [3]. Besides, NK also can activate the production of tissue plasminogen activator (t-PA), a single-chain protein weight of 70 kDa, which catalyzes the conversion of inactive plasminogen to active plasmin for clot break down [4,5]. Furthermore, NK enhances its fibrinolysis activity by cleaving the fibrinolysis inhibitor PAI-1 [6]. As it has been proved to be a potent thrombolytic enzyme both in vitro and in vivo [4,7], this activity has been suggested to be able to prevent and treat thromboembolic diseases. Also, long-term consumption of NK can prevent the occurrence of thromboembolic diseases [8,9,10]. At present, NK is mainly applied as a functional food ingredient, and its potential application as a drug is being investigated [11]. Compared to the clinical thrombolytic drugs (prolinase and streptokinase), NK possesses several advantages, such as being safe, low cost, all-natural supplement [12], and easy oral administration. Therefore, development of nattokinase-related products has been attracting intense interest worldwide [13].
The fermented soybean product, natto, is the main source for obtaining purified NK. A similar enzyme has been extracted from other fermented soybean-based foods, such as Thai thua nao [14], Chinese douchi [15], and Korean doen-jang [16]. Resembling fibrinolytic enzymes [17] are also obtained from other traditional fermented foods such as Chinese douche, Korean doen-jang, Korean Chungkook-jang soy sauce, and Thua nao from northern Thailand [13,14,16]. Therefore, many researchers had focused on the isolation and screening of microorganisms for enzyme production with high fibrinolytic activity [18,19], purifying and characterizing the newly found enzyme [15,20,21]. Currently, NK production is commercially available on the market [22]. However, its application is limited due to the strains sources and the low yield [11]. In recent years, many researchers have obtained hundreds of nattokinase-produced wild-type strains and nattokinase genetically engineered strains [22], but the NK activity and yield were not high enough. Most nattokinase-producing strains were isolated from the Japanese food natto or other fermented soybean-based foods [8], in contrast, few studies have been reported wild-type strains isolated from the environments (such as the soil), or concerning culture medium optimization using statistical experimental methods [23,24]. Hence, screening high-yielding wild-type strains of NK is a new approach to solve the urgent problem in both academic and commercial areas.
In this study, a wild-type nattokinase-producing strain, B. subtilis subsp. natto WTC016, was isolated from soil and identified based on morphological and physiological tests [5], as well as phylogenetic analysis of 16S rRNA and gyrA. In addition, the growth curve of WTC016 in liquid Luria-Bertani (LB) medium was determined, and optimal fermentation condition was screened out. This work laid the foundation for efficiently producing high amounts of NK using Bacillus subtilis subsp. natto WTC016.
2. Materials and Methods
2.1. Microorganism
B. subtilis subsp. natto strain WTC016 was isolated from soil samples [25] collected from Lingjia Mountain, Wuhan, Hubei, China.
2.2. Culture Media
The Luria-Bertani (LB) liquid medium: peptone 10 g/L, yeast extract 5 g/L, NaCl 10 g/L, pH 7.0 ~ 7.2, sterilized at 121 °C for 30 min, adding 15% (w/v) agar for solid medium [26].
2.3. Chemicals
The Pfu Taq DNA polymerase, DNA Marker and other molecular biological reagents were performed by Beijing TransGen Biotech Co., Ltd. Beijing, China. Chemical reagents for measuring NK activity such as fibrinogen, thrombin, urokinase, and barbital sodium and purity analysis were purchased from Sinopharm Chemical Reagents Co., Ltd. Beijing Liuyi Instrument Factory produced electrophoresis instrument (DYY-11).
2.4. Strains Isolation
A total of 5 g of soil sample was added into LB liquid medium that contained 0.25 mol/L NaAC. Sterile glass beads were added to shake and mix thoroughly. The sample was kept in 75 °C water for 10 min. The sample was centrifuged at 4000 rpm for 2 min (Eppendorf 5415D, Hamburg, Germany), and then the supernatant was diluted to 0.1 OD at 600 nm and plated on ICPM solid medium [27]. This was cultured at 30 °C for 36 h, then the morphology was observed under a 1000× microscope after simple staining.
2.5. Physiological and Biochemical Identification
The physiological and biochemical identification of WTC016 was determined, and Bacillus subtilis subsp. natto BEST195 and Bacillus subtilis subsp. subtilis 168 were used as reference strains [28,29].
2.6. Phylogenetic Tree Analysis
The primers used for cloning the 16s rRNA gene were 27F (AGAGTTTGATCCTGGCTCAG) and 1492R (TACGYTACCTTGTTACGACT) [29]. The primers used for cloning the DNA gyrase subunit A (gyrA) gene were ATGAGTGAACAAAACACAC and TCACACTTCTTCTTGTTCTT [30]. The total DNA of WTC016 was extracted following the methods as previous report [31]. The 50 μL PCR reaction system incorporated: WTC016 DNA 1 μL, primers 2 μL, dNTP 2 μL, Pfu DNA polymerase 1 μL, 5 × buffer 10 μL, and ddH2O were added to bring the final volume to 50 μL. The PCR was carried out in PCR Instrument (Bio-Rad MyCycler, Hercules, CA, USA) with following condition: 94 °C for 5 min; 95 °C for 30 s, 55 °C for 30 s, 72 °C for 2 min, 30 cycles; 72 °C for 10 min; 25 °C for 1 min. After the amplification was completed, the fragment was detected by agarose gel electrophoresis, recovered and sequenced, and then the phylogenetic tree was constructed using MEGA 5.1 software [32].
2.7. Determination of Growth Curve
A single colony was picked up and transferred to fresh liquid LB medium and incubated at 30 °C, 250 rpm overnight until the bacteria reached the stationary phase. The colony was then inoculated with 1 mL inoculum (for 100 mL LB liquid medium) and incubated at 30 °C, 250 rpm for another 24 h. The OD600 was measured every 1 h.
2.8. NK Activity Measurement
Fibrin Plate: The fibrinogen mixture consisted of 1 mL fibrinogen (5 mg/mL), 1 mL agarose solution (1%), 1 unit thrombin, and Tris–HCl buffer (pH 7.0) was poured into each well of the 12-well plate and incubated for 1 h at room temperature. The test sample was placed on the surface and incubated overnight at 37 °C [33].
Standard Curves of Urokinase Activity: The urokinase was diluted to a concentration of 100, 150, 200, 250, 300, 350, 400, and 450 IU/mL. Each 100 mL was spotted on a new prepared fibrin plate, allowed to stand for 10 min, and incubated at 37 °C for 18 h. The diameter of the lysed areas was then measured [34]. Plasminogen (with no plasmin) and urokinase were used as a negative and positive control, respectively [33]. Then to determine the urokinase enzyme concentration, the area of the transparent circles was calculated [34].
NK Activity Determination: The strain WTC016 was cultured overnight to the stationary phase (OD600 = 2.0), inoculated 0.5 mL to 50 mL liquid media and was shaken at 250 rpm, at 30 °C. Every 4 h, the sample was centrifuged at 12,000 rpm for 5 min. Then, a 100 μL of the supernatant was collected and plated on the fibrin plate and incubated for 18 h at 37 °C. The area of transparent circles was determined, which then was used to measure the enzyme concentration (NK equivalent to urokinase concentration in the fermentation broth).
2.9. Single-Factor Experiment to Optimize Fermentation Condition
Four single factors, including fermentation temperature, initial pH value, inoculum size, and loading volume were determined.
Fermentation temperature: The strain WTC016 was cultured overnight to the stationary phase (OD600 = 2.0), and then inoculated as 1% inoculum in 50 mL of liquid LB broth with 1% inoculum in 250 mL flask and fermented at different temperature conditions starting from 26, 28, 30, 32, 34, 36, 38, and 40 °C, and shaken at 250 rpm for 36 h. The NK concentration was then measured after the fermentation.
Initial pH Value: The strain was cultured overnight to the stationary phase (OD600 = 2.0) and then inoculated as 1% inoculum in 50 mL of liquid LB broth with initial pH of 5.0, 6.0, 7.0, 8.0, 9.0, 10.0, and 11.0, shaking at 30 °C and 250 rpm for 36 h in the 250 mL flask The NK concentration was measured after fermentation.
Inoculum Size: The strain was cultured overnight to the stationary phase (OD600 = 2.0) and then inoculated in 50 mL of liquid LB broth, shaking at 30 °C and 250 rpm for 36 h in the 250 mL flask. The inoculum size was varied as 0.5%, 1%, 2%, 3%, 4%, 5%, 6%, 7%, and 8% (V/V). The NK concentration was then measured after fermentation.
Loading Volume: The strain was cultured overnight to the stationary phase (OD600 = 2.0) and then inoculated as 1% inoculum in liquid LB broth, shaking at 30 °C and 250 rpm for 36 h in the 250 mL flask. The loading volume of the liquid LB broth was varied as 30, 40, 50, 60, 70, 80, 90, and 100 mL. The NK concentration was then measured after fermentation.
2.10. Orthogonal Test to Optimize Fermentation Condition
According to the above single factor results, the four factors of fermentation temperature, initial pH value, inoculum size, and loading volume were subjected to an L9 (34) orthogonal test [18,35]. The experimental design was shown in Table 1.

Table 1.
Test factors and levels of the orthogonal test design.
2.11. Data Analysis
One-way ANOVA was performed using SPSS 19.0 software, while the results were performed using Origin software, and each experiment was repeated at least three times.
3. Results and Discussion
3.1. Isolation and Morphological Characteristics of the WTC016
WTC016 was isolated from soil samples from Lingjia Mountain (Wuhan, Hubei). After being plated on LB medium and cultured for 24 h at 30 °C, the colonies of this strain were round, milky white, with a slightly toothed surface on the edges, opaque, moist, and slightly elevated (Figure 1). Based on simple staining, the bacteria cell was rod-shaped, with a length of 2.0–5.0 μm and a width of 0.5–0.8 μm (Figure 2A) after being cultured 24 h. After cultured for 72 h, WTC016 was observed to produce spores (Figure 2B). Based on the morphological characteristics of colonies and bacterial vegetative cells/spores described above, WTC016 was identified to be a Bacillus strain.
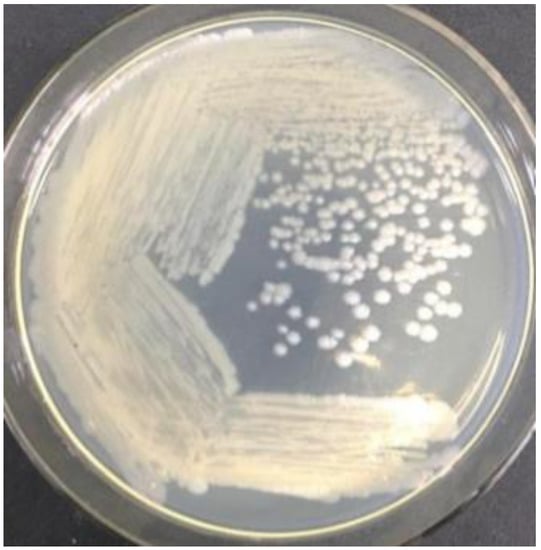
Figure 1.
The colony morphology of WTC016.
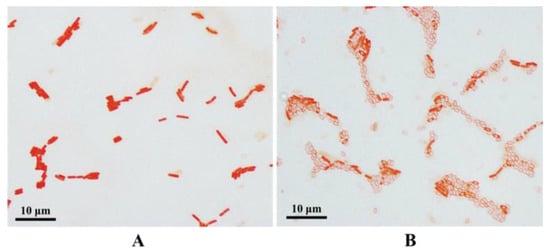
Figure 2.
The morphology of vegetative cells (A) and spores (B) of WTC016.
3.2. Molecular Identification of WTC016 Based on Phylogenetic Analysis of 16S rRNA and gyrA Genes
The phylogenetic tree based on partial 16S rRNA gene showed that WTC016 was most similar with B. subtilis subsp. natto BEST195, and affiliated with the B. subtilis group (Figure 3), which including B. subtilis and closely related taxa [36,37]. As 16sRNA analyses were difficult to differentiate the species of B. subtilis group, partial sequence of gyrA gene, which codes for DNA gyrase subunit A and have been used for accurately classifying the species in B. subtilis group (including B. subtilis) [38], was selected for further molecular identification. The partial gyrA sequence of WTC016 was 100% identical with that of B. subtilis subsp. natto BEST195. In addition, the phylogenetic tree of the partial gyrA sequences classified the WTC016 into B. subtilis (Figure 4)

Figure 3.
Phylogenetic tree based on the partial 16S rRNA of WTC016 and other homologous Bacillus strains. The phylogenetic tree was constructed using the maximum likelihood method by MEGA5 [32]. The Gen-Bank accession numbers of all the sequences are indicated in parentheses. The bar represents 0.02 substitutions per site. Dermabacter hominis (D. hominis) strain NCFB 2769 was used as the outgroup.
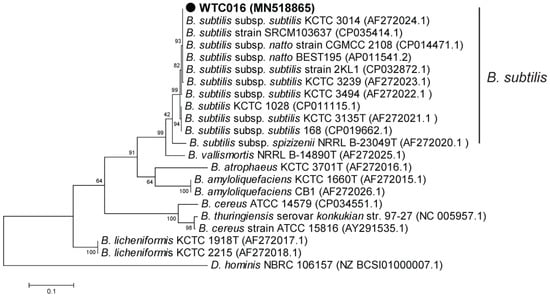
Figure 4.
Phylogenetic tree based on the partial gyrA of WTC016 and other homologous Bacillus strains. The phylogenetic tree was constructed using the maximum likelihood method by MEGA5 [32]. The Gen-Bank accession numbers of all the sequences are indicated in parentheses. The bar represents 0.01 substitutions per site. Dermabacter hominis (D. hominis) strain NCFB 2769 was used as the outgroup.
3.3. Physiological and Biochemical Identification of WTC016
According to physiological and biochemical identification (Table 2), WTC016 and Bacillus subtilis subsp. natto BEST195 were consistent in all 26 physiological and biochemical indicators, including Gram staining, sugar utilization, and other physiological and biochemical indicators. Meanwhile, WTC016, Bacillus subtilis subsp. natto BEST195 and Bacillus subtilis 168 were only different in the indicator of biotin-deficiency, which was indicated as the biotin-auxotroph in WTC016 and Bacillus subtilis subsp. natto BEST195, while non-necessity of biotin in B. subtilis 168 (Table 2). Based on this essential feature in distinguishing Bacillus subtilis subsp. natto from Bacillus subtilis [30], as well as morphological characteristics of colony and bacterial cell and the result of molecular identification, WTC016 strain was identified into Bacillus subtilis subsp. natto and named as Bacillus subtilis subsp. natto WTC016.

Table 2.
Physiological and biochemical identification of WTC016 *.
3.4. Determination of the Growth Curve and NK Yield of WTC016
The growth curve indicated that the growth of WTC016 entered the logarithmic growth phase after 3 h at 30 °C and with 250 rpm shaking in the liquid LB medium. Then the growth of the strain reached the stationary phase at 15 h after 12 h logarithmic growth (Figure 5A). At the early stage of fermentation, the amount of NK gradually increased with the increase of fermentation time. At the 26 h, the NK production reached a peak value of 1356.60 IU/mL, which followed with a constant-phase (Figure 5B).
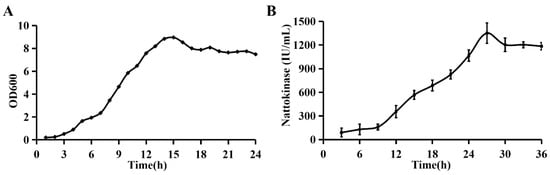
Figure 5.
Determination of the growth curve and nattokinase (NK) yield at different growth stages of WTC016. (A) The growth curve of WTC016 and (B) the dynamic of NK production throughout the cultivation of WTC016 in liquid Luria-Bertani (LB) medium.
The results indicate that the soil-sourced WTC016 possesses high level of NK productivity, and belongs to Bacillus subtilis subsp. natto. This is quite different from most reports on the discovery of fibrinolytic Bacillus subtilis group spp. strains with NK activity from the fermented food [14,15,16,18,39,40]. So far, very few Bacillus subtilis group spp. strains producing NK were isolated from the soils [24]. However, considering that the Bacillus spp. strains can distribute extensively in varied environments and possess multiple biofunctions to use the organic resources, including lignocellulose and fibrin [41,42,43,44], we predicted the nattokinase-producing Bacillus subtilis spp. strains should also widely distributed in different environments, especially in the soils. Our discovery of the soil-sourced WTC016 has partially supported this prediction.
3.5. Single-Factor Experiment to Optimize Fermentation Conditions for NK Production
The NK production increased gradually at the fermentation temperature of 26–30 °C, and reached the highest value at 30 °C, then gradually decreased along with the increase of the temperature. Thus, it can be suggested that the optimal fermentation temperature of WTC016 was 30 °C (Figure 6A), which is consistent with other strains of B. subtilis subsp. natto [18]. As the optimal temperature for the cultivation of most Bacillus strains [45], 30 °C should be best for the microbial metabolism and the activities of related enzymes, which may include NK. In addition, the temperature also affected the oxygen concentration and mass transfer rate in the fermentation broth, and, therefore, influenced the microbial metabolism [2].
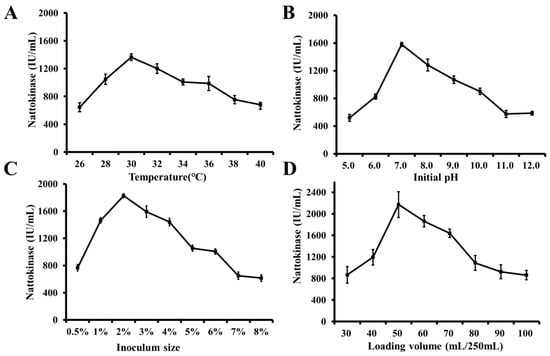
Figure 6.
Optimization of fermentation condition for producing nattokinase of WTC016. (A–D) Effects of temperature (A), initial pH initial (B), inoculum size (C), and loading volume (D), on the NK activity of WTC016.
The results presented in Figure 6B confirmed that pH 7.0 is the most conducive for the production of NK in the fermentation broth. This was consistent with the previous report stating that the enzyme activity of NK was generally stable in neutral conditions, and readily inactivated under both acidic and alkaline conditions [46]. It may be decided by that: (1) The pH of the medium can affect the growth of bacteria, the accumulation of metabolites, as well as the stability of metabolites during the fermentation process; (2) the pH of the medium can also influence the dissociation degree of the enzyme, the charge condition, the structure, and function of the protein. All these factors then alter enzyme activity.
As one of critical factor for microorganism fermentation [47], variation of the inoculum size has a significant influence on the NK production, which indicated the highest level at 2% (V/V) of inoculum size (Figure 6C) for the liquid fermentation in flask. It is consistent with that in another strain of Bacillus subtilis subsp. natto [48], while there was no correspondence with 5% (V/V) of optimal inoculum size in a 7.5 L bioreactor for the liquid fermentation of glucose oxidase in Aspergillus niger W-47, which may be because of the variation of the targeted enzymatic activities, strain species, and fermentation containers [47].
In terms of the liquid loading volume, NK production in the fermentation broth was highest at 50 mL in the 250 mL flask (Figure 6D), which is similar with the previous report [49]. Bacillus natto is an aerobic microorganism and therefore requires large amounts of oxygen to grow and produce NK. An insufficient amount of dissolved oxygen would directly hinder the growth of strains and therefore would yield little NK. In the flask culture, the amount of dissolved oxygen in the fermentation broth was inversely related to the volume of liquid in the flask. This makes the oxygen concentration in the fermentation broth a limiting factor for the synthesis of products, which causes the significant enzymatic decrease when the liquid volume was greater than 50 mL in 250 mL of flask.
3.6. Orthogonal Array Test to Optimize Fermentation Conditions for NK Production
Based on the single-factor experiment for optimizing each critical factor for fermentation, further statistical analysis for optimizing fermentation condition with multiple critical factors, such as the orthogonal array, was performed [18,19]. According to Table 1, four factors, including temperature, initial pH value, inoculum size, and liquid loading volume, were selected for orthogonal array experiment, and each of these factors contained three levels.
The optimal fermentation conditions are 30 °C, pH 7.0, 2% of inoculum size, and 60 mL of loading volume in the 250 mL flask, with which indicated the highest NK production as 3015 ± 116 IU/mL (Table 3). Moreover, the NK production reached a higher value as 3284 ± 58 IU/mL while we repeated the experiment under such fermentation conditions later. This is 2.74 fold higher than that of the original liquid fermentation of WTC016, which indicates the optimization of the fermentation conditions significant enhances NK production of this fibrinolytic strain. In addition, this extremely high level of NK activity/productivity is much higher than that of the other Bacillus spp. strains in the liquid fermentation in the shaking flask (130–1300 IU/mL) [18,24,48]. This implies the significant potential of WTC016 for NK production, which is greatly valuable for the development of dietary supplement and functional food, and the improvement of thrombosis and blood pressure [50].

Table 3.
Orthogonal array test to optimize fermentation condition for NK production.
4. Conclusions
In the present research, a fibrinolytic bacterium, WTC016, was isolated from soil samples from Lingjia Mountain (Wuhan, Hubei), and identified as Bacillus subtilis subsp. natto based on morphological, physiological and phylogenetic analysis. WTC016 not only indicated the high level of NK production, but also present a significant enhancement of NK production to extremely high levels under optimized condition of liquid fermentation in small-capacity chamber, such as the shaking flask. It implies this fibrinolytic strain, with significant potential for dietary supplement and functional food, as well as the therapy of thromboembolic diseases, which deserves further investigation, including the nattokinase-coding gene analysis and modification, could be used for the development of liquid or solid batch fermentation in high-capacity bioreactor for NK production. In addition, as an interesting example of the soil-sourced Bacillus spp. strain with extremely high NK production, the discovery of WTC016 implies the extensive distribution of fibrinolytic bacteria, especially the Bacillus subtilis subsp. natto strains, in the soil, which indicates the soil as a treasury of nattokinase-producing bacteria, including Bacillus spp. strains.
Author Contributions
S.J. and Z.C. performed the data curation, formal analysis, software data analysis, resources, and writing the original draft of the manuscript; C.W. and Y.L. reviewed and validated; M.F.F. reviewed, revised, and edited the manuscript; Z.Z. performed the funding acquisition, together with J.L. validated the methodology scheme, supervision, and the project administration, and finished most of the revision work. All listed authors discussed the results and approved the final manuscript. (to be updated by system).
Funding
This work was supported by the National Natural Science Foundation of China (31600006, 31750110464), the China Postdoctoral Science Foundation (2016M602316), the Natural Science Foundation of Hubei Province (2018CFB529), the Research Fund for the Doctoral of Wuhan Polytechnic (2016BS005), Huazhong Agricultural University, Talented Young Scientist Program (TYSP Grant No.42000481-7), and the College Excellent Youth Science and Technology Innovation Team Project of Hubei Province (T201535).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sumi, H.; Imai, M.; Naito, S.; Yatagai, C.; Yanagisawa, Y.; Yoshida, E. Nattokinase as thermally stable and broad spectrum enzyme. J. Thromb. Haemost. 2010, 8, 62. [Google Scholar]
- Unrean, P.; Nguyen, N.H.A. Metabolic pathway analysis and kinetic studies for production of nattokinase in bacillus subtilis. Bioproc. Biosyst. Eng. 2013, 36, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Furie, B.; Furie, B.C. Mechanisms of disease: Mechanisms of thrombus formation. N. Engl. J. Med. 2008, 359, 938–949. [Google Scholar] [CrossRef]
- Fujita, M.; Ito, Y.; Hong, K.; Nishimuro, S. Characterization of nattokinase-degraded products from human fibrinogen or cross-linked fibrin. Fibrinolysis 1995, 9, 157–164. [Google Scholar] [CrossRef]
- Ku, T.W.; Tsai, R.L.; Pan, T.M. A simple and cost-saving approach to optimize the production of subtilisin nat by submerged cultivation of bacillus subtilis natto. J. Agric. Food Chem. 2009, 57, 292–296. [Google Scholar] [CrossRef]
- Urano, T.; Ihara, H.; Umemura, K.; Suzuki, Y.; Oike, M.; Akita, S.; Tsukamoto, Y.; Suzuki, I.; Takada, A. The profibrinolytic enzyme subtilisin nat purified from bacillus subtilis cleaves and inactivates plasminogen activator inhibitor type 1. J. Biol. Chem. 2001, 276, 24690–24696. [Google Scholar] [CrossRef] [PubMed]
- Sumi, H.; Hamada, H.; Nakanishi, K.; Hiratani, H. Enhancement of the fibrinolytic-activity in plasma by oral-administration of nattokinase. Acta Haematol. Basel 1990, 84, 139–143. [Google Scholar] [CrossRef]
- Dabbagh, F.; Negahdaripour, M.; Berenjian, A.; Behfar, A.; Mohammadi, F.; Zamani, M.; Irajie, C.; Ghasemi, Y. Nattokinase: Production and application. Appl. Microbiol. Biotechnol. 2014, 98, 9199–9206. [Google Scholar] [CrossRef]
- Wan, R.; Hong, T.; Tariq, Y.; Chang, A. Pharmacotherapy of vitreomacular traction. Curr. Pharm. Des. 2018, 24, 4874–4881. [Google Scholar] [CrossRef]
- Raskob, G.E.; Angchaisuksiri, P.; Blanco, A.N.; Buller, H.; Gallus, A.; Hunt, B.J.; Hylek, E.M.; Kakkar, A.; Konstantinides, S.V.; McCumber, M.; et al. Thrombosis a major contributor to global disease burden. Arterioscl. Throm. Vas. 2014, 34, 2363–2371. [Google Scholar] [CrossRef]
- Peng, Y.; Yang, X.J.; Zhang, Y.Z. Microbial fibrinolytic enzymes: An overview of source, production, properties, and thrombolytic activity in vivo. Appl. Microbiol. Biotechnol. 2005, 69, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.Q.; Yao, J.; Sparks, S.; Wang, K.Y. Nattokinase: An oral antithrombotic agent for the prevention of cardiovascular disease. Int. J. Mol. Sci. 2017, 18, 523. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.T.; Luo, M.F.; Xie, Y.C.; Yang, L.R.; Li, H.J.; Xu, L.; Liu, H.Z. Strain screening, fermentation, separation, and encapsulation for production of nattokinase functional food. Appl. Biochem. Biotechnol. 2012, 168, 1753–1764. [Google Scholar] [CrossRef] [PubMed]
- Inatsu, Y.; Nakamura, N.; Yuriko, Y.; Fushimi, T.; Watanasiritum, L.; Kawamoto, S. Characterization of bacillus subtilis strains in thua nao, a traditional fermented soybean food in northern thailand. Lett. Appl. Microbiol. 2006, 43, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Huang, Q.; Zhang, R.H.; Zhang, Y.Z. Purification and characterization of a fibrinolytic enzyme produced by bacillus amyloliquefaciens dc-4 screened from douchi, a traditional chinese soybean food. Comp. Biochem. Phys. B 2003, 134, 45–52. [Google Scholar] [CrossRef]
- Kim, W.; Choi, K.; Kim, Y.; Park, H.; Choi, J.; Lee, Y.; Oh, H.; Kwon, I.; Lee, S. Purification and characterization of a fibrinolytic enzyme produced from bacillus sp strain ck 11-4 screened from chungkook-jang. Appl. Environ. Microbiol. 1996, 62, 2482–2488. [Google Scholar]
- Yao, Z.; Liu, X.M.; Shim, J.M.; Lee, K.W.; Kim, H.J.; Kim, J.H. Properties of a fibrinolytic enzyme secreted by bacillus amyloliquefaciens rsb34, isolated from doenjang. J. Microbiol. Biotechnol. 2017, 27, 9–18. [Google Scholar] [CrossRef]
- Suwanmanon, K.; Hsieh, P.C. Isolating bacillus subtilis and optimizing its fermentative medium for gaba and nattokinase production. Cyta-J. Food 2014, 12, 282–290. [Google Scholar] [CrossRef]
- Mahajan, P.M.; Gokhale, S.V.; Lele, S.S. Production of nattokinase using bacillus natto nrrl 3666: Media optimization, scale up, and kinetic modeling. Food Sci. Biotechnol. 2010, 19, 1593–1603. [Google Scholar] [CrossRef]
- Chang, C.T.; Fan, M.H.; Kuo, F.C.; Sung, H.Y. Potent fibrinolytic enzyme from a mutant of bacillus subtilis imr-nk1. J. Agric. Food Chem. 2000, 48, 3210–3216. [Google Scholar] [CrossRef]
- Agrebi, R.; Haddar, A.; Hmidet, N.; Jellouli, K.; Manni, L.; Nasri, M. Bsf1 fibrinolytic enzyme from a marine bacterium bacillus subtilis a26: Purification, biochemical and molecular characterization. Process Biochem. 2009, 44, 1252–1259. [Google Scholar] [CrossRef]
- Ni, H.; Guo, P.C.; Jiang, W.L.; Fan, X.M.; Luo, X.Y.; Li, H.H. Expression of nattokinase in escherichia coli and renaturation of its inclusion body. J. Biotechnol. 2016, 231, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.T.; Chiang, C.J.; Chao, Y.P. Medium optimization for the production of recombinant nattokinase by bacillus subtilis using response surface methodology. Biotechnol. Prog. 2007, 23, 1327–1332. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.G.; Xing, J.M.; Chang, T.S.; Ma, Z.Y.; Liu, H.Z. Optimization of nutritional conditions for nattokinase production by bacillus natto nlsse using statistical experimental methods. Process Biochem. 2005, 40, 2757–2762. [Google Scholar] [CrossRef]
- Tan, Y.X.; Mok, W.K.; Lee, J.; Kim, J.; Chen, W.N. Solid state fermentation of brewers’ spent grains for improved nutritional profile using bacillus subtilis wx-17. Fermentation 2019, 5, 52. [Google Scholar] [CrossRef]
- Kuchen, B.; Maturano, Y.P.; Mestre, M.V.; Combina, M.; Toro, M.E.; Vazquez, F. Selection of native non-saccharomyces yeasts with biocontrol activity against spoilage yeasts in order to produce healthy regional wines. Fermentation 2019, 5, 60. [Google Scholar] [CrossRef]
- Zhou, Y.; Ye, W.X.; Zhou, Y.; Zhu, C.G.; Sun, M.; Yu, Z.N. Ethanol tolerance, yield of melanin, swarming motility and growth are correlated with the expression levels of aiia gene in bacillus thuringiensis. Enzym. Microb. Technol. 2006, 38, 967–974. [Google Scholar] [CrossRef]
- Li, S.; Du, S.; Li, C. Screening and identification of the antagonistic strain dl-59 of b. Velezensis against a. Brassicae and biocontrol efficiency. Front. Agric.China 2012, 5, 581–587. [Google Scholar] [CrossRef]
- Boone, D.R.; Castenholz, R.W.; Garrity, G.M. Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Springer: New York, NY, USA, 2001. [Google Scholar]
- Kubo, Y.; Rooney, A.P.; Tsukakoshi, Y.; Nakagawa, R.; Hasegawa, H.; Kimura, K. Phylogenetic analysis of bacillus subtilis strains applicable to natto (fermented soybean) production. Appl. Environ. Microbiol. 2011, 77, 6463–6469. [Google Scholar] [CrossRef]
- Mccorkle, G. Molecular-cloning-a laboratory manual-maniatis, t, fritsch, ef, sambrook, j. Am. Sci. 1983, 71, 418. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. Mega5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.L.; Wang, Y.P.; Xiao, Y.; Wang, Y.; Wu, J.; Liu, C.B.; Ye, H.H.; Li, F.L.; Yu, H.N.; Lai, R. A bi-functional anti-thrombosis protein containing both direct-acting fibrin(ogen)olytic and plasminogen-activating activities. PLoS ONE 2011, 6, e17519. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.M.; Feng, C.; Zhong, J.; Huan, L.D. Roles of s3 site residues of nattokinase on its activity and substrate specificity. J. Biochem. 2007, 142, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Azin, M.; Moravej, R.; Zareh, D. Production of xylanase by trichoderma longibrachiatum on a mixture of wheat bran and wheat straw: Optimization of culture condition by taguchi method. Enzym. Microb. Technol. 2007, 40, 801–805. [Google Scholar] [CrossRef]
- Berendsen, E.M.; Zwietering, M.H.; Kuipers, O.P.; Wells-Bennik, M.H.J. Two distinct groups within the bacillus subtilis group display significantly different spore heat resistance properties. Food Microbiol. 2015, 45, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Peltier, G.L.; Schroeder, F.R. The relation between proteolytic and amylolytic enzyme production by isolates of the bacillus-subtilis group. J. Bacteriol. 1949, 57, 127–130. [Google Scholar] [PubMed]
- Chun, J.; Bae, K.S. Phylogenetic analysis of bacillus subtilis and related taxa based on partial gyra gene sequences. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2000, 78, 123–127. [Google Scholar] [CrossRef]
- Kim, S.H.; Choi, N.S. Purification and characterization of subtilisin dj-4 secreted by bacillus sp strain dj-4 screened from doen-jang. Biosci. Biotechnol. Biochem. 2000, 64, 1722–1725. [Google Scholar] [CrossRef]
- Vaithilingam, M.; Chandrasekaran, S.D.; Gupta, S.; Paul, D.; Sahu, P.; Selvaraj, J.N.; Babu, V. Extraction of nattokinase enzyme from bacillus cereus isolated from rust. Natl. Acad. Sci. Lett. India 2016, 39, 263–267. [Google Scholar] [CrossRef]
- Altayar, M.; Sutherland, A.D. Bacillus cereus is common in the environment but emetic toxin producing isolates are rare. J. Appl. Microbiol. 2006, 100, 7–14. [Google Scholar] [CrossRef]
- Nicholson, W.L. Roles of bacillus endospores in the environment. Cell. Mol. Life Sci. 2002, 59, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Gong, G.; Kim, S.; Lee, S.M.; Woo, H.M.; Park, T.H.; Um, Y. Complete genome sequence of bacillus sp 275, producing extracellular cellulolytic, xylanolytic and ligninolytic enzymes. J. Biotechnol. 2017, 254, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Raza, M.F.; Zheng, Z.Q.; Zhang, X.H.; Dong, X.X.; Zhang, H.Y. Complete genome sequence of bacillus velezensis zy-1-1 reveals the genetic basis for its hemicellulosic/cellulosic substrate-inducible xylanase and cellulase activities. 3 Biotech 2018, 8, 465. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.H.; Song, J.Y.; Kim, K.M.; Kim, M.K.; Lee, I.Y.; Kim, S.B.; Kim, H.S.; Han, N.S.; Lee, B.H.; Kim, B.S. Production of nattokinase by batch and fed-batch culture of bacillus subtilis. N. Biotechnol. 2010, 27, 341–346. [Google Scholar] [CrossRef]
- Zheng, Z.L.; Zuo, Z.Y.; Liu, Z.G.; Tsai, K.C.; Liu, A.F.; Zou, G.L. Construction of a 3d model of nattokinase, a novel fibrinolytic enzyme from bacillus natto. A novel nucleophilic catalytic mechanism for nattokinase. A novel nucleophilic catalytic mechanism for nattokinase. J. Mol. Graph. Model. 2005, 23, 373–380. [Google Scholar] [CrossRef]
- Ul Haq, I.; Nawaz, A.; Mukhtar, H.; Asad-Ur-Rehman. Optimization of inoculum volume, fermentation medium and aeration rate for the production of glucose oxidase by uv mutant strain of aspergillus niger an-14. Pak. J. Bot. 2015, 47, 329–332. [Google Scholar]
- Xie, Q.; Guo, Y. The optimization of fermentation conditions of nattokinase. J. South China Univ. Technol. (Nat. Sci.) 1999, 27, 127–131. [Google Scholar]
- Hu, L.L.; Li, J.; Zhang, Y.F.; Yang, W.Z. Optimization of liquid fermentation conditions of nattokinase. Chem. Bioeng. 2011, 28, 42–45. [Google Scholar]
- Berenjian, A.; Mahanama, R.; Kavanagh, J.; Dehghani, F.; Ghasemi, Y. Nattokinase production: Medium components and feeding strategy studies. Chem. Ind. Chem. Eng. Q. 2014, 20, 541–547. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).