Advances in the Study of Candida stellata
Abstract
1. Characteristics of the Genus Candida
2. Ecological and Physiological Properties of Genus Candida
3. Methods of Isolation and Identification of Genus Candida
4. Characteristics of Candida stellata
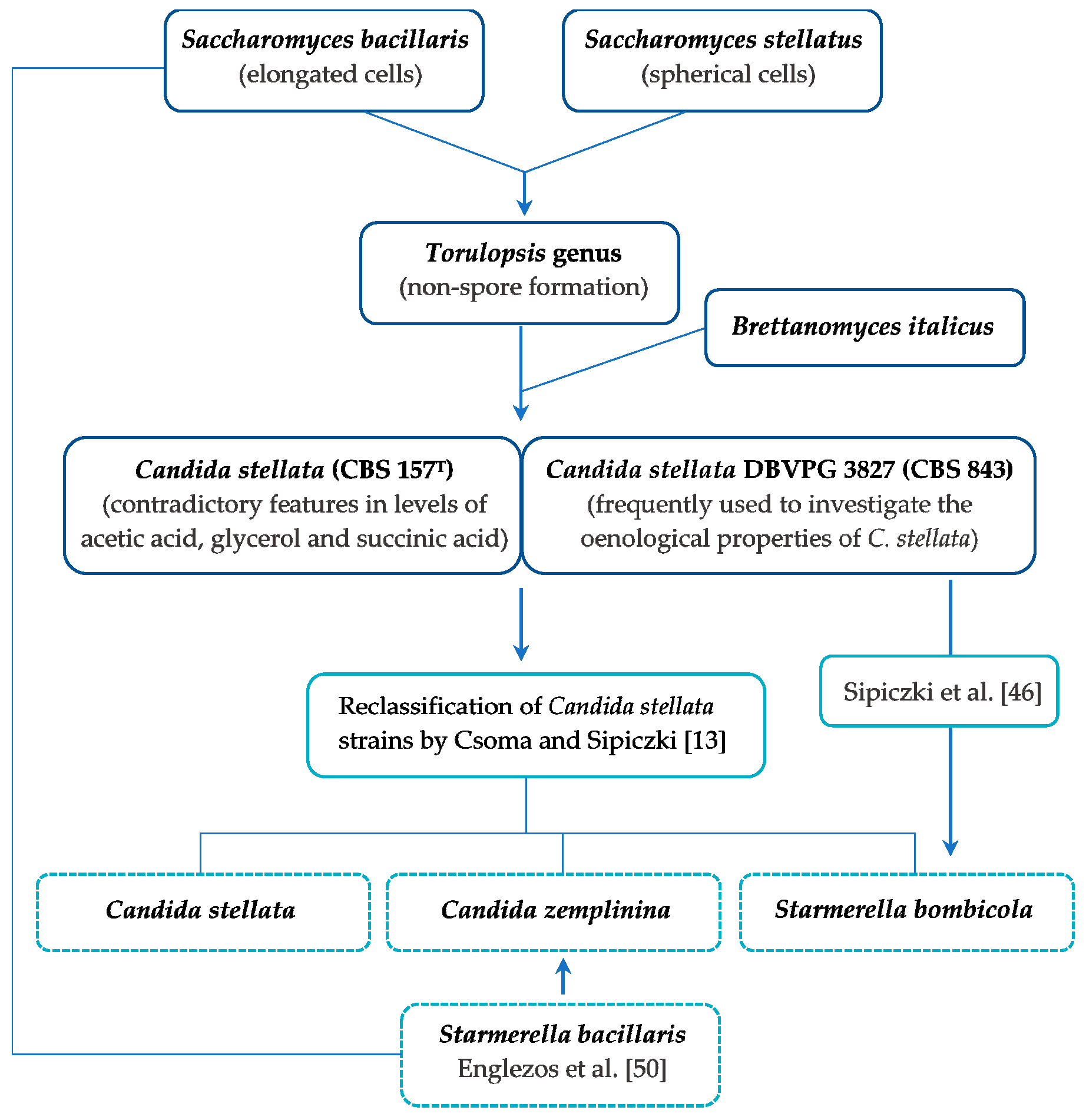
5. Taxonomic Reclassification of Candida stellata
6. Characteristics of Candida zemplinina sp. nov. Sipiczki
7. Characteristics of Starmerella bombicola
8. Characteristics of Starmerella bacillaris (synonym C. zemplinina)
9. Metabolic Features and By-Products from Candida stellata Activity
9.1. Fructophilic Character
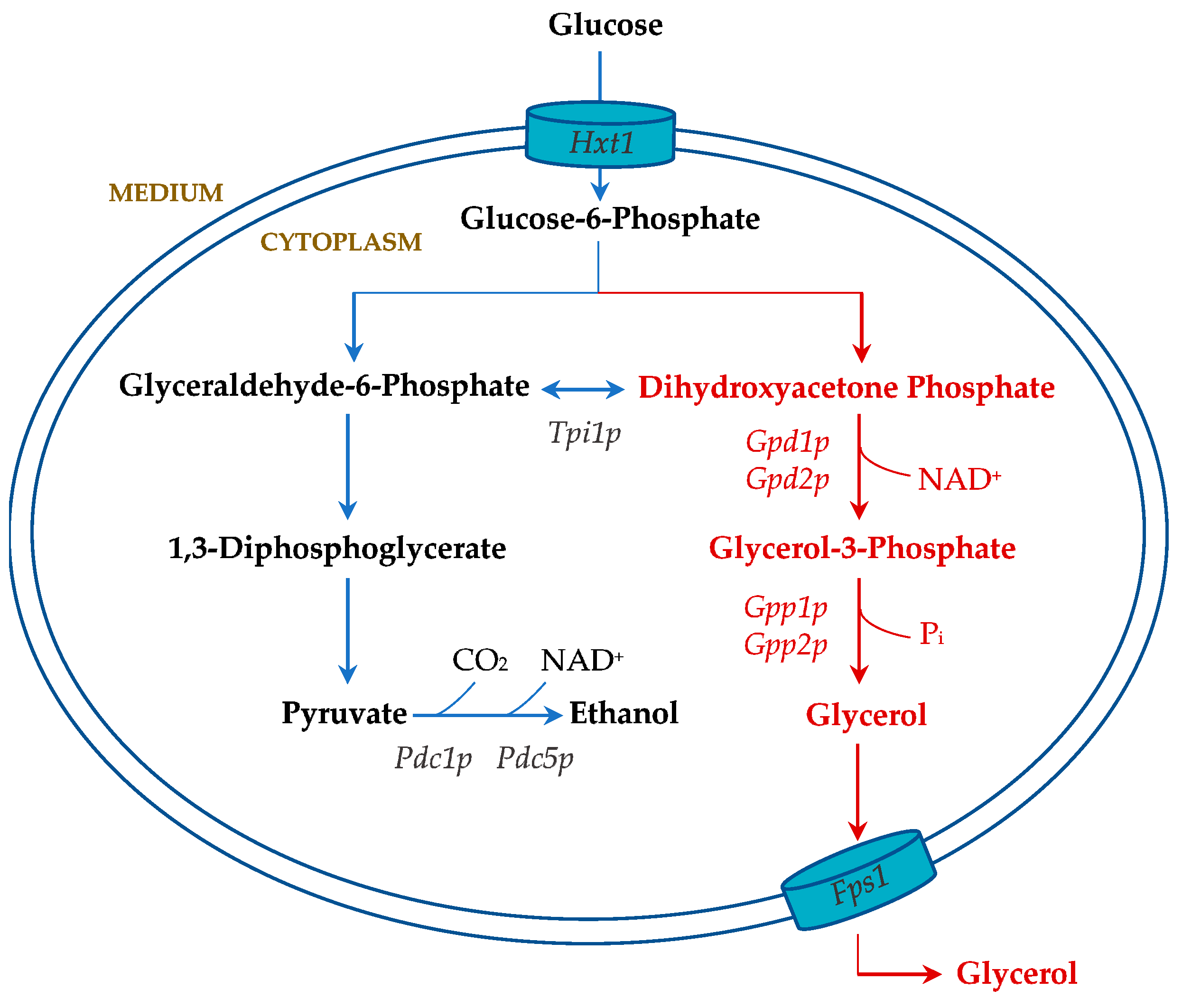
9.2. Alternative Carbon Metabolism: Glycerol Production
9.3. Biotechnological Application of Extracellular Enzymes Secreted by Candida stellata
9.3.1. Pectinases
9.3.2. Proteases
9.3.3. Cellulases and Hemicellulases
9.3.4. Glycosidases
9.3.5. Invertases
9.4. Production of Sophorolipids Biosurfactants by Candida
10. Co-Fermentations between Candida stellata and Saccharomyces cerevisiae: A Way against Standardized Wines
- Effect on some analytical compounds as increased glycerol concentration, enhanced total acidity, and reduced acetic acid concentration of wine.
- Enhancement of desirable aromatic compounds (esters, volatile thiols).
- Reduction of final ethanol content of the wine.
- Improvement of complexity and overall quality of wine.
- Larger release of polysaccharides (mannoproteins).
11. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lachance, M.A.; Boekhout, T.; Scorzetti, G.; Fell, J.W.; Kurtzman, C.P. Candida Berkhout (1923). In The Yeasts; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier, B.V.: Amsterdam, The Netherlands, 2011; pp. 987–1278. ISBN 9780444521491. [Google Scholar]
- Hommel, R.K. Candida: Introduction. In Encyclopedia of Food Microbiology: Second Edition; Batt, C.A., Tortorello, M.L., Eds.; Elsevier: London, UK, 2014; pp. 367–373. ISBN 9780123847331. [Google Scholar]
- Van Bogaert, I.N.A.; Zhang, J.; Soetaert, W. Microbial synthesis of sophorolipids. Process Biochem. 2011, 46, 821–833. [Google Scholar] [CrossRef]
- Strauss, M.L.A.; Jolly, N.P.; Lambrechts, M.G.; van Rensburg, P. Screening for the production of extracellular hydrolytic enzymes by non-Saccharomyces wine yeasts. J. Appl. Microbiol. 2001, 91, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Günther, C.S.; Goddard, M.R. Do yeasts and Drosophila interact just by chance? Fungal Ecol. 2018. [Google Scholar] [CrossRef]
- Pretorius, I.S.; van der Westhuizen, T.J.; Augustyn, O.P.H. Yeast biodiversity in vineyards and wineries and its importance to the South African wine industry. S. Afr. J. Enol. Vitic. 1999, 20, 61–75. [Google Scholar] [CrossRef]
- Mills, D.A.; Johannsen, E.A.; Cocolin, L. Yeast diversity and persistence in botrytis-affected wine fermentations. Appl. Environ. Microbiol. 2002, 68, 4884–4893. [Google Scholar] [CrossRef] [PubMed]
- Mora, J.; Mulet, A. Effects of some treatments of grape juice on the population and growth of yeast species during fermentation. Am. J. Enol. Vitic. 1991, 42, 133–136. [Google Scholar]
- Antunovics, Z.; Csoma, H.; Sipiczki, M. Molecular and genetic analysis of the yeast flora of botrytized Tokaj wines. Bull. O.I.V. 2003, 76, 380–397. [Google Scholar]
- Soden, A.; Francis, I.L.; Oakey, H.; Henschke, P.A. Effects of co-fermentation with Candida stellata and Saccharomyces cerevisiae on the aroma and composition of Chardonnay wine. Aust. J. Grape Wine Res. 2000, 6, 21–30. [Google Scholar] [CrossRef]
- Sipiczki, M. Candida zemplinina sp. nov., an osmotolerant and psychrotolerant yeast that ferments sweet botrytized wines. Int. J. Syst. Evol. Microbiol. 2003, 53, 2079–2083. [Google Scholar] [CrossRef] [PubMed]
- Sipiczki, M. Species identification and comparative molecular and physiological analysis of Candida zemplinina and Candida stellata. J. Basic Microbiol. 2004, 44, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Csoma, H.; Sipiczki, M. Taxonomic reclassification of Candida stellata strains reveals frequent occurrence of Candida zemplinina in wine fermentation. FEMS Yeast Res. 2008, 8, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Boulton, R.B.; Singleton, V.L.; Bisson, L.F.; Kunkee, R.E. Principles and Practices of Winemaking; Chapman & Hall: New York, NY, USA, 1996. [Google Scholar]
- Fuselsang, K.C. Wine Microbiology; Chapman & Hall: New York, NY, USA, 1997. [Google Scholar]
- Pramateftaki, P.V.; Lanaridis, P.; Typas, M.A. Molecular identification of wine yeasts at species or strain level: A case study with strains from two vine-growing areas of Greece. J. Appl. Microbiol. 2000, 89, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Valente, P.; Gouveia, F.C.; De Lemos, G.A.; Pimentel, D.; Van Elsas, J.D.; Mendonça-Hagler, L.C.; Hagler, A.N. PCR amplification of the rDNA internal transcribed spacer region for differentiation of Saccharomyces cultures. FEMS Microbiol. Lett. 1996, 137, 253–256. [Google Scholar] [CrossRef] [PubMed]
- Guillamón, M.; Sabat, J.; Barrio, E.; Cano, J.; Querol, A. Rapid identification of wine yeast species based on RFLP analysis of the ribosomal internal transcribed spacer (ITS) region. Arch. Microbiol. 1998, 169, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Esteve-Zarzoso, B.; Belloch, C.; Uruburu, F.; Querol, A. Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Bacteriol. 1999, 49, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Cocolin, L.; Bisson, L.F.; Mills, D.A. Direct profiling of the yeast dynamics in wine fermentations. FEMS Microbiol. Lett. 2000, 189, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Cocolin, L.; Heisey, A.; Mills, D.A. Direct identification of the indigenous yeasts in commercial wine fermentations. Am. J. Enol. Vitic. 2001, 52, 49–53. [Google Scholar]
- López, V.; Fernández-Espinar, M.T.; Barrio, E.; Ramón, D.; Querol, A. A new PCR-based method for monitoring inoculated wine fermentations. Int. J. Food Microbiol. 2003, 81, 63–71. [Google Scholar] [CrossRef]
- Cocolin, L.; Mills, D.A. Wine yeast inhibition by sulfur dioxide: A comparison of culture-dependent and independent methods. Am. J. Enol. Vitic. 2003, 54, 125–130. [Google Scholar]
- Vitzthum, F.; Bernhagen, J. SYBR Green I: An ultrasensitive fluorescent dye for double-stranded DNA quantification in solution and other applications. Recent Res. Devel. Anal. Biochem. 2002, 2, 65–93. [Google Scholar]
- Phister, T.G.; Mills, D. A Real-time PCR assay for detection and enumeration of Dekkera bruxellensis in wine. Appl. Environ. Microbiol. 2003, 69, 7430–7434. [Google Scholar] [CrossRef] [PubMed]
- García, M.; Esteve-Zarzoso, B.; Crespo, J.; Cabellos, J.M.; Arroyo, T. Yeast monitoring of wine mixed or sequential fermentations made by native strains from D.O. “Vinos de Madrid” using real-time quantitative PCR. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Minarik, E.; Hanikova, A. Die hefeflora konzentrierer traubenermoste und deren einfluss auf die stabilitat der weine. Wein-Wissen 1982, 3, 187–192. [Google Scholar]
- Rosini, G.; Federici, F.; Martini, A. Yeast flora of grape berries during ripening. Microb. Ecol. 1982, 8, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Fleet, G.H. Evolution of yeasts and lactic acid bacteria during fermentation and storage of Bordeaux wines. Appl. Environ. Microbiol. 1984, 48, 1034–1038. [Google Scholar] [PubMed]
- Pardo, I.; Garcia, M.I.; Zuniga, M.; Uruburu, F. Dynamics of microbial populations during fermentation of wine from the Utiel-Requena region of Spain. Appl. Environ. Microbiol. 1989, 50, 539–541. [Google Scholar]
- Holloway, P.; van Twest, R.A.; Subden, R.E.; Lachance, M.A. A strain of Candida stellata of special interest to oenologists. Food Res. Int. 1992, 25, 147–149. [Google Scholar] [CrossRef]
- Constanti, M.; Poblet, M.; Arola, L.; Mas, A.; Guillamón, J.M. Analysis of yeast populations during alcoholic fermentation in a newly established winery. Am. J. Enol. Vitic. 1997, 48, 339–344. [Google Scholar]
- Fernández, M.T.; Úbeda, J.F.; Briones, A.I. Comparative study of non-Saccharomyces microflora of musts in fermentation, by physiological and molecular methods. FEMS Microbiol. Lett. 1999, 173, 223–229. [Google Scholar] [CrossRef]
- Torija, M.J.; Rozès, N.; Poblet, M.; Guillamón, J.M.; Mas, A. Yeast population dynamics in spontaneous fermentations: Comparison between two different wine-producing areas over a period of three years. Antonie Van Leeuwenhoek 2001, 79, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Clemente-Jiménez, J.M.; Mingorance-Cazorla, L.; Martínez-Rodríguez, S.; Las Heras-Vázquez, F.J.; Rodríguez-Vico, F. Molecular characterization and oenological properties of wine yeasts isolated during spontaneous fermentation of six varieties of grape must. Food Microbiol. 2004, 21, 149–155. [Google Scholar] [CrossRef]
- Combina, M.; Elía, A.; Mercado, L.; Catania, C.; Ganga, A.; Martínez, C. Dynamics of indigenous yeast populations during spontaneous fermentation of wines from Mendoza, Argentina. Int. J. Food Microbiol. 2005, 99, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Divol, B.; Lonvaud-Funel, A. Evidence for viable but nonculturable yeasts in botrytis-affected wine. J. Appl. Microbiol. 2005, 99, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Hierro, N.; González, Á.; Mas, A.; Guillamón, J.M. Diversity and evolution of non-Saccharomyces yeast populations during wine fermentation: Effect of grape ripeness and cold maceration. FEMS Yeast Res. 2006, 6, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Mora, J.; Barbas, J.I.; Mulet, A. Growth of yeast species during the fermentation of musts inoculated with Kluyveromyces thermotolerans and Saccharomyces cerevisiae. Am. J. Enol. Vitic. 1990, 41, 156–159. [Google Scholar]
- Povhe Jemec, K.; Raspor, P. Initial Saccharomyces cerevisiae concentration in single or composite cultures dictates bioprocess kinetics. Food Microbiol. 2005, 22, 293–300. [Google Scholar] [CrossRef]
- Xufre, A.; Albergaria, H.; Inácio, J.; Spencer-Martins, I.; Gírio, F. Application of fluorescence in situ hybridisation (FISH) to the analysis of yeast population dynamics in winery and laboratory grape must fermentations. Int. J. Food Microbiol. 2006, 108, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.S. Wine Science-Principles, Practice, Perception, 2nd ed.; Academic Press: San Diego, CA, USA, 2000. [Google Scholar]
- Šipiczki, M.; Ciani, M.; Csoma, H. Taxonomic reclassification of Candida stellata DBVPG 3827. Folia Microbiol. (Praha) 2005, 50, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Englezos, V.; Giacosa, S.; Rantsiou, K.; Rolle, L.; Cocolin, L. Starmerella bacillaris in winemaking: Opportunities and risks. Curr. Opin. Food Sci. 2017, 17, 30–35. [Google Scholar] [CrossRef]
- Soles, R.M.; Ough, C.S.; Kunkee, R.E. Ester concentration differences in wine fermented by various species and strains of yeasts. Am. J. Enol. Vitic. 1982, 33, 94–98. [Google Scholar]
- Ciani, M.; Ferraro, L. Enhanced glycerol content in wines made with immobilized Candida stellata cells. Appl. Environ. Microbiol. 1996, 62, 128–132. [Google Scholar] [PubMed]
- Ciani, M.; Maccarelli, F. Oenological properties of non-Saccharomyces yeasts associated with wine-making. World J. Microbiol. Biotechnol. 1998, 14, 199–203. [Google Scholar] [CrossRef]
- Rosa, C.A.; Lachance, M.A. The yeast genus Starmerella gen. nov. and Starmerella bombicola sp. nov., the teleomorph of Candida bombicola (Spencer, Gorin & Tullock) Meyer & Yarrow. Int. J. Syst. Bacteriol. 1998, 48, 1413–1417. [Google Scholar] [CrossRef] [PubMed]
- Barbe, J.C.; De Revel, G.; Joyeux, A.; Bertrand, A.; Lonvaud-Funel, A. Role of botrytized grape micro-organisms in SO2 binding phenomena. J. Appl. Microbiol. 2001, 90, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Coton, E.; Coton, M.; Levert, D.; Casaregola, S.; Sohier, D. Yeast ecology in French cider and black olive natural fermentations. Int. J. Food Microbiol. 2006, 108, 130–135. [Google Scholar] [CrossRef] [PubMed]
- McNeill, J.; Barrie, F.; Buck, W.; Demoulin, V.; Greuter, W.; Hawkworth, D.L.; Herendeen, P.; Knapp, S.; Marhold, K.; Prado, J.; et al. International Code of Nomenclature for Algae, Fungi and Plants; Regnum Vegetabile: Melbourne, Australia, 2012. [Google Scholar]
- Duarte, F.L.; Pimentel, N.H.; Teixeira, A.; Fonseca, A. Saccharomyces bacillaris is not a synonym of Candida stellata: Reinstatement as Starmerella bacillaris comb. nov. Antonie van Leeuwenhoek 2012, 102, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, D.S.; Hønholt, S.; Tano-Debrah, K.; Jespersen, L. Yeast populations associated with Ghanaian cocoa fermentations analysed using denaturing gradient gel electrophoresis (DGGE). Yeast 2005, 22, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, D.S.; Teniola, O.D.; Ban-Koffi, L.; Owusu, M.; Andersson, T.S.; Holzapfel, W.H. The microbiology of Ghanaian cocoa fermentations analysed using culture-dependent and culture-independent methods. Int. J. Food Microbiol. 2007, 114, 168–186. [Google Scholar] [CrossRef] [PubMed]
- Lachance, M.A.; Starmer, W.T.; Rosa, C.A.; Bowles, J.M.; Barker, J.S.F.; Janzen, D.H. Biogeography of the yeasts of ephemeral flowers and their insects. FEMS Yeast Res. 2001, 1, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, V.; Malfeito-Ferreira, M. Spoilage yeasts in the wine industry. Int. J. Food Microbiol. 2003, 86, 23–50. [Google Scholar] [CrossRef]
- Puig, S.; Querol, A.; Barrio, E.; Pérez-Ortín, J.E. Mitotic recombination and genetic changes in Saccharomyces cerevisiae during wine fermentation. Appl. Environ. Microbiol. 2000, 66, 8–13. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef] [PubMed]
- Yarrow, D.; Meyer, S.A. Proposal for amendment of the diagnosis of the genus Candida Berkhout nom. cons. Int. J. Syst. Bacteriol. 1978, 28, 611–615. [Google Scholar] [CrossRef]
- de Koster, C.G.; Heerma, W.; Pepermans, H.A.M.; Groenewegen, A.; Peters, H.; Haverkamp, J. Tandem mass spectrometry and nuclear magnetic resonance spectroscopy studies of Candida bombicola sophorolipid and product formed on hydrolysis by cutinase. Anal. Biochem. 1995, 230, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Pfliegler, W.P.; Horváth, E.; Kállai, Z.; Sipiczki, M. Diversity of Candida zemplinina isolates inferred from RAPD, micro/minisatellite and physiological analysis. Microbiol. Res. 2014, 169, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Englezos, V.; Rantsiou, K.; Torchio, F.; Rolle, L.; Gerbi, V.; Cocolin, L. Exploitation of the non-Saccharomyces yeast Starmerella bacillaris (synonym Candida zemplinina) in wine fermentation: Physiological and molecular characterizations. Int. J. Food Microbiol. 2015, 199, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Tofalo, R.; Schirone, M.; Torriani, S.; Rantsiou, K.; Cocolin, L.; Perpetuini, G.; Suzzi, G. Diversity of Candida zemplinina strains from grapes and Italian wines. Food Microbiol. 2012, 29, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Magyar, I.; Tóth, T. Comparative evaluation of some oenological properties in wine strains of Candida stellata, Candida zemplinina, Saccharomyces uvarum and Saccharomyces cerevisiae. Food Microbiol. 2011, 28, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Solieri, L.; Landi, S.; De Vero, L.; Giudici, P. Molecular assessment of indigenous yeast population from traditional balsamic vinegar. J. Appl. Microbiol. 2006, 101, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Ciani, M.; Ferraro, L. Combined use of immobilized Candida stellata cells and Saccharomyces cerevisiae to improve the quality of wines. J. Appl. Microbiol. 1998, 85, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Ciani, M.; Ferraro, L.; Fatichenti, F. Influence of glycerol production on the aerobic and anaerobic growth of the wine yeast Candida stellata. Enzyme Microb. Technol. 2000, 27, 698–703. [Google Scholar] [CrossRef]
- Milanovic, V.; Ciani, M.; Oro, L.; Comitini, F. Starmerella bombicola influences the metabolism of Saccharomyces cerevisiae at pyruvate decarboxylase and alcohol dehydrogenase level during mixed wine fermentation. Microb. Cell Fact. 2012, 3, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Alves-Araújo, C.; Pacheco, A.; Almeida, M.J.; Spencer-Martins, I.; Leão, C.; Sousa, M.J. Sugar utilization patterns and respiro-fermentative metabolism in the baker’s yeast Torulaspora delbrueckii. Microbiology 2007, 153, 898–904. [Google Scholar] [CrossRef] [PubMed]
- Pina, C.; Gonçalves, P.; Prista, C.; Loureiro-Dias, M.C. Ffz1, a new transporter specific for fructose from Zygosaccharomyces bailii. Microbiology 2004, 150, 2429–2433. [Google Scholar] [CrossRef] [PubMed]
- Leandro, M.J.; Cabral, S.; Prista, C.; Loureiro-Dias, M.C.; Sychrová, H. The high-capacity specific fructose facilitator ZrFfz1 is essential for the fructophilic behavior of Zygosaccharomyces rouxii CBS 732T. Eukaryot. Cell 2014, 13, 1371–1379. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, C.; Wisecaver, J.H.; Kominek, J.; Salema-Oom, M.; Leandro, M.J.; Shen, X.-X.; Opulente, D.; Zhou, X.; Peris, D.; Kurtzman, C.P.; et al. Evidence for loss and adaptive reacquisition of alcoholic fermentation in an early-derived fructophilic yeast lineage. Elife 2018, 7, e33034. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, C.; Coelho, M.A.; Salema-Oom, M.; Gonçalves, P. Stepwise functional evolution in a fungal sugar transporter family. Mol. Biol. Evol. 2016, 33, 352–366. [Google Scholar] [CrossRef] [PubMed]
- Flores, C.L.; Rodriguez, C.; Petit, T.; Gancedo, C. Carbohydrate and energy-yielding metabolism in non-conventional yeasts. FEMS Microbiol. Rev. 2000, 24, 507–529. [Google Scholar] [CrossRef]
- Mortensen, A.; Aguilar, F.; Crebelli, R.; Di Domenico, A.; Dusemund, B.; Frutos, M.J.; Galtier, P.; Gott, D.; Gundert-Remy, U.; Leblanc, J.; et al. Re-evaluation of glycerol (E 422) as a food additive. EFSA J. 2017, 15. [Google Scholar] [CrossRef]
- Eriksson, P.; Andre, L.; Ansell, R.; Blomberg, A.; Alder, L. Cloning and characterization of GPD2, a second gene encoding sn-glycerol 3-phosphate dehydrogenase (NAD+) in Saccharomyces cerevisiae, and its comparison with GPD1. Mol. Microbiol. 1995, 17, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, S. Osmotic stress signalling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002, 66, 300–372. [Google Scholar] [CrossRef] [PubMed]
- Dihazi, H.; Kessler, R.; Eschrich, K. High osmolarity glycerol (HOG) pathway-induced phosphorylation and activation of 6-phosphofructo-2-kinase are essential for glycerol accumulation and yeast cell proliferation under hyperosmotic stress. J. Biol. Chem. 2004, 279, 23961–23968. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Peña, J.M.; García, R.; Nombela, C.; Arroyo, J. The high-osmolarity glycerol (HOG) and cell wall integrity (CWI) signalling pathways interplay: A yeast dialogue between MAPK routes. Yeast 2010, 27, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Ciani, M. Wine vinegar production using base wines made with different yeast species. J. Sci. Food Agric. 1998, 78, 290–294. [Google Scholar] [CrossRef]
- Scanes, K.T.; Hohmann, S.; Prior, B.A. Glycerol production by the yeast Saccharomyces cerevisiae and its relevance to wine: A review. S. Afr. J. Enol. Vitic. 1998, 19, 17–24. [Google Scholar] [CrossRef]
- Prior, B.A.; Toh, T.H.; Jolly, N.; Baccari, C.; Mortimer, R.K. Impact of yeast breeding for elevated glycerol production on fermentative activity and metabolite formation in Chardonnay wine. S. Afr. J. Enol. Vitic. 2000, 21, 92–99. [Google Scholar] [CrossRef]
- Pretorius, I.S. Tailoring wine yeast for the new millennium: Novel approaches to the ancient art of winemaking. Yeast 2000, 16, 675–729. [Google Scholar] [CrossRef]
- Noble, A.C.; Bursick, G.F. The contribution of glycerol to perceived viscosity and sweetness in white wine. Am. J. Enol. Vitic. 1984, 35, 110–112. [Google Scholar]
- Fleet, G.H. Wine. In Food Microbiology: Fundamentals and Frontiers; Doyle, M.P., Beuchat, L.R., Eds.; ASM Press: Washington, DC, USA, 2007; pp. 863–890. [Google Scholar]
- Ferraro, L.; Fatichenti, F.; Ciani, M. Pilot scale vinification process using immobilized Candida stellata cells and Saccharomyces cerevisiae. Process Biochem. 2000, 35, 1125–1129. [Google Scholar] [CrossRef]
- García, M.; Arroyo, T.; Crespo, J.; Cabellos, J.M.; Esteve-Zarzoso, B. Use of native non-Saccharomyces strain: A. new strategy in D.O. “Vinos de Madrid” (Spain) wines elaboration. Eur. J. Food Sci. Technol. 2017, 5, 1–31. [Google Scholar]
- Gobbi, M.; De Vero, L.; Solieri, L.; Comitini, F.; Oro, L.; Giudici, P.; Ciani, M. Fermentative aptitude of non-Saccharomyces wine yeast for reduction in the ethanol content in wine. Eur. Food Res. Technol. 2014, 239, 41–48. [Google Scholar] [CrossRef]
- Nigam, P.S. Microbial enzymes with special characteristics for biotechnological applications. Biomolecules 2013, 3, 597–611. [Google Scholar] [CrossRef] [PubMed]
- Anbu, P.; Gopinath, S.C.B.; Chaulagain, B.P.; Lakshmipriya, T. Microbial enzymes and their applications in industries and medicine 2016. Biomed. Res. Int. 2017. [Google Scholar] [CrossRef] [PubMed]
- Pando Bedriñana, R.; Lastra Queipo, A.; Suárez Valles, B. Screening of enzymatic activities in non-Saccharomyces cider yeasts. J. Food Biochem. 2012, 36, 683–689. [Google Scholar] [CrossRef]
- Maturano, Y.P.; Rodríguez, L.A.; Toro, M.E.; Nally, M.C.; Vallejo, M.; Castellanos de Figueroa, L.I.; Combina, M.; Vazquez, F. Multi-enzyme production by pure and mixed cultures of Saccharomyces and non-Saccharomyces yeasts during wine fermentation. Int. J. Food Microbiol. 2012, 155, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Esteve-Zarzoso, B.; Manzanares, P.; Ramón, D.; Querol, A. The role of non-Saccharomyces yeasts in industrial winemaking. Int. Microbiol. 1998, 1, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Claus, H.; Mojsov, K. Enzymes for wine fermentation: Current and perspective applications. Fermentation 2018, 4, 52. [Google Scholar] [CrossRef]
- Carpita, N.C.; Gibeaut, D.M. Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993, 3, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Canal-Llaubères, R.-M. Enzymes in winemaking. In Wine Microbiology and Biotechnology; Fleet, G.H., Ed.; Harwood Academic Publishers: Chur, Switzerland, 1993; pp. 477–506. [Google Scholar]
- Hadfield, K.A.; Bennett, A.B. Polygalacturonases: Many genes in search of a function. Plant Physiol. 1998, 117, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Grassin, C.; Fauquembergue, P. Application of pectinases in beverages. In Pectin and pectinases; Visser, J., Voragen, A.G.J., Eds.; Elsevier: Amsterdam, The Netherlands, 1996; pp. 453–462. [Google Scholar]
- Boccas, F.; Roussos, S.; Gutiérrez, M.; Serrano, L.; Viniegra, G.G. Production of pectinase from coffee pulp in solid-state fermentation system-selection of wild fungal isolate of high potency by a simple 3-step screening technique. J. Food Sci. Technol. 1994, 31, 22–26. [Google Scholar]
- Evans, J.D.; Akin, D.E.; Foulk, J.A. Flax-retting by polygalacturonase-containing enzyme mixtures and effects on fiber properties. J. Biotechnol. 2002, 97, 223–231. [Google Scholar] [CrossRef]
- Fernández, M.; Úbeda, J.F.; Briones, A.I. Typing of non-Saccharomyces yeasts with enzymatic activities of interest in wine-making. Int. J. Food Microbiol. 2000, 59, 29–36. [Google Scholar] [CrossRef]
- Merín, M.G.; Martín, M.C.; Rantsiou, K.; Cocolin, L.; De Ambrosini, V.I.M. Characterization of pectinase activity for enology from yeasts occurring in Argentine Bonarda grape. Braz. J. Microbiol. 2015, 46, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Cordero-Bueso, G.; Esteve-Zarzoso, B.; Cabellos, J.M.; Gil-Díaz, M.; Arroyo, T. Biotechnological potential of non-Saccharomyces yeasts isolated during spontaneous fermentations of Malvar (Vitis vinifera cv. L.). Eur. Food Res. Technol. 2013, 236, 193–207. [Google Scholar] [CrossRef]
- García, M.; Apolinar-Valiente, R.; Williams, P.; Esteve-Zarzoso, B.; Arroyo, T.; Crespo, J.; Doco, T. Polysaccharides and oligosaccharides produced on Malvar wines elaborated with Torulaspora delbrueckii CLI 918 and Saccharomyces cerevisiae CLI 889 native yeasts from D.O. “Vinos de Madrid”. J. Agric. Food Chem. 2017, 65, 6656–6664. [Google Scholar] [CrossRef]
- Andorrà, I.; Berradre, M.; Rozès, N.; Mas, A.; Guillamón, J.M.; Esteve-Zarzoso, B. Effect of pure and mixed cultures of the main wine yeast species on grape must fermentations. Eur. Food Res. Technol. 2010, 231, 215–224. [Google Scholar] [CrossRef]
- Theron, L.W.; Divol, B. Microbial aspartic proteases: Current and potential applications in industry. Appl. Microbiol. Biotechnol. 2014, 98, 8853–8868. [Google Scholar] [CrossRef] [PubMed]
- Dizy, M.; Bisson, L.F. Proteolytic activity of yeast strains during grape juice fermentation. Am. J. Enol. Vitic. 2000, 51, 155–167. [Google Scholar]
- Romero-Cascales, I.; Fernández-Fernández, J.I.; Ros-García, J.M.; López-Roca, J.M.; Gómez-Plaza, E. Characterisation of the main enzymatic activities present in six commercial macerating enzymes and their effects on extracting colour during winemaking of Monastrell grapes. Int. J. Food Sci. Technol. 2008, 43, 1295–1305. [Google Scholar] [CrossRef]
- Capozzi, V.; Garofalo, C.; Chiriatti, M.A.; Grieco, F.; Spano, G. Microbial terroir and food innovation: The case of yeast biodiversity in wine. Microbiol. Res. 2015, 181, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Thongekkaew, J.; Kongsanthia, J. Screening and identification of cellulase producing yeast from Rongkho Forest, Ubon Ratchathani University. Bioeng. Biosci. 2016, 4, 29–33. [Google Scholar] [CrossRef]
- Williams, P.J.; Cynkar, W.; Francis, I.L.; Gray, J.D.; Iland, P.G.; Coombe, B.G. Quantification of glycosides in grapes, juices, and wines through a determination of glycosyl glucose. J. Agric. Food Chem. 1995, 43, 121–128. [Google Scholar] [CrossRef]
- Winterhalter, P.; Skouroumounis, G.K. Glycoconjugated aroma compounds: Occurrence, role and biotechnological transformation. Adv. Biochem. Eng. Biotechnol. 1997, 55, 73–105. [Google Scholar] [PubMed]
- Rosi, I.; Vinella, M.; Domizio, P. Characterization of β-glucosidase activity in yeasts of oenological origin. J. Appl. Bacteriol. 1994, 77, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Cordero Otero, R.R.; Ubeda Iranzo, J.F.; Briones-Perez, A.I.; Potgieter, N.; Villena, M.A.; Pretorius, I.S.; van Rensburg, P. Characterization of the β-glucosidase activity produced by enological strains of non-Saccharomyces yeasts. Food Microbiol. Saf. 2003, 68, 2564–2569. [Google Scholar] [CrossRef]
- Hock, R.; Benda, I.; Schreier, P. Formation of terpenes by yeasts during alcoholic fermentation. Zeitschrift für Leb. Und-forsch. 1984, 179, 450–452. [Google Scholar] [CrossRef]
- Jolly, N.P.; Augustyn, O.H.P.; Pretorius, I.S. The effect of non-Saccharomyces yeasts on fermentation and wine quality. S. Afr. J. Enol. Vitic. 2003, 24, 55–62. [Google Scholar] [CrossRef]
- Acosta, N.; Beldarraín, A.; Rodríguez, L.; Alonso, Y. Characterization of recombinant invertase expressed in methylotrophic yeasts. Biotechnol. Appl. Biochem. 2000, 32, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Huang, C. Modeling of ethanol fermentation using Zymomonas mobilis ATCC 10988 grown on the media containing glucose and fructose. Biochem. Eng. J. 2000, 4, 217–227. [Google Scholar] [CrossRef]
- Uma, C.; Gomathi, D.; Ravikumar, G.; Kalaiselvi, M.; Palaniswamy, M. Production and properties of invertase from a Cladosporium cladosporioides in SmF using pomegranate peel waste as substrate. Asian Pac. J. Trop. Biomed. 2012, S605–S611. [Google Scholar] [CrossRef]
- Pataro, C.; Guerra, J.B.; Gomes, F.C.O.; Neves, M.J.; Pimentel, P.F.; Rosa, C.A. Trehalose accumulation, invertase activity and physiological characteristics of yeasts isolates from 24 h fermentative cycles during the production of artisanal Brazilian cachaça. Braz. J. Microbiol. 2002, 33, 202–208. [Google Scholar] [CrossRef]
- Gargel, C.A.; Baffi, M.A.; Gomes, E.; Da-Silva, R. Invertase from a Candida stellata strain isolated from grape: Production and physico-chemical characterization. J. Microbiol. Biotechnol. Food Sci. 2014, 4, 24–28. [Google Scholar] [CrossRef]
- Van Bogaert, I.N.A.; Saerens, K.; De Muynck, C.; Develter, D.; Soetaert, W.; Vandamme, E.J. Microbial production and application of sophorolipids. Appl. Microbiol. Biotechnol. 2007, 76, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Price, N.P.J.; Ray, K.J.; Kuo, T.M. Production of sophorolipid biosurfactants by multiple species of the Starmerella (Candida) bombicola yeast clade. FEMS Microbiol. Lett. 2010, 311, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Parekh, V.J.; Pandit, A.B. Optimization of fermentative production of sophorolipid biosurfactant by Starmerella bombicola NRRL Y-17069 using response surface methodology. Int. J. Pharm. Biol. Sci. 2011, 1, 103–116. [Google Scholar]
- Pekin, G.; Vardar-Sukan, F.; Kosaric, N. Production of sophorolipids from Candida bombicola ATCC 22214 using Turkish corn oil and honey. Eng. Life Sci. 2005, 5, 357–362. [Google Scholar] [CrossRef]
- Cavalero, D.A.; Cooper, D.G. The effect of medium composition on the structure and physical state of sophorolipids produced by Candida bombicola ATCC 22214. J. Biotechnol. 2003, 103, 31–41. [Google Scholar] [CrossRef]
- De Waele, S.; Vandenberghe, I.; Laukens, B.; Planckaert, S.; Verweire, S.; Van Bogaert, I.N.A.; Soetaert, W.; Devreese, B.; Ciesielska, K. Optimized expression of the Starmerella bombicola lactone esterase in Pichia pastoris through temperature adaptation, codon-optimization and co-expression with HAC1. Protein Expr. Purif. 2018, 143, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Andorrà, I.; Berradre, M.; Mas, A.; Esteve-Zarzoso, B.; Guillamón, J.M. Effect of mixed culture fermentations on yeast populations and aroma profile. LWT-Food Sci. Technol. 2012, 49, 8–13. [Google Scholar] [CrossRef]
- Sadoudi, M.; Tourdot-Maréchal, R.; Rousseaux, S.; Steyer, D.; Gallardo-Chacón, J.J.; Ballester, J.; Vichi, S.; Guérin-Schneider, R.; Caixach, J.; Alexandre, H. Yeast-yeast interactions revealed by aromatic profile analysis of Sauvignon Blanc wine fermented by single or co-culture of non-Saccharomyces and Saccharomyces yeasts. Food Microbiol. 2012, 32, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Uscanga, B.; François, J.M. A study of the yeast cell wall composition and structure in response to growth conditions and mode of cultivation. Lett. Appl. Microbiol. 2003, 37, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Doco, T.; Quellec, N.; Moutounet, M.; Pellerin, P. Polysaccharide patterns during the ageing of Carignan noir red wines. Am. J. Enol. Vitic. 1999, 50, 25–32. [Google Scholar]
- Dufrechou, M.; Doco, T.; Poncet-Legrand, C.; Sauvage, F.X.; Vernhet, A. Protein/Polysaccharide interactions and their impact on haze formation in white wines. J. Agric. Food Chem. 2015, 63, 10042–10053. [Google Scholar] [CrossRef] [PubMed]
- Marchal, R.; Jeandet, P. Use of enological additives for colloid and tartrate salt stabilization in white wines and for improvement of sparkling wine foaming properties. In Wine Chemistry and Biochemistry; Moreno-Arribas, M.V., Polo, M.C., Eds.; Springer: New York, NY, USA, 2009; pp. 127–158. ISBN 9780387741161. [Google Scholar]
- Chalier, P.; Angot, B.; Delteil, D.; Doco, T.; Gunata, Z. Interactions between aroma compounds and whole mannoprotein isolated from Saccharomyces cerevisiae strains. Food Chem. 2007, 100, 22–30. [Google Scholar] [CrossRef]
- Pérez-Magariño, S.; Martínez-Lapuente, L.; Bueno-Herrera, M.; Ortega-Heras, M.; Guadalupe, Z.; Ayestarán, B. Use of commercial dry yeast products rich in mannoproteins for white and rosé sparkling wine elaboration. J. Agric. Food Chem. 2015, 63, 5670–5681. [Google Scholar] [CrossRef] [PubMed]
- Vidal, S.; Francis, L.; Williams, P.; Kwiatkowski, M.; Gawel, R.; Cheynier, V.; Waters, E. The mouth-feel properties of polysaccharides and anthocyanins in a wine like medium. Food Chem. 2004, 85, 519–525. [Google Scholar] [CrossRef]
- Guilloux-Benatier, M.; Guerreau, J.; Feuillat, M. Influence of initial colloid content on yeast macromolecule production and on the metabolism of wine microorganisms. Am. J. Enol. Vitic. 1995, 46, 486–492. [Google Scholar]
- Domizio, P.; Romani, C.; Comitini, F.; Gobbi, M.; Lencioni, L.; Mannazu, I.; Ciani, M. Potential spoilage non-Saccharomyces yeasts in mixed cultures with Saccharomyces cerevisiae. Ann. Microbiol. 2011, 61, 137–144. [Google Scholar] [CrossRef]
- Giovani, G.; Rosi, I.; Bertuccioli, M. Quantification and characterization of cell wall polysaccharides released by non-Saccharomyces yeast strains during alcoholic fermentation. Int. J. Food Microbiol. 2012, 160, 113–118. [Google Scholar] [CrossRef] [PubMed]
- González-Royo, E.; Pascual, O.; Kontoudakis, N.; Esteruelas, M.; Esteve-Zarzoso, B.; Mas, A.; Canals, J.M.; Zamora, F. Oenological consequences of sequential inoculation with non-Saccharomyces yeasts (Torulaspora delbrueckii or Metschnikowia pulcherrima) and Saccharomyces cerevisiae in base wine for sparkling wine production. Eur. Food Res. Technol. 2015, 240, 999–1012. [Google Scholar] [CrossRef]
- Suzuki, M.; Suh, S.O.; Sugita, T.; Nakase, T.A. phylogenetic study on galactose-containing Candida species based on 18S ribosomal DNA sequences. J. Gen. Appl. Microbiol. 1999, 45, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Cominiti, F.; Gobbi, M.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 2011, 28, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Domizio, P.; Liu, Y.; Bisson, L.F.; Barile, D. Use of non-Saccharomyces wine yeasts as novel sources of mannoproteins in wine. Food Microbiol. 2014, 43, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Ballou, C. Structure and biosynthesis of the mannan component of the yeast cell envelope. Adv. Microb. Physiol. 1976, 14, 93–158. [Google Scholar] [CrossRef] [PubMed]

| C. stellata (CBS 157) | C. zemplinina | S. bombicola | |
|---|---|---|---|
| Growth in high sugar concentration | + | + | + |
| Growth in botrytized grape berries | − | ++ | + |
| Growth in presence of 1% of acetic acid | − | + | v 1 |
| Formation of ascospores | − | − | + |
| Banding pattern (electrophoretic karyotype) | 3 | 3 | 2 |
| Chromosomal polymorphism | yes | no | no |
| % D1/D2 sequence in difference with C. stellata (CBS 157) | 8.1 | nd 2 | |
| MboI and DraI digestion distingue between the species | yes | yes | |
| CfoI, HaeIII, and HinfI digestion distingue between the species | yes | yes |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
García, M.; Esteve-Zarzoso, B.; Cabellos, J.M.; Arroyo, T. Advances in the Study of Candida stellata. Fermentation 2018, 4, 74. https://doi.org/10.3390/fermentation4030074
García M, Esteve-Zarzoso B, Cabellos JM, Arroyo T. Advances in the Study of Candida stellata. Fermentation. 2018; 4(3):74. https://doi.org/10.3390/fermentation4030074
Chicago/Turabian StyleGarcía, Margarita, Braulio Esteve-Zarzoso, Juan Mariano Cabellos, and Teresa Arroyo. 2018. "Advances in the Study of Candida stellata" Fermentation 4, no. 3: 74. https://doi.org/10.3390/fermentation4030074
APA StyleGarcía, M., Esteve-Zarzoso, B., Cabellos, J. M., & Arroyo, T. (2018). Advances in the Study of Candida stellata. Fermentation, 4(3), 74. https://doi.org/10.3390/fermentation4030074