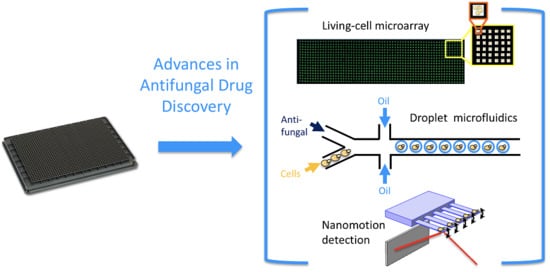
Micro- and Nanoscale Approaches in Antifungal Drug Discovery
Abstract
1. Introduction
2. Emerging Fungal Diseases and Antifungal Drugs
3. Omics-Based Antifungal Drug Discovery Approaches
4. Micro- and Nanoscale Approaches
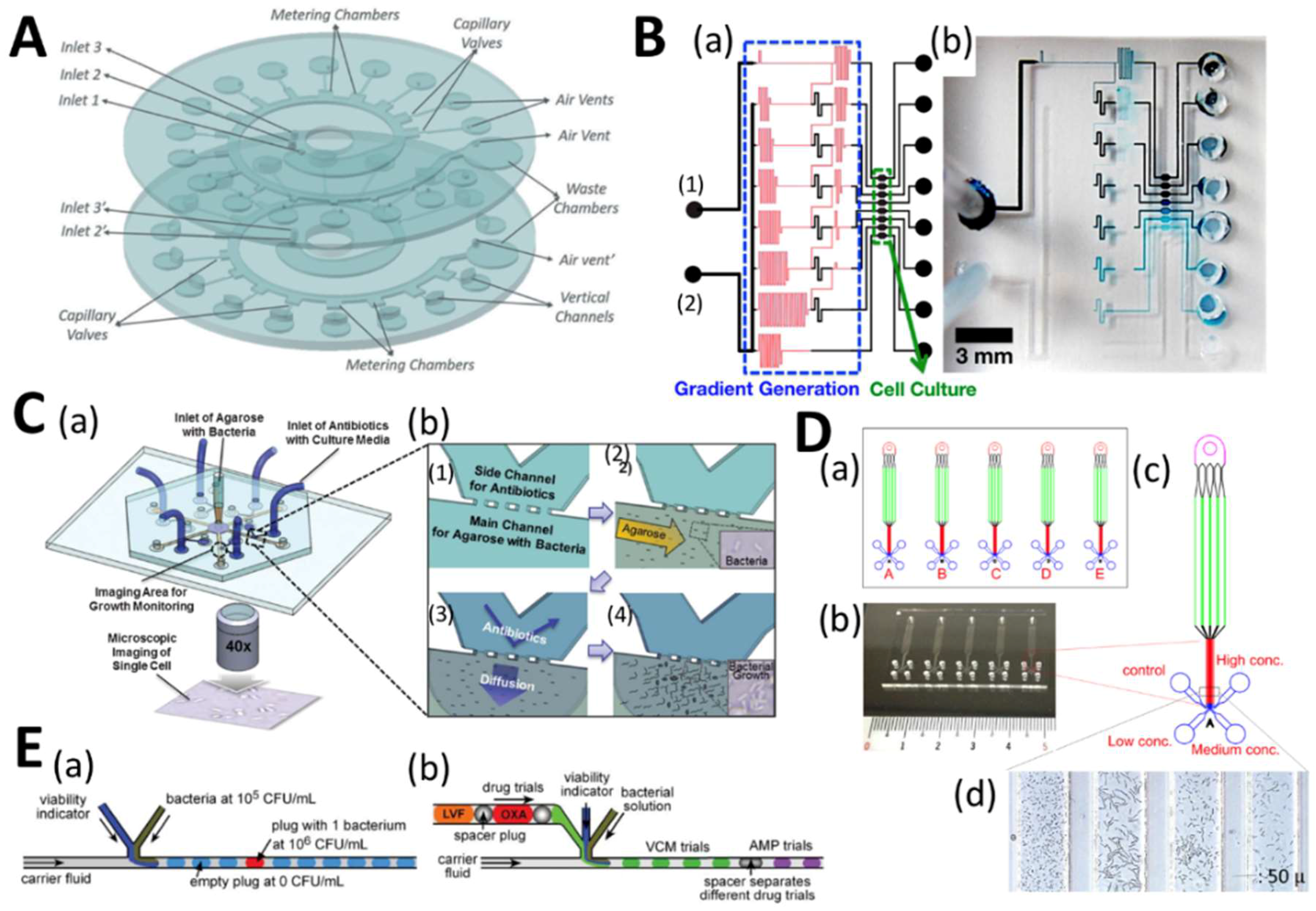
4.1. Microfluidic High-Throughput Antifungal Drug Discovery
4.2. Microfluidics for Antifungal Susceptibility Testing
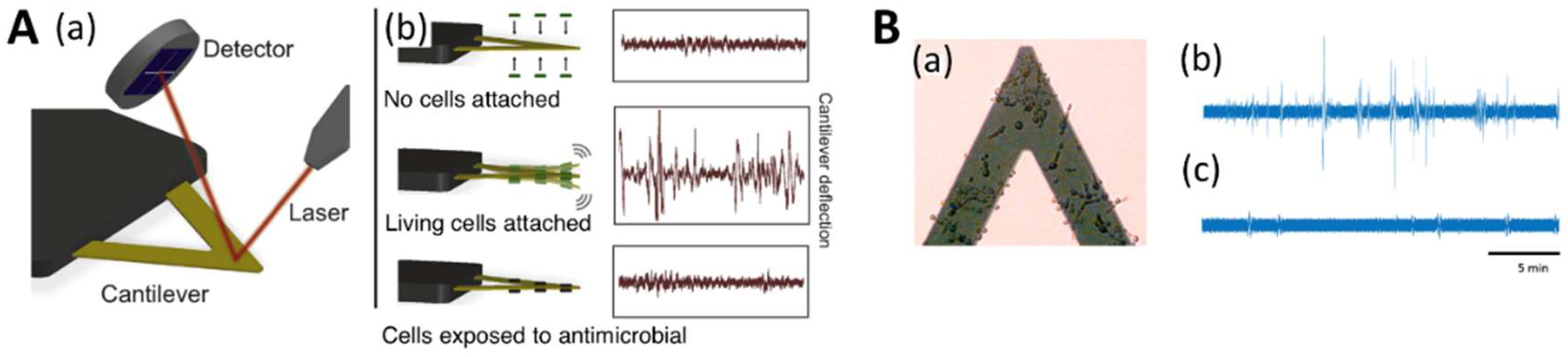
4.3. Nanomotion Analysis for Evaluating Antifungal Suscessibility
4.4. Antifungal Nanoparticles
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Vos, T.; Flaxman, A.D.; Naghavi, M.; Lozano, R.; Michaud, C.; Ezzati, M.; Shibuya, K.; Salomon, J.A.; Abdalla, S.; Aboyans, V.; et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990–2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2163–2196. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Levitz, S.M. Tackling human fungal infections. Science 2012, 336, 647. [Google Scholar] [CrossRef] [PubMed]
- Richardson, M.D. Changing patterns and trends in systemic fungal infections. J. Antimicrob. Chemother. 2005, 56 (Suppl. 1), i5–i11. [Google Scholar] [CrossRef] [PubMed]
- Huttunen, R.; Syrjänen, J. Obesity and the risk and outcome of infection. Int. J. Obes. 2013, 37, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Rytter, M.J.; Kolte, L.; Briend, A.; Friis, H.; Christensen, V.B. The immune system in children with malnutrition—A systematic review. PLoS ONE 2014, 9, e105017. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.K.; Zambruni, M.; Melby, C.L.; Melby, P.C. Impact of childhood malnutrition on host defense and infection. Clin. Microbiol. Rev. 2017, 30, 919–971. [Google Scholar] [CrossRef] [PubMed]
- Denning, D.W.; Bromley, M.J. Infectious Disease. How to bolster the antifungal pipeline. Science 2015, 347, 1414–1416. [Google Scholar] [CrossRef] [PubMed]
- Lemke, A.; Kiderlen, A.F.; Kayser, O. Amphotericin B. Appl. Microbiol. Biotechnol. 2005, 68, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Bellmann, R. Pharmacodynamics and pharmacokinetics of antifungals for treatment of invasive aspergillosis. Curr. Pharm. Des. 2013, 19, 3629–3647. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A. Antifungal drug resistance: Mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 2012, 125 (Suppl. 1), S3–S13. [Google Scholar] [CrossRef] [PubMed]
- Pianalto, K.M.; Alspaugh, J.A. New Horizons in Antifungal Therapy. J. Fungi. 2016, 2, 26. [Google Scholar] [CrossRef] [PubMed]
- Nivoix, Y.; Ubeaud-Sequier, G.; Engel, P.; Levêque, D.; Herbrecht, R. Drug-drug interactions of triazole antifungal agents in multimorbid patients and implications for patient care. Curr. Drug. Metab. 2009, 10, 395–409. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.E. Current concepts in antifungal pharmacology. Mayo Clin. Proc. 2011, 86, 805–817. [Google Scholar] [CrossRef] [PubMed]
- Campoy, S.; Adrio, J.L. Antifungals. Mayo Clin. Proc. 2017, 133, 86–96. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, M.W.; Walsh, T.J. Drugs currently under investigation for the treatment of invasive candidiasis. Expert. Opin. Investig. Drugs 2017, 26, 825–831. [Google Scholar] [CrossRef] [PubMed]
- Perfect, J.R. The antifungal pipeline: A reality check. Nat. Rev. Drug Discov. 2017, 16, 603–616. [Google Scholar] [CrossRef] [PubMed]
- Fuentefria, A.M.; Pippi, B.; Dalla Lana, D.F.; Donato, K.K.; de Andrade, S.F. Antifungals discovery: An insight into new strategies to combat antifungal resistance. Lett. Appl. Microbiol. 2018, 66, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Weig, M.; Brown, A.J. Genomics and the development of new diagnostics and anti-Candida drugs. Trends Microbiol. 2007, 15, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Payne, D.J.; Gwynn, M.N.; Holmes, D.J.; Pompliano, D.L. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007, 6, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Roemer, T.; Boone, C. Systems-level antimicrobial drug and drug synergy discovery. Nat. Chem. Biol. 2013, 9, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Schenone, M.; Dančík, V.; Wagner, B.K.; Clemons, P.A. Target identification and mechanism of action in chemical biology and drug discovery. Nat. Chem. Biol. 2013, 9, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Nucci, M.; Marr, K.A. Emerging fungal diseases. Clin. Infect. Dis. 2005, 41, 521–526. [Google Scholar] [CrossRef] [PubMed]
- Castón-Osorio, J.J.; Rivero, A.; Torre-Cisneros, J. Epidemiology of invasive fungal infection. Int. J. Antimicrob. Agents 2008, 32 (Suppl. 2), S103–S109. [Google Scholar] [CrossRef]
- Miceli, M.H.; Díaz, J.A.; Lee, S.A. Emerging opportunistic yeast infections. Lancet Infect. Dis. 2011, 11, 142–151. [Google Scholar] [CrossRef]
- Galimberti, R.; Torre, A.C.; Baztán, M.C.; Rodriguez-Chiappetta, F. Emerging systemic fungal infections. Clin. Dermatol. 2012, 30, 633–650. [Google Scholar] [CrossRef] [PubMed]
- Enoch, D.A.; Yang, H.; Aliyu, S.H.; Micallef, C. The Changing Epidemiology of Invasive Fungal Infections. In Human Fungal Pathogen Identification. Methods in Molecular Biology; Lion, T., Ed.; Humana Press: New York, NY, USA, 2017; Volume 1508, pp. 17–65. ISBN 978-1-4939-6513-7. [Google Scholar]
- Schwartz, I.S.; Patterson, T.F. The Emerging Threat of antifungal resistance in transplant infectious diseases. Curr. Infect. Dis. Rep. 2018, 20, 2. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J.; Gibbs, D.L.; Newell, V.A.; Ellis, D.; Tullio, V.; Rodloff, A.; Fu, W.; Ling, T.A. Global Antifungal Surveillance Group. Results from the ARTEMIS DIS Global Antifungal Surveillance Study, 1997 to 2007: A 10.5-year analysis of susceptibilities of Candida Species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J. Clin. Microbiol. 2010, 48, 1366–1377. [Google Scholar] [CrossRef] [PubMed]
- Khawcharoenporn, T.; Apisarnthanarak, A.; Mundy, L.M. Non-neoformans cryptococcal infections: A systematic review. Infection 2007, 35, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Enache-Angoulvant, A.; Hennequin, C. Invasive Saccharomyces infection: A comprehensive review. Clin. Infect. Dis. 2005, 41, 1559–1568. [Google Scholar] [CrossRef] [PubMed]
- Anoop, V.; Rotaru, S.; Shwed, P.S.; Tayabali, A.F.; Arvanitakis, G. Review of current methods for characterizing virulence and pathogenicity potential of industrial Saccharomyces cerevisiae strains towards humans. FEMS Yeast Res. 2015, 15, fov057-2. [Google Scholar] [CrossRef] [PubMed]
- Lass-Flörl, C.; Cuenca-Estrella, M. Changes in the epidemiological landscape of invasive mould infections and disease. J. Antimicrob. Chemother. 2017, 72 (Suppl. 1), i5–i11. [Google Scholar] [CrossRef] [PubMed]
- Ribes, J.A.; Vanover-Sams, C.L.; Baker, D.J. Zygomycetes in human disease. Clin. Microbiol. Rev. 2000, 13, 236–301. [Google Scholar] [CrossRef] [PubMed]
- Kontoyiannis, D.P.; Marr, K.A.; Park, B.J.; Alexander, B.D.; Anaissie, E.J.; Walsh, T.J.; Ito, J.; Andes, D.R.; Baddley, J.W.; Brown, J.M.; et al. Prospective surveillance for invasive fungal infections in hematopoietic stem cell transplant recipients, 2001–2006: Overview of the Transplant-Associated Infection Surveillance Network (TRANSNET) Database. Clin. Infect. Dis. 2010, 50, 1091–1100. [Google Scholar] [CrossRef] [PubMed]
- Roilides, E.; Zaoutis, T.E.; Walsh, T.J. Invasive zygomycosis in neonates and children. Clin. Microbiol. Infect. 2009, 15 (Suppl. 5), 50–54. [Google Scholar] [CrossRef] [PubMed]
- Lanternier, F.; Lortholary, O. Zygomycosis and diabetes mellitus. Clin. Microbiol. Infect. 2009, 15 (Suppl. 5), 21–25. [Google Scholar] [CrossRef] [PubMed]
- Nucci, M.; Anaissie, E. Fusarium infections in immunocompromised patients. Clin. Microbiol. Rev. 2007, 20, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Tortorano, A.M.; Richardson, M.; Roilides, E.; Diepeningen, A.V.; Caira, M.; Munoz, P.; Johnson, E.; Meletiadis, J.; Pana, Z.D.; Lackner, M.; et al. ESCMID & ECMM Joint Guidelines on Diagnosis and Management of Hyalohyphomycosis: Fusarium spp., Scedosporium spp., and others. Clin. Microbiol. Infect. 2014, 20 (Suppl. 3), 27–46. [Google Scholar] [PubMed]
- Guarro, J.; Kantarcioglu, A.S.; Horré, R.; Luis Rodriguez-Tudela, J.; Cuenca Estrella, M.; Berenguer, J.; Sybren De Hoog, G. Scedosporium apiospermum: Changing clinical spectrum of a therapy-refractory opportunist. Med. Mycol. 2006, 44, 295–327. [Google Scholar] [CrossRef] [PubMed]
- Montesinos, I.; Dodemont, M.; Lagrou, K.; Jacobs, F.; Etienne, I.; Denis, O. New case of azole-resistant Aspergillus fumigatus due to TR46/Y121F/T289A mutation in Belgium. J. Antimicrob. Chemother. 2014, 69, 3439–3440. [Google Scholar] [CrossRef] [PubMed]
- Buil, J.B.; Meis, J.F.; Melchers, W.J.; Verweij, P.E. Are the TR46/Y121F/T289A Mutations in Azole-Resistant Aspergillosis Patient Acquired or Environmental? Antimicrob. Agents Chemother. 2016, 60, 3259–3260. [Google Scholar] [CrossRef] [PubMed]
- Ostrosky-Zeichner, L.; Casadevall, A.; Galgiani, J.N.; Odds, F.C.; Rex, J.H. An insight into the antifungal pipeline: Selected new molecules and beyond. Nat. Rev. Drug Discov. 2010, 9, 719–727. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, P.; Ferrari, S.; Coste, A.T. Antifungal resistance and new strategies to control fungal infections. Int. J. Microbiol. 2012, 2012, 713687. [Google Scholar] [CrossRef] [PubMed]
- Ballard, S.A.; Lodola, A.; Tarbit, M.H. A comparative study of 1-substituted imidazole and 1,2,4-triazole antifungal compounds as inhibitors of testosterone hydroxylations catalysed by mouse hepatic microsomal cytochromes P-450. Biochem. Pharmacol. 1988, 37, 4643–4651. [Google Scholar] [CrossRef]
- DiDomenico, B. Novel antifungal drugs. Curr. Opin. Microbiol. 1999, 2, 509–515. [Google Scholar] [CrossRef]
- Arévalo, M.P.; Carrillo-Muñoz, A.J.; Salgado, J.; Cardenes, D.; Brió, S.; Quindós, G.; Espinel-Ingroff, A. Antifungal activity of the echinocandin anidulafungin (VER002, LY-303366) against yeast pathogens: A comparative study with M27-A microdilution method. J. Antimicrob. Chemother. 2003, 51, 163–166. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Sheehan, D.; Puzniak, L.; Schlamm, H.; Ghannoum, M.A. Echinocandins: Are they all the same? J. Chemother. 2011, 23, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Firooz, A.; Nafisi, S.; Maibach, H.I. Novel drug delivery strategies for improving econazole antifungal action. Int. J. Pharm. 2015, 495, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Denning, D.W.; Hope, W.W. Therapy for fungal diseases: Opportunities and priorities. Trends Microbiol. 2010, 18, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Terra, L.; Abreu, P.A.; Teixeira, V.L.; Paixão, I.C.; Pereira, R.; Leal, B.; Lourenço, A.L.; Rampelotto, P.H.; Castro, H.C. Mycoses and Antifungals: Reviewing the basis of a current problem that still is a biotechnological target for marine products. Front. Mar. Sci. 2014, 1, 12. [Google Scholar] [CrossRef]
- Vengurlekar, S.; Sharma, R.; Trivedi, P. Efficacy of some natural compounds as antifungal agents. Pharmacogn. Rev. 2012, 6, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Moya, F.; Colom-Valiente, M.F.; Martinez-Peinado, P.; Martinez-Lopez, J.E.; Puelles, E.; Sempere-Ortells, J.M.; Lopez-Llorca, L.V. Carbon and nitrogen limitation increase chitosan antifungal activity in Neurospora crassa and fungal human pathogens. Fungal. Biol. 2015, 119, 154–169. [Google Scholar] [CrossRef] [PubMed]
- Abbaszadeh, S.; Sharifzadeh, A.; Shokri, H.; Khosravi, A.R.; Abbaszadeh, A. Antifungal efficacy of thymol, carvacrol, eugenol and menthol as alternative agents to control the growth of food-relevant fungi. J. Mycol. Med. 2014, 24, e51–e56. [Google Scholar] [CrossRef] [PubMed]
- Barros, F.M.C.; Pippi, B.; Dresch, R.R.; Dauber, B.; Luciano, S.C.; Apel, M.A.; Fuentefria, A.M.; Von Poser, G.L. Antifungal and antichemotactic activities and quantification of phenolic compounds in lipophilic extracts of Hypericum spp. native to South Brazil. Ind. Crops Prod. 2013, 44, 294–299. [Google Scholar] [CrossRef]
- Danielli, L.J.; Dos Reis, M.; Bianchini, M.; Camargo, G.S.; Bordignon, S.A.L.; Guerreiro, I.K.; Fuentefria, A.; Apel, M.A. Antidermatophytic activity of volatile oil and nanoemulsion of Stenachaenium megapotamicum (Spreng.) Baker. Ind. Crops Prod. 2013, 50, 23–28. [Google Scholar] [CrossRef]
- Machado, G.R.; Pippi, B.; Dalla Lana, D.F.; Amaral, A.P.S.; Teixeira, M.L.; De Souza, K.C.B.; Fuentefria, A.M. Reversal of fluconazole resistance induced by a synergistic effect with Acca sellowiana in Candida glabrata strains. Pharm. Biol. 2016, 54, 2410–2419. [Google Scholar] [CrossRef] [PubMed]
- Bicanic, T.; Muzoora, C.; Brouwer, A.E.; Meintjes, G.; Longley, N.; Taseera, K.; Rebe, K.; Loyse, A.; Jarvis, J.; Bekker, L.G.; et al. Independent association between rate of clearance of infection and clinical outcome of HIV-associated cryptococcal meningitis: Analysis of a combined cohort of 262 patients. Clin. Infect. Dis. 2009, 49, 702–709. [Google Scholar] [CrossRef] [PubMed]
- Pierce, C.G.; Uppuluri, P.; Tristan, A.R.; Wormley, F.L., Jr.; Mowat, E.; Ramage, G.; Lopez-Ribot, J.L. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat. Protoc. 2008, 3, 1494–1500. [Google Scholar] [CrossRef] [PubMed]
- LaFleur, M.D.; Lucumi, E.; Napper, A.D.; Diamond, S.L.; Lewis, K.N. Novel high-throughput screen against Candida albicans identifies antifungal potentiators and agents effective against biofilms. J. Antimicrob. Chemother. 2011, 66, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, M.C.; de la Cruz, M.; Cantizani, J.; Moreno, C.; Tormo, J.R.; Mellado, E.; De Lucas, J.R.; Asensio, F.; Valiante, V.; Brakhage, A.A.; et al. A new approach to drug discovery: High-throughput screening of microbial natural extracts against Aspergillus fumigatus using resazurin. J. Biomol. Screen 2012, 17, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Rabjohns, J.L.A.; Park, Y.D.; Dehdashti, J.; Henderson, C.; Zelazny, A.; Metallo, S.J.; Zheng, W.; Williamson, P.R. A high-throughput screening assay for fungicidal compounds against Cryptococcus neoformans. J. Biomol. Screen 2014, 19, 270–277. [Google Scholar] [CrossRef] [PubMed]
- DiDone, L.; Scrimale, T.; Baxter, B.K.; Krysan, D.J. A high-throughput assay of yeast cell lysis for drug discovery and genetic analysis. Nat. Protoc. 2010, 5, 1107–1114. [Google Scholar] [CrossRef] [PubMed]
- Cameron-Clarke, A.; Hulse, G.A.; Clifton, L.; Cantrell, I.C. The Use of Adenylate Kinase Measurement to Determine Causes of Lysis in Lager Yeast. J Am. Soc. Brew. Chem. 2003, 61, 152–156. [Google Scholar] [CrossRef]
- Roemer, T.; Krysan, D.J. Antifungal drug development: Challenges, unmet clinical needs, and new approaches. Cold Spring Harb. Perspect. Med. 2014, 4, A019703. [Google Scholar] [CrossRef] [PubMed]
- Perlstein, E.O.; Ruderfer, D.M.; Roberts, D.C.; Schreiber, S.L.; Kruglyak, L. Genetic basis of individual differences in the response to small-molecule drugs in yeast. Nat. Genet. 2007, 39, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Pierce, S.E.; Fung, E.L.; Jaramillo, D.F.; Chu, A.M.; Davis, R.W.; Nislow, C.; Giaever, G. A unique and universal molecular barcode array. Nat. Methods 2006, 3, 601–603. [Google Scholar] [CrossRef] [PubMed]
- Muroi, M.; Kazami, S.; Noda, K.; Kondo, H.; Takayama, H.; Kawatani, M.; Usui, T.; Osada, H. Application of proteomic profiling based on 2D-DIGE for classification of compounds according to the mechanism of action. Chem. Biol. 2010, 17, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.D.; Sibley, G.E.; Beckmann, N.; Dobb, K.S.; Slater, M.J.; McEntee, L.; du Pré, S.; Livermore, J.; Bromley, M.J.; Wiederhold, N.P.; et al. F901318 represents a novel class of antifungal drug that inhibits dihydroorotate dehydrogenase. Proc. Natl. Acad. Sci. USA 2016, 133, 12809–12814. [Google Scholar] [CrossRef] [PubMed]
- Winzeler, E.A.; Shoemaker, D.D.; Astromoff, A.; Liang, H.; Anderson, K.; Andre, B.; Bangham, R.; Benito, R.; Boeke, J.D.; Bussey, H.; et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 1999, 285, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Tong, A.H.; Evangelista, M.; Parsons, A.B.; Xu, H.; Bader, G.D.; Pagé, N.; Robinson, M.; Raghibizadeh, S.; Hogue, C.W.; Bussey, H.; et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 2001, 294, 2364–2368. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; Baryshnikova, A.; Bellay, J.; Kim, Y.; Spear, E.D.; Sevier, C.S.; Ding, H.; Koh, J.L.; Toufighi, K.; Mostafavi, S.; et al. The genetic landscape of a cell. Science 2010, 327, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Hillenmeyer, M.E.; Fung, E.; Wildenhain, J.; Pierce, S.E.; Hoon, S.; Lee, W.; Proctor, M.; St Onge, R.P.; Tyers, M.; Koller, D.; et al. The chemical genomic portrait of yeast: Uncovering a phenotype for all genes. Science 2008, 320, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, J.B.; Schoeberl, B.; Nielsen, U.B.; Sorger, P.K. Systems biology and combination therapy in the quest for clinical efficacy. Nat. Chem. Biol. 2006, 2, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Lehár, J.; Stockwell, B.R.; Giaever, G.; Nislow, C. Combination chemical genetics. Nat. Chem. Biol. 2008, 4, 674–681. [Google Scholar] [CrossRef] [PubMed]
- Borisy, A.A.; Elliott, P.J.; Hurst, N.W.; Lee, M.S.; Lehar, J.; Price, E.R.; Serbedzija, G.; Zimmermann, G.R.; Foley, M.A.; Stockwell, B.R.; et al. Systematic discovery of multicomponent therapeutics. Proc. Natl. Acad. Sci. USA 2003, 100, 7977–7982. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yan, K.; Zhang, Y.; Huang, R.; Bian, J.; Zheng, C.; Sun, H.; Chen, Z.; Sun, N.; An, R.; et al. High-throughput synergy screening identifies microbial metabolites as combination agents for the treatment of fungal infections. Proc. Natl. Acad. Sci. USA 2007, 104, 4606–4611. [Google Scholar] [CrossRef] [PubMed]
- Zhai, B.; Zhou, H.; Yang, L.; Zhang, J.; Jung, K.; Giam, C.Z.; Xiang, X.; Lin, X. Polymyxin B, in combination with fluconazole, exerts a potent fungicidal effect. J. Antimicrob. Chemother. 2010, 65, 931–938. [Google Scholar] [CrossRef] [PubMed]
- Spitzer, M.; Griffiths, E.; Blakely, K.M.; Wildenhain, J.; Ejim, L.; Rossi, L.; De Pascale, G.; Curak, J.; Brown, E.; Tyers, M.; et al. Cross-species discovery of syncretic drug combinations that potentiate the antifungal fluconazole. Mol. Syst. Biol. 2011, 7, 499. [Google Scholar] [CrossRef] [PubMed]
- Cowen, L.E.; Singh, S.D.; Köhler, J.R.; Collins, C.; Zaas, A.K.; Schell, W.A.; Aziz, H.; Mylonakis, E.; Perfect, J.R.; Whitesell, L.; Lindquist, S. Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc. Natl. Acad. Sci. USA 2009, 106, 2818–2823. [Google Scholar] [CrossRef] [PubMed]
- Epp, E.; Vanier, G.; Harcus, D.; Lee, A.Y.; Jansen, G.; Hallett, M.; Sheppard, D.C.; Thomas, D.Y.; Munro, C.A.; Mullick, A.; et al. Reverse genetics in Candida albicans predicts ARF cycling is essential for drug resistance and virulence. PLoS Pathog. 2010, 6, e1000753. [Google Scholar] [CrossRef] [PubMed]
- Sharom, J.R.; Bellows, D.S.; Tyers, M. From large networks to small molecules. Curr. Opin. Chem. Biol. 2004, 8, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Jansen, G.; Lee, A.Y.; Epp, E.; Fredette, A.; Surprenant, J.; Harcus, D.; Scott, M.; Tan, E.; Nishimura, T.; Whiteway, M.; et al. Chemogenomic profiling predicts antifungal synergies. Mol. Syst. Biol. 2009, 5, 338. [Google Scholar] [CrossRef] [PubMed]
- Lehár, J.; Krueger, A.S.; Avery, W.; Heilbut, A.M.; Johansen, L.M.; Price, E.R.; Rickles, R.J.; Short, G.F., 3rd; Staunton, J.E.; Jin, X.; et al. Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat. Biotechnol. 2009, 27, 659–666. [Google Scholar] [CrossRef]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Squires, T.M.; Quake, S.R. Microfluidics: Fluid physics at the nanoliter scale. Rev. Mod. Phys. 2005, 77, 977–1026. [Google Scholar] [CrossRef]
- Dertinger, S.K.W.; Chiu, D.T.; Jeon, N.L.; Whitesides, G.M. Generation of Gradients Having Complex Shapes Using Microfluidic Networks. Anal. Chem. 2001, 73, 1240–1246. [Google Scholar] [CrossRef]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Knight, J.B.; Vishwanath, A.; Brody, J.P.; Austin, R.H. Hydrodynamic focusing on a silicon chip: Mixing nanoliters in microseconds. Phys. Rev. Lett. 1998, 80, 3863–3866. [Google Scholar] [CrossRef]
- Pollack, L.; Tate, M.W.; Finnefrock, A.C.; Kalidas, C.; Trotter, S.; Darnton, N.C.; Lurio, L.; Austin, R.H.; Batt, C.A.; Gruner, S.M.; et al. Time resolved collapse of a folding protein observed with small angle X-ray scattering. Phys. Rev. Lett. 2001, 86, 4962–4965. [Google Scholar] [CrossRef] [PubMed]
- Wen, N.; Zhao, Z.; Fan, B.; Chen, D.; Men, D.; Wang, J.; Chen, J. Development of Droplet Microfluidics Enabling High-Throughput Single-Cell Analysis. Molecules 2016, 21, 881. [Google Scholar] [CrossRef] [PubMed]
- Clausell-Tormos, J.; Lieber, D.; Baret, J.C.; El-Harrak, A.; Miller, O.J.; Frenz, L.; Blouwolff, J.; Humphry, K.J.; Köster, S.; Duan, H.; et al. Droplet-based microfluidic platforms for the encapsulation and screening of Mammalian cells and multicellular organisms. Chem. Biol. 2008, 15, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Wu, X.; Young, A.T.; Haynes, C.L. Microfluidics-based in vivo mimetic systems for the study of cellular biology. Acc. Chem. Res. 2014, 47, 1165–1173. [Google Scholar] [CrossRef] [PubMed]
- Mehling, M.; Tay, S. Microfluidic cell culture. Curr. Opin. Biotechnol. 2014, 25, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Velve-Casquillas, G.; le Berre, M.; Piel, M.; Tran, P.T. Microfluidic tools for cell biological research. Nano Today 2010, 5, 28–47. [Google Scholar] [CrossRef] [PubMed]
- Castel, D.; Pitaval, A.; Debily, M.A.; Gidrol, X. Cell microarrays in drug discovery. Drug Discov. Today 2006, 11, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.S.; Davis, M.M. Molecular and functional analysis using live cell microarrays. Curr. Opin. Chem. Biol. 2006, 10, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Yarmush, M.L.; King, K.R. Living-cell microarrays. Annu. Rev. Biomed. Eng. 2009, 11, 235–257. [Google Scholar] [CrossRef] [PubMed]
- Willaert, R.; Sahli, H. On-chip living-cell microarrays for network biology. In Bioinformatics—Trends and Methodologies; Mahdavi, M.A., Ed.; InTech-Open Access: PublisherRejeka, Croatia, 2011; pp. 609–630. ISBN 978-953-307-282-1. [Google Scholar]
- Willaert, R.G.; Goossens, K. Microfluidic bioreactors for cellular microarrays. Fermentation 2015, 1, 38–78. [Google Scholar] [CrossRef]
- Jonczyk, R.; Kurth, T.; Lavrentieva, A.; Walter, J.G.; Scheper, T.; Stahl, F. Living cell microarrays: An overview of concepts. Microarrays 2016, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Charvin, G.; Cross, F.R.; Siggia, E.D. Forced periodic expression of G1 cyclins phase-locks the budding yeast cell cycle. Proc. Natl. Acad. Sci. USA 2009, 106, 6632–6637. [Google Scholar] [CrossRef] [PubMed]
- Bean, J.M.; Siggia, E.D.; Cross, F.R. Coherence and timing of cell cycle start examined at single-cell resolution. Mol. Cell 2006, 21, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Lidstrom, M.E.; Konopka, M.C. The role of physiological heterogeneity in microbial population behavior. Nat. Chem. Biol. 2010, 6, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Martins, B.M.; Locke, J.C. Microbial individuality: How single-cell heterogeneity enables population level strategies. Curr. Opin. Microbiol. 2015, 24, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, K.; Oehling, V.; Dusny, C.; Schmid, A. Beyond the bulk: Disclosing the life of single microbial cells. FEMS Microbiol. Rev. 2017, 41, 751–780. [Google Scholar] [CrossRef] [PubMed]
- Pierce, C.G.; Srinivasan, A.; Uppuluri, P.; Ramasubramanian, A.K.; López-Ribot, J.L. Antifungal therapy with an emphasis on biofilms. Curr. Opin. Pharmacol. 2013, 13, 726–730. [Google Scholar] [CrossRef] [PubMed]
- Borghi, E.; Borgo, F.; Morace, G. Fungal Biofilms: Update on Resistance. Adv. Exp. Med. Biol. 2016, 931, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Allan, R.N.; Howlin, R.P.; Stoodley, P.; Hall-Stoodley, L. Targeting microbial biofilms: Current and prospective therapeutic strategies. Nat. Rev. Microbiol. 2017, 15, 740–755. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wang, Y.; Liu, N.; Dong, G.; Sheng, C. Tackling Fungal Resistance by Biofilm Inhibitors. J. Med. Chem. 2017, 60, 2193–2211. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, A.; Uppuluri, P.; Lopez-Ribot, J.; Ramasubramanian, A.K. Development of a high-throughput Candida albicans biofilm chip. PLoS ONE 2011, 6, e19036. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, A.; Lopez-Ribot, J.L.; Ramasubramanian, A.K. Candida albicans biofilm chip (CaBChip) for high-throughput antifungal drug screening. J. Vis. Exp. 2012, 18, e3845. [Google Scholar] [CrossRef]
- Srinivasan, A.; Leung, K.P.; Lopez-Ribot, J.L.; Ramasubramanian, A.K. High-throughput nano-biofilm microarray for antifungal drug discovery. MBio 2013, 4, E00331-13. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, A.; Torres, N.S.; Leung, K.P.; Lopez-Ribot, J.L.; Ramasubramanian, A.K. nBioChip, a Lab-on-a-Chip Platform of Mono- and Polymicrobial Biofilms for High-Throughput Downstream Applications. mSphere 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Bharracharyya, M.K.; Dong, L. Plant Pathogen Spores Grow in Microfluidic Droplets: A High-Throughput Approach to Antifungal Drug Screening. In Proceedings of the IEEE Conference paper Transducers’ 11, Beijing, China, 5–9 June 2011. [Google Scholar]
- Michael, C.A.; Dominey-Howes, D.; Labbate, M. The antimicrobial resistance crisis: Causes, consequences, and management. Front. Public Health 2014, 2, 145. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, J.H.; Ferraro, M.J. Antimicrobial susceptibility testing: A review of general principles and contemporary practices. Clin. Infect. Dis. 2009, 49, 1749–1755. [Google Scholar] [CrossRef] [PubMed]
- Barenfanger, J.; Drake, C.; Kacich, G. Clinical and financial benefits of rapid bacterial identification and antimicrobial susceptibility testing. J. Clin. Microbiol. 1999, 37, 1415–1418. [Google Scholar] [PubMed]
- Arikan, S. Current status of antifungal susceptibility testing methods. Med. Mycol. 2007, 45, 569–587. [Google Scholar] [CrossRef] [PubMed]
- Daniels, R. Surviving the first hours in sepsis: Getting the basics right (an intensivist’s perspective). J. Antimicrob. Chemother. 2011, 66, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Alcazar-Fuoli, L.; Mellado, E. Current status of antifungal resistance and its impact on clinical practice. Br. J. Haematol. 2014, 166, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Syal, K.; Mo, M.; Yu, H.; Iriya, R.; Jing, W.; Guodong, S.; Wang, S.; Grys, T.E.; Haydel, S.E.; Tao, N. Current and emerging techniques for antibiotic susceptibility tests. Theranostics 2017, 7, 1795–1805. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. Approved Standard M27-A3, 3rd ed.; CLSI: Wayne, PA, USA, 2008. [Google Scholar]
- Arendrup, M.C.; Cuenca-Estrella, M.; Lass-Florl, C.; Hope, W.; Eucast, A. EUCAST technical note on the EUCAST definitive document EDef 7.2: Method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts EDef 7.2 (EUCAST-AFST). Clin. Microbiol. Infect. 2012, 18, E246–E247. [Google Scholar] [CrossRef] [PubMed]
- Pulido, M.R.; García-Quintanilla, M.; Martín-Peña, R.; Cisneros, J.M.; McConnell, M.J. Progress on the development of rapid methods for antimicrobial susceptibility testing. J. Antimicrob. Chemother. 2013, 68, 2710–2717. [Google Scholar] [CrossRef] [PubMed]
- Posteraro, B.; Torelli, R.; De Carolis, E.; Posteraro, P.; Sanguinetti, M. Antifungal susceptibility testing: Current role from the clinical laboratory perspective. Mediterr. J. Hematol. Infect. Dis. 2014, 6, e2014030. [Google Scholar] [CrossRef] [PubMed]
- Subcommittee on Antifungal Susceptibility Testing of the ESCMID European Committee for Antimicrobial Susceptibility Testing. EUCAST Technical Note on the method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for conidia-forming moulds. Clin. Microbiol. Infect. 2008, 14, 982–984. [Google Scholar]
- Vale-Silva, L.A.; Buchta, V. Antifungal susceptibility testing by flow cytometry: Is it the future? Mycoses 2006, 49, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Vale-Silva, L.A.; Pinto, P.; Lopes, V.; Ramos, H.; Pinto, E. Comparison of the Etest and a rapid flow cytometry-based method with the reference CLSI broth microdilution protocol M27-A3 for the echinocandin susceptibility testing of Candida spp. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Vella, A.; De Carolis, E.; Vaccaro, L.; Posteraro, P.; Perlin, D.S.; Kostrzewa, M.; Posteraro, B.; Sanguinetti, M. Rapid antifungal susceptibility testing by matrix-assisted laser desorption ionization time of flight mass spectrometry analysis. J. Clin. Microbiol. 2013, 51, 2964–2969. [Google Scholar] [CrossRef] [PubMed]
- Furustrand Tafin, U.; Clauss, M.; Hauser, P.M.; Bille, J.; Meis, J.F.; Trampuz, A. Isothermal microcalorimetry: A novel method for real- time determination of antifungal susceptibility of Aspergillus species. Clin. Microbiol. Infect. 2012, 18, E241–E245. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.; McBeth, C.; Kalashnikov, M.; Boardman, A.K.; Sharon, A.; Sauer-Budge, A.F. Microfluidic advances in phenotypic antibiotic susceptibility testing. Biomed. Microdevices 2016, 18, 103. [Google Scholar] [CrossRef] [PubMed]
- Mittman, S.A.; Huard, R.C.; Della-Latta, P.; Whittier, S. Comparison of BD phoenix to vitek 2, microscan MICroSTREP, and Etest for antimicrobial susceptibility testing of Streptococcus pneumoniae. J. Clin. Microbiol. 2009, 47, 3557–3561. [Google Scholar] [CrossRef] [PubMed]
- Chatzigeorgiou, K.S.; Sergentanis, T.N.; Tsiodras, S.; Hamodrakas, S.J.; Bagos, P.G. Phoenix 100 versus Vitek 2 in the identification of gram-positive and gram-negative bacteria: A comprehensive meta-analysis. J. Clin. Microbiol. 2011, 49, 3284–3291. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Suh, S.J.; Hamon, M.; Hong, J.W. Determination of antibiotic EC50 using a zero-flow microfluidic chip based growth phenotype assay. Biotechnol. J. 2015, 10, 1783–1791. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Zhao, X. Rapid identification and susceptibility testing of uropathogenic microbes via immunosorbent ATP-bioluminescence assay on a microfluidic simulator for antibiotic therapy. Anal. Chem. 2015, 87, 2410–2418. [Google Scholar] [CrossRef] [PubMed]
- Puchberger-Enengl, D.; van den Driesche, S.; Krutzler, C.; Keplinger, F.; Vellekoop, M.J. Hydrogel-based microfluidic incubator for microorganism cultivation and analyses. Biomicrofluidics 2015, 9, 014127. [Google Scholar] [CrossRef] [PubMed]
- Bouquet, O.; Kocsis, B.; Kilár, F.; Lóránd, T.; Kustos, I. Amphotericin B and fluconazole susceptibility of Candida species determined by cell-chip technology. Mycoses 2012, 55, e90–e96. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Lu, Y.; Sin, M.L.; Mach, K.E.; Zhang, D.D.; Gau, V.; Liao, J.C.; Wong, P.K. Antimicrobial susceptibility testing using high surface-to-volume ratio microchannels. Anal. Chem. 2010, 82, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Sinn, I.; Kinnunen, P.; Albertson, T.; McNaughton, B.H.; Newton, D.W.; Burns, M.A.; Kopelman, R. Asynchronous magnetic bead rotation (AMBR) biosensor in microfluidic droplets for rapid bacterial growth and susceptibility measurements. Lab Chip 2011, 11, 2604–2611. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Gao, J.; Zhang, D.D.; Gau, V.; Liao, J.C.; Wong, P.K. Single cell antimicrobial susceptibility testing by confined microchannels and electrokinetic loading. Anal. Chem. 2013, 85, 3971–3976. [Google Scholar] [CrossRef] [PubMed]
- Mohan, R.; Mukherjee, A.; Sevgen, S.E.; Sanpitakseree, C.; Lee, J.; Schroeder, C.M.; Kenis, P.J. A multiplexed microfluidic platform for rapid antibiotic susceptibility testing. Biosens. Bioelectron. 2013, 49, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhen, L.; Liu, J.; Wu, J. Rapid antibiotic susceptibility testing in a microfluidic pH sensor. Anal. Chem. 2013, 85, 2787–2794. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Mu, X.; Guo, Z.; Hao, H.; Zhang, C.; Zhao, Z.; Wang, Q. A novel microbead-based microfluidic device for rapid bacterial identification and antibiotic susceptibility testing. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 2223–2230. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.C.; Cestellos-Blanco, S.; Inoue, K.; Zare, R.N. Miniaturized Antimicrobial Susceptibility Test by Combining Concentration Gradient Generation and Rapid Cell Culturing. Antibiotics 2015, 4, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Derzsi, L.; Kaminski, T.S.; Garstecki, P. Antibiograms in five pipetting steps: Precise dilution assays in sub-microliter volumes with a conventional pipette. Lab Chip 2016, 16, 893–901. [Google Scholar] [CrossRef] [PubMed]
- Leonard, H.; Halachmi, S.; Ben-Dov, N.; Nativ, O.; Segal, E. Unraveling Antimicrobial Susceptibility of Bacterial Networks on Micropillar Architectures Using Intrinsic Phase-Shift Spectroscopy. ACS Nano 2017, 11, 6167–6177. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Huang, X.; Chu, Q.; Ning, X.; Wang, Y.; Kong, S.K.; Zhang, X.; Wang, G.; Ho, H.P. A linear concentration gradient generator based on multi-layered centrifugal microfluidics and its application in antimicrobial susceptibility testing. Lab Chip 2018, 17. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Qiu, Y.; Glidle, A.; McIlvenna, D.; Luo, Q.; Cooper, J.; Shi, H.C.; Yin, H. Gradient microfluidics enables rapid bacterial growth inhibition testing. Anal. Chem. 2014, 86, 3131–3137. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.P.; Richmond, D.L.; Brennan-Krohn, T.; Elliott, H.L.; Kirby, J.E. Development of MAST: A Microscopy-Based Antimicrobial Susceptibility Testing Platform. SLAS Technol. 2017, 22, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Yoo, J.; Kim, K.J.; Kim, E.G.; Park, K.O.; Kim, H.; Kim, H.; Jung, H.; Kim, T.; Choi, M.; et al. Rapid drug susceptibility test of Mycobacterium tuberculosis using microscopic time-lapse imaging in an agarose matrix. Appl. Microbiol. Biotechnol. 2016, 100, 2355–2365. [Google Scholar] [CrossRef] [PubMed]
- Boedicker, J.Q.; Li, L.; Kline, T.R.; Ismagilov, R.F. Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab Chip 2008, 8, 1265–1272. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Jung, Y.G.; Kim, J.; Kim, S.; Jung, Y.; Na, H.; Kwon, S. Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Lab Chip 2013, 13, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Kalashnikov, M.; Lee, J.C.; Campbell, J.; Sharon, A.; Sauer-Budge, A.F. A microfluidic platform for rapid, stress-induced antibiotic susceptibility testing of Staphylococcus aureus. Lab Chip 2012, 12, 4523–4532. [Google Scholar] [CrossRef] [PubMed]
- Price, C.S.; Kon, S.E.; Metzger, S. Rapid antibiotic susceptibility phenotypic characterization of Staphylococcus aureus using automated microscopy of small numbers of cells. J. Microbiol. Methods 2014, 98, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Yoo, J.; Lee, M.; Kim, E.G.; Lee, J.S.; Lee, S.; Joo, S.; Song, S.H.; Kim, E.C.; Lee, J.C.; et al. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci. Transl. Med. 2014, 6, 267ra174. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; An, Y.; Hjort, K.; Hjort, K.; Sandegren, L.; Wu, Z. Time lapse investigation of antibiotic susceptibility using a microfluidic linear gradient 3D culture device. Lab Chip 2014, 14, 3409–3418. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Sakakihara, S.; Grushnikov, A.; Kikuchi, K.; Noji, H.; Yamaguchi, A.; Iino, R.; Yagi, Y.; Nishino, K. A Microfluidic Channel Method for Rapid Drug-Susceptibility Testing of Pseudomonas aeruginosa. PLoS ONE 2016, 11, e0148797. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.M.; Thundat, T. Microcantilever biosensors. Methods 2005, 37, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Fritz, J. Cantilever biosensors. Analyst 2008, 133, 855–863. [Google Scholar] [CrossRef] [PubMed]
- Willaert, R.; Kasas, S.; Devreese, B.; Dietler, G. Yeast nanobiotechnology. Fermentation 2016, 2, 18. [Google Scholar] [CrossRef]
- Braun, T.; Ghatkesar, M.K.; Backmann, N.; Grange, W.; Boulanger, P.; Letellier, L.; Lang, H.P.; Bietsch, A.; Gerber, C.; Hegner, M. Quantitative time-resolved measurement of membrane protein-ligand interactions using microcantilever array sensors. Nat. Nanotechnol. 2009, 4, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Godin, M.; Tabard-Cossa, V.; Miyahara, Y.; Monga, T.; Williams, P.J.; Beaulieu, L.Y.; Bruce Lennox, R.; Grutter, P. Cantilever-based sensing: The origin of surface stress and optimization strategies. Nanotechnology 2010, 21, 75501. [Google Scholar] [CrossRef] [PubMed]
- Godin, M.; Delgado, F.F.; Son, S.; Grover, W.H.; Bryan, A.K.; Tzur, A.; Jorgensen, P.; Payer, K.; Grossman, A.D.; Kirschner, M.W.; et al. Using buoyant mass to measure the growth of single cells. Nat. Methods 2010, 7, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Ndieyira, J.W.; Watari, M.; Barrera, A.D.; Zhou, D.; Vögtli, M.; Batchelor, M.; Cooper, M.A.; Strunz, T.; Horton, M.A.; Abell, C.; et al. Nanomechanical detection of antibiotic-mucopeptide binding in a model for superbug drug resistance. Nat. Nanotechnol. 2008, 3, 691–696. [Google Scholar] [CrossRef] [PubMed]
- Lang, H.P.; Baller, M.K.; Berger, R.; Gerber, C.; Gimzewski, J.K.; Battiston, F.M.; Fornaro, P.; Ramseyer, J.P.; Meyer, E.; Guntherodt, H.J. An artificial nose based a micromechanical cantilever array. Anal. Chim. Acta 1999, 393, 59–65. [Google Scholar] [CrossRef]
- Braun, T.; Barwich, V.; Ghatkesar, M.K.; Bredekamp, A.H.; Gerber, C.; Hegner, M.; Lang, H.P. Micromechanical mass sensors for biomolecular detection in a physiological environment. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2005, 72, 031907. [Google Scholar] [CrossRef] [PubMed]
- Hosaka, S.; Chiyoma, T.; Ikeuchi, A.; Okano, H.; Sone, H.; Izumi, T. Possibility of a femtogram mass biosensor using a self-sensing cantilever. Curr. Appl. Phys. 2006, 6, 384–388. [Google Scholar] [CrossRef]
- Liu, Y.; Schweizerb, L.M.; Wanga, W.; Reubena, R.L.; Schweizer, M.; Shu, W. Label-free and real-time monitoring of yeast cell growth by the bending of polymer microcantilever biosensors. Sens. Actuator B-Chem. 2013, 178, 621–626. [Google Scholar] [CrossRef]
- Bryan, A.K.; Goranov, A.; Amon, A.; Manalis, S.R. Measurement of mass, density, and volume during the cell cycle of yeast. Proc. Natl. Acad. Sci. USA 2010, 107, 999–1004. [Google Scholar] [CrossRef] [PubMed]
- Cermak, N.; Olcum, S.; Delgado, F.F.; Wasserman, S.C.; Payer, K.R.; Murakami, M.A.; Knudsen, S.M.; Kimmerling, R.J.; Stevens, M.M.; Kikuchi, Y.; et al. High-throughput measurement of single-cell growth rates using serial microfluidic mass sensor arrays. Nat. Biotechnol. 2016, 34, 1052–1059. [Google Scholar] [CrossRef] [PubMed]
- Burg, T.P.; Godin, M.; Knudsen, S.M.; Shen, W.; Carlson, G.; Foster, J.S.; Babcock, K.; Manalis, S.R. Weighing of biomolecules, single cells and single nanoparticles in fluid. Nature 2007, 446, 1066–1069. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Jang, J.; Irimia, D.; Sturgis, J.; Lee, J.; Robinson, J.P.; Toner, M.; Bashir, R. ‘Living cantilever arrays’ for characterization of mass of single live cells in fluids. Lab Chip 2008, 8, 1034–1041. [Google Scholar] [CrossRef] [PubMed]
- Bryan, A.K.; Hecht, V.C.; Shen, W.; Payer, K.; Grover, W.H.; Manalis, S.R. Measuring single cell mass, volume, and density with dual suspended microchannel resonators. Lab Chip 2014, 14, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Nugaeva, N.; Gfeller, K.Y.; Backmann, N.; Lang, H.P.; Düggelin, M.; Hegner, M. Micromechanical cantilever array sensors for selective fungal immobilization and fast growth detection. Biosens. Bioelectron. 2005, 21, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Stupar, P. Atomic Force Microscopy of Biological Systems: Quantitative Imaging and Nanomotion Detection. Ph.D. Thesis, EPFL Scientific Publications, Lausanne, Switzerland, 2018. [Google Scholar]
- Longo, G.; Alonso-Sarduy, L.; Rio, L.M.; Bizzini, A.; Trampuz, A.; Notz, J.; Dietler, G.; Kasas, S. Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nat. Nanotechnol. 2013, 8, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Stupar, P.; Opota, O.; Longo, G.; Prod’hom, G.; Dietler, G.; Greub, G.; Kasas, S. Nanomechanical sensor applied to blood culture pellets: A fast approach to determine the antibiotic susceptibility against agents of bloodstream infections. Clin. Microbiol. Infect. 2017, 23, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Stupar, P.; Yvanoff, C.; Chomicki, W.; Dietler, G.; Kasas, S.; Willaert, R. Exploring Nanoscale Motions of Yeast Cells. In Proceedings of the XIX Annual Linz Winter Workshop, Linz, Austria, 3–6 February 2017. [Google Scholar]
- Vanden Boer, P.; Stupar, P.; Chomicki, W.; Dietler, G.; Kasas, S.; Willaert, R. Nanomotion Detection of Single Yeast Cell Growth. In Proceedings of the XX Annual Linz Winter Workshop, Linz, Austria, 2–5 February 2018. [Google Scholar]
- Morones, J.R.; Elechiguerra, J.L.; Camacho, A.; Ramirez, J.T. The bactericidal effect of silver nanoparticles. Nanotechnology 2005, 16, 2346–2353. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Kuk, E.; Yu, K.N.; Kim, J.H.; Park, S.J.; Lee, H.J.; Kim, S.H.; Park, Y.K.; Park, Y.H.; Hwang, C.Y.; et al. Antimicrobial effects of silver nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2007, 3, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Ahamed, M.; Alsalhi, M.S.; Siddiqui, M.K. Silver nanoparticle applications and human health. Clin. Chim. Acta 2010, 411, 1841–1848. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Schleusener, H.J. Nanosilver: A nanoproduct in medical application. Toxicol. Lett. 2008, 176, 1e12. [Google Scholar] [CrossRef] [PubMed]
- Rai, M.; Yadav, A.; Gade, A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol. Adv. 2009, 27, 76–83. [Google Scholar] [CrossRef] [PubMed]
- De Jong, W.H.; Van Der Ven, L.T.; Sleijffers, A.; Park, M.V.; Jansen, E.H.; Van Loveren, H.; Vandebriel, R.J. Systemic and immunotoxicity of silver nanoparticles in an intravenous 28 days repeated dose toxicity study in rats. Biomaterials 2013, 34, 8333–8343. [Google Scholar] [CrossRef] [PubMed]
- Durán, N.; Durán, M.; de Jesus, M.B.; Seabra, A.B.; Fávaro, W.J.; Nakazato, G. Silver nanoparticles: A new view on mechanistic aspects on antimicrobial activity. Nanomedicine 2016, 12, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Ahamed, M.; Karns, M.; Goodson, M.; Rowe, J.; Hussain, S.M.; Schlager, J.J.; Hong, Y. DNA damage response to different surface chemistry of silver nanoparticles in mammalian cells. Toxicol. Appl. Pharmacol. 2008, 233, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Kawata, K.; Osawa, M.; Okabe, S. In vitro toxicity of silver nanoparticles at noncytotoxic doses to HepG2 human hepatoma cells. Environ. Sci. Technol. 2009, 43, 6046–6051. [Google Scholar] [CrossRef] [PubMed]
- AshaRani, P.V.; Low Kah Mun, G.; Hande, M.P.; Valiyaveettil, S. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano 2009, 3, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Panáček, A.; Kvítek, L.; Smékalová, M.; Večeřová, R.; Kolář, M.; Röderová, M.; Dyčka, F.; Šebela, M.; Prucek, R.; Tomanec, O.; et al. Bacterial resistance to silver nanoparticles and how to overcome it. Nat. Nanotechnol. 2018, 13, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Ammar, H.A.; El-Desouky, T.A. Green synthesis of nanosilver particles by Aspergillus terreus HA1N and Penicillium expansum HA2N and its antifungal activity against mycotoxigenic fungi. J. Appl. Microbiol. 2016, 121, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; He, D.; Gao, S.; Wang, D.; Yokoyama, K.; Wang, L. Biosynthesis of silver nanoparticles by the fungus Arthroderma fulvum and its antifungal activity against genera of Candida, Aspergillus and Fusarium. Int. J. Nanomedicine 2016, 11, 1899–1906. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.T.; Jameel, N. Antifungal Activity of Silver Nanoparticles Produced from Fungus, Penicillium fellutanum at Different pH. J. Microb. Biochem. Technol. 2016, 8, 5. [Google Scholar] [CrossRef]
- Fukushima, K.; Liu, S.; Wu, H.; Engler, A.C.; Coady, D.J.; Maune, H.; Pitera, J.; Nelson, A.; Wiradharma, N.; Venkataraman, S.; et al. Supramolecular high-aspect ratio assemblies with strong antifungal activity. Nat. Commun. 2013, 4, 2861. [Google Scholar] [CrossRef] [PubMed]
- Esteban-Tejeda, L.; Malpartida, F.; Esteban-Cubillo, A.; Pecharromán, C.; Moya, J.S. Antibacterial and antifungal activity of a soda-lime glass containing copper nanoparticles. Nanotechnology 2009, 20, 505701. [Google Scholar] [CrossRef] [PubMed]
- Paulo, C.S.; Vidal, M.; Ferreira, L.S. Antifungal nanoparticles and surfaces. Biomacromolecules 2010, 11, 2810–2817. [Google Scholar] [CrossRef] [PubMed]
- Perfect, J.R. Fungal virulence genes as targets for antifungal chemotherapy. Antimicrob. Agents Chemoth. 1996, 40, 1577–1583. [Google Scholar]
- Kitamura, A.; Someya, K.; Hata, M.; Nakajima, R.; Takemura, M. Discovery of a small-molecule inhibitor of β-1,6-glucan synthesis. Antimicrob. Agents Chemoth. 2009, 53, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Baxter, B.K.; DiDone, L.; Ogu, D.; Schor, S.; Krysan, D.J. Identification, in vitro activity and mode of action of phosphoinositide-dependent-1 kinase inhibitors as antifungal molecules. ACS Chem. Biol. 2011, 6, 502–510. [Google Scholar] [CrossRef] [PubMed]
| Class Compound | MoA | Spectrum of Activity | Comment |
|---|---|---|---|
| Polyenes | |||
| Amphotericin B | Selective binding to ergosterol cause the formation of pores in the membrane. | Treatment of deep mycoses, candidiasis, cryptococcosis, histoplasmosis, blastomycosis, paracoccodioidomycosis, cocciodioidomycosis, aspergillosis, extracutaneous sporotrichichosis, and some cases of mucormycosis, hyalohyphomycosis, and phaeohyphomycosis, S. cerevisiae. | Fungicidal, broad spectrum, intravenous, little resistance observed, significant nephrotoxicity, indirectly affects action of many drugs. |
| Nystatin/Nyotran | Selective binding to ergosterol cause the formation of pores in the membrane. | Candidiasis | Nyotran is a liposomal formulation of nystatin with lowered toxicity. |
| Natamycin | Binds to ergosterol in the plasma membrane, preventing ergosterol-dependent fusion of vacuoles, membrane fusion, and fission. | Keratinophilic fungi, corneal infections | |
| Azoles | |||
| Fluconazole | Selective inhibition of fungal cytochrome P450-dependent lanosterol-14-α-demethylase. | Candida immitis, C. neoformans, Paracoccidioides brasiliensis; lower activity against Aspergillus, Fusarium, Scedosporium, Penicillium species and other filamentous fungi. | Fluconazole resistant C. albicans and non-albicans strains increasing. |
| Itraconazole | Selective inhibition of fungal cytochrome P450-dependent lanosterol-14-α-demethylase. | Most Candida species, P. brasiliensis, H. capsulatum, Blastomyces dermatitidis, Aspergillus fumigatus, A. niger, Penicillium marneffei. | Better than fluconazole in the treatment of cocciodioido-mycosis, not reaching the central nervous system; numerous drug interactions due to inhibition of CYP 3A4. |
| Voriconazole | Selective inhibition of fungal cytochrome P450-dependent lanosterol-14-α-demethylase. | Candida species, including C. krusei, S. cerevisiae. | More potent than fluconazole; very rapid metabolism in children; numerous drug interactions due to inhibition of CYP 3A4. |
| Econazole | Selective inhibition of fungal cytochrome P450-dependent lanosterol-14-α-demethylase. | Trichophyton rubrum, T. Mentagrophytes, Epidermophyton floccosum. | Is an immidazole antifungal for the treatment of tinea pedis and crusis, pityriasis versicolor. |
| Tioconazole | Selective inhibition of fungal cytochrome P450-dependent lanosterol-14-α-demethylase. | C. albicans, Trichophyton sp., Epidermophyton sp. | Is an immidazole antifungal for topical treatment of superficial mycoses (ringworm, jock itch, athlete’s foot, tinea versicolor. |
| Echinocandins | |||
| Caspofungin | Fungal β-1,3-glucan synthase inhibitors. | Candida species, Aspergillus species. | Fungicidal for Candida, fungistatic for Aspergillus; modest efficacy as first-line agent for invasive aspergillosis; intravenous formulation only; interacts with ciclosporin and rifampicin. |
| Anidulafungin | Fungal β-1,3-glucan synthase inhibitors. | Candida species, Aspergillus species. | Fungicidal for Candida, fungistatic for Aspergillus; licensed for the treatment of invasive and esophageal candidiasis. |
| Micafungin | Fungal β-1,3-glucan synthase inhibitors. | Candida species, Aspergillus species. | Fungicidal for Candida, fungistatic for Aspergillus; licensed invasive and esophageal candidiasis. |
| 5-Fluoropyrimidine | |||
| 5-Fluorocystosine | Selective conversion of toxic intermediate (5-fluorouridine). | Cryptococcosis, candidiasis, chromoblastomycosis; high MIC for some strains of Aspergillus, Penicillium and several Zygomycetes, except for chromoblastomycosis. | 5-Fluorocystosine is always used in combination with amphotericin B. |
| High-Throughput Technology | Application | Microorganism | Characteristics | References |
|---|---|---|---|---|
| Cellular microarray | Antifungal biofilm screening | Candida albicans | Cells robotically printed, 768 (48 × 16 array) cultures of 50 nL biofilms in collagen. | [111,112] |
| Cellular microarray | Antifungal biofilm screening | C. albicans | Cells robotically printed, 1200 (60 × 20 array) cultures of 30 nL biofilms in alginate. | [113] |
| Cellular microarray | Antibiotic and antifungal biofilm screening | C. albicans, Staphylococcus aureus, Pseudomonas aeruginosa | Robotically printing of mono- and polymicrobial biofilms, 576 (48 × 12 array) cultures of 30 nL biofilms in alginate. | [114] |
| Droplet microfluidics | Antifungal drug screening | Phytophthora sojae | The plant pathogen spores and the drug were encapsulated in liquid droplets. Phenotypic responses to the drug at different concentrations were microscopically quantified. | [115] |
| Microorganism | Measurement Principle | Description | Reference |
|---|---|---|---|
| Fungi | |||
| Candida strains | Fluorescence-based distinction between living and dead cells. | Cell Chip kit 1 used as cell sorter and the determination of fluorescence histograms of previously labelled cells. | [138] |
| Candida albicans | Immunosorbent ATP-bioluminescence assay. | The microfluidic device employs a fiberglass membrane sandwiched between two polypropylene components, with capture antibodies immobilised on the membrane. Cells immobilised in alginate hydrogel. | [136] |
| Saccharomyces cerevisiae | Fluorescence staining and imaging after incubation. | Cell seeding and diffusive medium supply is provided by phase-guide technology, enabling operation of continuous culturing. | [137] |
| Bacteria | |||
| Escherichia coli | Optical imaging of single cell growth (number of cells). | Growth of cells in channels, with a large surface-to-volume ratio. | [139] |
| E. coli | Magnetic bead rotation, which is inversely proportional to bacterial mass. | Droplet microfluidics where single cells are entrapped in liquid drops. | [140] |
| E. coli | Optical imaging of single cell growth (number of cells). | Individual cells grow in gas permeable (PDMS) microchannels, with dimensions comparable to a single cell. | [141] |
| E. coli | Fluorescence imaging of cells that express green fluorescent protein. | Growth of cells in 12 sets of quadruplicate microfluidic chambers. Quantification of the effect of four antibiotics and their combinations. | [142] |
| E. coli | Reflectometric interference spectroscopy of pH-sensitive chitosan hydrogel measured the accumulation of metabolic products. | Growth of cells in microfluidic channels. | [143] |
| E. coli | Fluorescence imaging of immunomicrobeads attached to the cells. | Growth in microfluidic chambers. | [144] |
| E. coli | Optical imaging. | The standard broth microdilution method was miniaturised in a microfluidic chip that generates an antibiotic concentration gradient and delivers antibiotic-containing culture media to eight 30-nL chambers for cell culture. | [145] |
| E. coli | Fluorescence imaging. | The microfluidic chip allows the carrying out of commonly executed antibiotic susceptibility assays in an array of nanoliter droplets. | [146] |
| E. coli | Optical phase-shift reflectometric interference spectroscopic measurements. | The use of biofunctionalised silicon micropillar arrays to provide both a preferable solid-liquid interface for bacteria networking and a simultaneous transducing element that monitors the response of bacteria when exposed to chosen antibiotics in real time. | [147] |
| E. coli | Spectral absorbance of cell suspensions. | An automated linear gradient generator based on centrifugal microfluidics. | [148] |
| E. coli, Nitrosomas europaea | Optical imaging of single cell growth (number of cells). | The cells grow in a layer of agarose upon which a gradient of the antibiotic is applied. | [149] |
| Enterobacter cloacae, E. coli, Klebsiella pneumoniae, P. aeruginosa, Acinetobacter baumannii | Optical imaging of bacterial replication. | A solid-phase microwell growth surface in a 384-well plate format was used, with inkjet printing–based application of both antimicrobials and bacteria at any desired concentrations. | [150] |
| Enterococcus faecalis, E. coli | Fluorescence staining and imaging after incubation. | Cell seeding and diffusive medium supply is provided by phase-guide technology, enabling operation of continuous culturing. | [137] |
| Mycobacterium tuberculosis | Optical imaging of single cell growth (number of cells). | Cells were immobilized in an agarose matrix, which was molded in a microfluidic chip. | [151] |
| Pseudomonas aeruginosa | Fluorescence imaging of GFP-expressing cells. | The microfluidic chip generates a logarithmic concentration gradient through semidirect dilution in a zero-flow condition and cells grow in nanoliter reactors. | [135] |
| Staphylococcus aureus | Fluorescence imaging of viability indicator. | Stochastic confinement of individual cells into liquid plugs (droplet microfluidics); distinction between sensitive and resistant bacteria. | [152] |
| S. aureus | Optical imaging of single cell growth (number of cells). | Antibiotics diffuse into a microfluidic channel containing the cells. | [153] |
| S. aureus | Fluorescence imaging for dead cells (rates of killing). | Cells are covalently bound to the bottom of the channels and fluid flow shear stress activation of pathways that are targets of antibiotics. | [154] |
| S. aureus | Optical imaging of single cell growth (number of cells). | Chip with 32 individual fluidic channels. | [155] |
| S. aureus, E. coli, K. pneumoniae, P. aeroginosa | Optical imaging of single cell growth (number of cells). | Antibiotics diffuse into a microfluidic agarose channels in 96-well format. | [156] |
| S. aureus, S. epidermitis, S. saprophyticus, E. coli, K. pneumonia, P. aeruginosa, P. mirabilis, Streptococcus pyrogenes, S. viridans | Immunosorbent ATP-bioluminescence assay. | The microfluidic device employs a fiberglass membrane sandwiched between two polypropylene components, with capture antibodies immobilised on the membrane. Cells immobilised in alginate hydrogel. | [136] |
| Salmonella thyphimurium, E. coli, S. aureus | Optical imaging of single cell growth (number of cells). | Cells immobilised in agarose with a gradient of the antibiotic in the gel slab. | [157] |
| P. aeruginosa | Optical imaging of single cell growth (number of cells). | Growth of cells in four parallel channels. | [158] |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Willaert, R.G. Micro- and Nanoscale Approaches in Antifungal Drug Discovery. Fermentation 2018, 4, 43. https://doi.org/10.3390/fermentation4020043
Willaert RG. Micro- and Nanoscale Approaches in Antifungal Drug Discovery. Fermentation. 2018; 4(2):43. https://doi.org/10.3390/fermentation4020043
Chicago/Turabian StyleWillaert, Ronnie G. 2018. "Micro- and Nanoscale Approaches in Antifungal Drug Discovery" Fermentation 4, no. 2: 43. https://doi.org/10.3390/fermentation4020043
APA StyleWillaert, R. G. (2018). Micro- and Nanoscale Approaches in Antifungal Drug Discovery. Fermentation, 4(2), 43. https://doi.org/10.3390/fermentation4020043