Starter Cultures for Sparkling Wine
Abstract
:1. Introduction
2. Sparkling Wine: Production Process, Legislation, and Classification
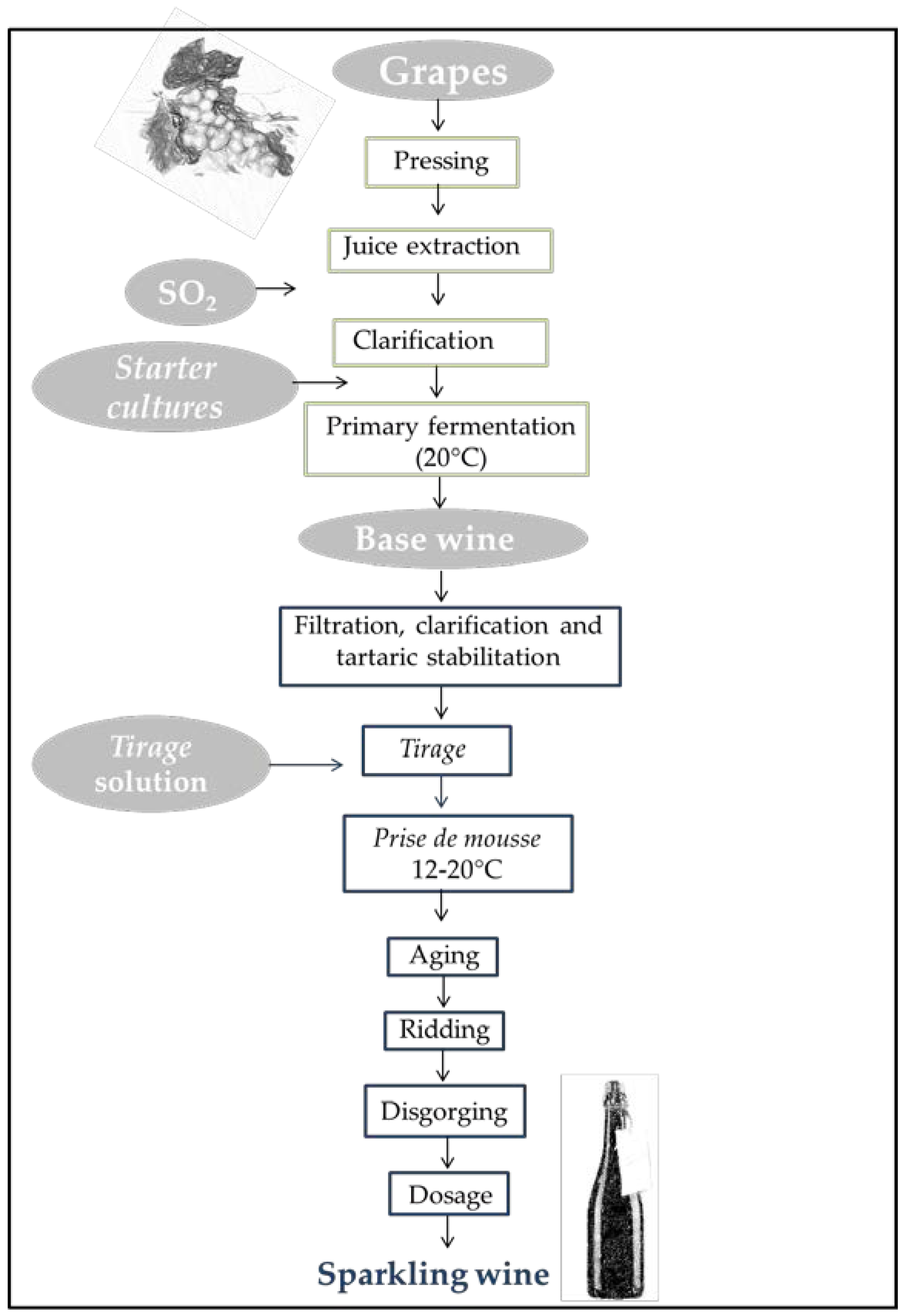
2.1. Production of Sparkling Wine Using the Charmat Method
2.2. Production of Sparkling Wine Using the Traditional Method
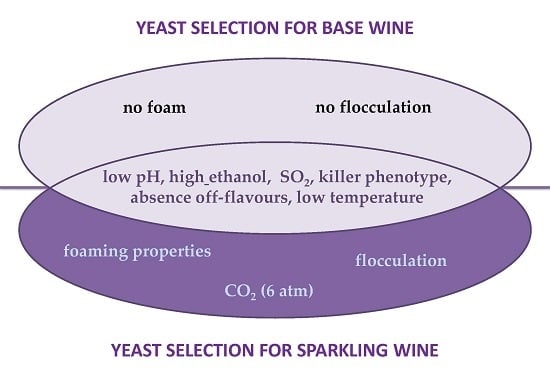
3. Yeast Characterization for Wine Base Production
3.1. Yeast Genotypic Characterization: Methods to Differentiate Saccharomyces Cerevisiae Strains
3.2. Yeast Technological and Qualitative Characterization for Starter Culture Production
4. Yeast Technological Characterization for Secondary Fermentation of Sparkling Wine Production
5. Sparkling Wine Production: Role of Non-Saccharomyces and Lactic Acid Bacteria
6. Safety Aspects Correlated to Base and Sparkling Wine
7. Biotechnological Applications
8. Conclusions
Acknowledgments
Conflicts of Interest
References
- Blasco, L.; Viñas, M.; Villa, T.G. Proteins influencing foam formation in wine and beer: The role of yeast. Int. Microbiol. 2011, 14, 61–71. [Google Scholar] [PubMed]
- Legras, J.-L.; Merdinoglu, D.; Cornuet, J.-M.; Karst, F. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol. Ecol. 2007, 16, 2091–2102. [Google Scholar] [CrossRef] [PubMed]
- Jolly, N.P.; Varela, C.; Pretorius, I.S. Not your ordinary yeast: Non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 2014, 14, 215–237. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, C.; El Khoury, M.; Lucas, P.; Bely, M.; Russo, P.; Spano, G.; Capozzi, V. Autochthonous starter cultures and indigenous grape variety for regional wine production. J. Appl. Microbiol. 2015, 118, 1395–1408. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, C.; Tristezza, M.; Grieco, F.; Spano, G.; Capozzi, V. From grape berries to wine: Population dynamics of cultivable yeasts associated to “Nero di Troia” autochthonous grape cultivar. World J. Microbiol. Biotechnol. 2016, 32, 59. [Google Scholar] [CrossRef] [PubMed]
- Capozzi, V.; Garofalo, C.; Chiriatti, M.A.; Grieco, F.; Spano, G. Microbial terroir and food innovation: The case of yeast biodiversity in wine. Microbiol. Res. 2015, 181, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Carrascosa, A.V.; Martinez-Rodiguez, A.; Cebollero, E.; Gonzalez, R. Saccharomyces Yeasts II: Secondary fermentation. In Molecular Wine Microbiology; Elsevier: London, UK, 2011; Volume 2, pp. 33–48. [Google Scholar]
- Cebollero, E.; Gonzalez, R. Induction of autophagy by second-fermentation yeasts during elaboration of sparkling wines. Appl. Environ. Microbiol. 2006, 72, 4121–4127. [Google Scholar] [CrossRef] [PubMed]
- González-Royo, E.; Pascual, O.; Kontoudakis, N.; Esteruelas, M.; Esteve-Zarzoso, B.; Mas, A.; Canals, J.M.; Zamora, F. Oenological consequences of sequential inoculation with non-Saccharomyces yeasts (Torulaspora delbrueckii or Metschnikowia pulcherrima) and Saccharomyces cerevisiae in base wine for sparkling wine production. Eur. Food Res. Technol. 2014, 240, 999–1012. [Google Scholar] [CrossRef]
- Ody-Brasier, A.; Vermeulen, F. The price you pay price-setting as a response to norm violations in the market for champagne grapes. Adm. Sci. Q. 2014, 59, 109–144. [Google Scholar] [CrossRef]
- Torresi, S.; Frangipane, M.T.; Anelli, G. Biotechnologies in sparkling wine production. Interesting approaches for quality improvement: A review. Food Chem. 2011, 129, 1232–1241. [Google Scholar] [CrossRef] [PubMed]
- Perpetuini, G.; Di Gianvito, P.; Arfelli, G.; Schirone, M.; Corsetti, A.; Tofalo, R.; Suzzi, G. Biodiversity of autolytic ability in flocculent Saccharomyces cerevisiae strains suitable for traditional sparkling wine fermentation. Yeast 2016, 33, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Magariño, S.; Martínez-Lapuente, L.; Bueno-Herrera, M.; Ortega-Heras, M.; Guadalupe, Z.; Ayestarán, B. Use of commercial dry yeast products rich in mannoproteins for white and rosé sparkling wine elaboration. J. Agric. Food Chem. 2015, 63, 5670–5681. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, S.J.; de Leonardis, A.; Lustrato, G.; Testa, B.; Iorizzo, M. Yeast autolysis in sparkling wine aging: Use of killer and sensitive Saccharomyces cerevisiae strains in co-culture. Recent Pat. Biotechnol. 2015, 9, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, H.; Guilloux-Benatier, M. Yeast autolysis in sparkling wine—A review. Aust. J. Grape Wine Res. 2006, 12, 119–127. [Google Scholar] [CrossRef]
- Tita, O.; Jascanu, V.; Tita, M.; Sand, C. Economical comparative analysis of different bottle fermentation methods. In Proceedings of the AVA 2003, International Conference on Agricultural Economics, Rural Development and Informatics in the New Millennium, Debrecen, Hungary, 1–2 April 2003.
- Pozo-Bayón, M.A.; Andujar-Ortiz, I.; Alcaide-Hidalgo, J.M.; Martín-Alvarez, P.J.; Moreno-Arribas, M.V. Characterization of commercial inactive dry yeast preparations for enological use based on their ability to release soluble compounds and their behavior toward aroma compounds in model wines. J. Agric. Food Chem. 2009, 57, 10784–10792. [Google Scholar] [CrossRef] [PubMed]
- Ganss, S.; Kirsch, F.; Winterhalter, P.; Fischer, U.; Schmarr, H.-G. Aroma changes due to second fermentation and glycosylated precursors in Chardonnay and Riesling sparkling wines. J. Agric. Food Chem. 2011, 59, 2524–2533. [Google Scholar] [CrossRef] [PubMed]
- Pozo-Bayón, M.A.; Hernández, M.T.; Martín-Alvarez, P.J.; Polo, M.C. Study of low molecular weight phenolic compounds during the aging of sparkling wines manufactured with red and white grape varieties. J. Agric. Food Chem. 2003, 51, 2089–2095. [Google Scholar] [CrossRef] [PubMed]
- Ribéreau-Gayon, P.; Dubourdieu, D.; Donèche, B.; Lonvaud, A. Trattato di Enologia I, Microbiologia del Vino; Edagricole: Bologna, Italy, 2004. [Google Scholar]
- Zoecklein, B. A Review of Méthode Champenoise Production, 2nd ed.; Virginia Tech: Blacksburg, VA, USA, 2002. [Google Scholar]
- Martínez-Rodríguez, A.J.; Pueyo, E. Sparkling Wines and Yeast Autolysis. In Wine Chemistry and Biochemistry; Moreno-Arribas, M.V., Polo, M.C., Eds.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Bidan, P.; Feuillat, M.; Moulin, J.P. Les vins mousseux. Bull. l’OIV 1986, 59, 663–664. [Google Scholar]
- Nunez, Y.P.; Carrascosa, A.V.; González, R.; Polo, M.C.; Martínez-Rodríguez, A.J. Effect of accelerated autolysis of yeast on the composition and foaming properties of sparkling wines elaborated by a champenoise method. J. Agric. Food Chem. 2005, 53, 7232–7237. [Google Scholar] [CrossRef] [PubMed]
- Luguera, C.; Moreno-Arribas, M.V.; Pueyo, E.; Bartolomé, B.; Polo, M.C. Fractionation and partial characterization of protein fractions present at different stages of the production of sparkling wines. Food Chem. 1998, 63, 465–471. [Google Scholar] [CrossRef]
- Nikolaou, E.; Soufleros, E.H.; Bouloumpasi, E.; Tzanetakis, N. Selection of indigenous Saccharomyces cerevisiae strains according to their oenological characteristics and vinification results. Food Microbiol. 2006, 23, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Šuranská, H.; Vránová, D.; Omelková, J. Isolation, identification and characterization of regional indigenous Saccharomyces cerevisiae strains. Braz. J. Microbiol. 2016, 47, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Mercado, L.; Sturm, M.E.; Rojo, M.C.; Ciklic, I.; Martínez, C.; Combina, M. Biodiversity of Saccharomyces cerevisiae populations in Malbec vineyards from the “Zona Alta del Río Mendoza” region in Argentina. Int. J. Food Microbiol. 2011, 151, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Pérez, F.; Regodón, J.; Valdés, M.; de Miguel, C.; Ramírez, M. Cycloheximide resistance as marker for monitoring yeasts in wine fermentations. Food Microbiol. 2000, 17, 119–128. [Google Scholar] [CrossRef]
- Ambrona, J.; Maqueda, M.; Zamora, E.; Ramírez, M. Sulfometuron Methyl Resistance as Genetic Marker for Monitoring Yeast Populations in Wine Fermentations. J. Agric. Food Chem. 2005, 53, 7438–7443. [Google Scholar] [CrossRef] [PubMed]
- Ambrona, J.; Vinagre, A.; Maqueda, M.; Ramírez, M. Rhodamine-pink as a genetic marker for yeast populations in wine fermentation. J. Agric. Food Chem. 2006, 54, 2977–2984. [Google Scholar] [CrossRef] [PubMed]
- Velázquez, R.; Zamora, E.; Álvarez, M.L.; Álvarez, M.L.; Ramírez, M. Using mixed inocula of new killer strains of Saccharomyces cerevisiae to improve the quality of traditional sparkling-wine. Food Microbiol. 2016, 59, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Martínez, C.; Cosgaya, P.; Vásquez, C.; Gac, S.; Ganga, A. High degree of correlation between 464 molecular polymorphism and geographic origin of wine yeast strains. J. Appl. Microbiol. 2007, 103, 2185–2195. [Google Scholar] [CrossRef] [PubMed]
- Schuller, D.; Valero, E.; Dequin, S.; Casal, M. Survey of molecular methods for the typing of wine yeast strains. FEMS Microbiol. Lett. 2004, 231, 19–26. [Google Scholar] [CrossRef]
- Querol, A.; Barrio, E.; Huerta, T.; Ramón, D. Molecular monitoring of wine fermentations conducted by active dry yeast strains. Appl. Environ. Microbiol. 1992, 58, 2948–2953. [Google Scholar] [PubMed]
- Guillamon, J.M.; Sabaté, J.; Barrio, E.; Cano, J.; Querol, A. Rapid identification of wine yeast species based on RFLP analysis of the ribosomal internal transcribed spacer (ITS) region. Arch. Microbiol. 1998, 169, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Espinar, M.T.; López, V.; Ramón, D.; Bartra, E.; Querol, A. Study of the authenticity of commercial wine yeast strains by molecular techniques. Int. J. Food Microbiol. 2001, 70, 1–10. [Google Scholar] [CrossRef]
- Torija, M.J.; Rozès, N.; Poblet, M.; Guillamón, J.M.; Mas, A. Yeast population dynamics in spontaneous fermentations: Comparison between two different wine-producing areas over a period of three years. Antonie Van Leeuwenhoek 2001, 79, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Blondin, B.; Vezinhet, F. Identification de souches de levures oenologiques par leurs caryotypes obtenus en électrophorèse en champ pulsé. Rev. Franç. Oenol. 1988, 28, 7–11. [Google Scholar]
- Ness, F.; Lavallée, F.; Dubourdieu, D.; Aigle, M.; Dulau, L. Identification of yeast strains using the polymerase chain reaction. J. Sci. Food Agric. 1993, 62, 89–94. [Google Scholar] [CrossRef]
- Quesada, M.P.; Cenis, J.L. Use of Random Amplified Polymorphic DNA (RAPD-PCR) in the Characterization of Wine Yeasts. Am. J. Enol. Vitic. 1995, 46, 204–208. [Google Scholar]
- Strand, M.; Prolla, T.A.; Liskay, R.M.; Petes, T.D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 1993, 365, 274–276. [Google Scholar] [CrossRef] [PubMed]
- Pérez, M.A.; Gallego, F.J.; Martínez, I.; Hidalgo, P. Detection, distribution and selection of microsatellites (SSRs) in the genome of the yeast Saccharomyces cerevisiae as molecular markers. Lett. Appl. Microbiol. 2001, 33, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Fay, J.C.; Benavides, J.A. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 2005, 1, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Guillamón, J.M.; Barrio, E.; Querol, A. Characterization of Wine Yeast Strains of the Saccharomyces genus on the basis of molecular markers: Relationships between genetic distance and geographic or ecological origin. Syst. Appl. Microbiol. 1996, 19, 122–132. [Google Scholar] [CrossRef]
- Capece, A.; Romaniello, R.; Siesto, G.; Pietrafesa, R.; Massari, C.; Poeta, C.; Romano, P. Selection of indigenous Saccharomyces cerevisiae strains for Nero d’Avola wine and evaluation of selected starter implantation in pilot fermentation. Int. J. Food Microbiol. 2010, 144, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Capece, A.; Romaniello, R.; Siesto, G.; Romano, P. Diversity of Saccharomyces cerevisiae yeasts associated to spontaneously fermenting grapes from an Italian “heroic vine-growing area”. Food Microbiol. 2012, 31, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Barrajón, N.; Arévalo-Villena, M.; Rodríguez-Aragón, L.J.; Briones, A. Ecological study of wine yeast in inoculated vats from La Mancha region. Food Control. 2009, 20, 778–783. [Google Scholar] [CrossRef]
- Maqueda, M.; Zamora, E.; Rodríguez-Cousiño, N.; Ramírez, M. Wine yeast molecular typing using a simplified method for simultaneously extracting mtDNA, nuclear DNA and virus dsRNA. Food Microbiol. 2010, 27, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, R.; Gómez, A.; Robles, V.; Rodríguez, P.; Cebollero, E.; Tabera, L.; Carrascosa, A.V.; Gonzalez, R. Multilocus sequence typing of oenological Saccharomyces cerevisiae strains. Food Microbiol. 2009, 26, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Tristezza, M.; Fantastico, L.; Vetrano, C.; Bleve, G.; Corallo, D.; Grieco, F.; Mita, G.; Grieco, F. Molecular and technological characterization of Saccharomyces cerevisiae strains isolated from natural fermentation of Susumaniello grape must in Apulia, Southern Italy. Int. J. Microbiol. 2014, 2014, 897427. [Google Scholar] [CrossRef] [PubMed]
- Tristezza, M.; Vetrano, C.; Bleve, G.; Grieco, F.; Tufariello, M.; Quarta, A.; Mita, G.; Spano, G.; Grieco, F. Autochthonous fermentation starters for the industrial production of Negroamaro wines. J. Ind. Microbiol. Biotechnol. 2011, 39, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Tristezza, M.; Vetrano, C.; Bleve, G.; Spano, G.; Capozzi, V.; Logrieco, A.; Mita, G.; Grieco, F. Biodiversity and safety aspects of yeast strains characterized from vineyards and spontaneous fermentations in the Apulia Region, Italy. Food Microbiol. 2013, 36, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Pretorius, I.S. Tailoring wine yeast for the new millennium: Novel approaches to the ancient art of winemaking. Yeast 2000, 16, 675–729. [Google Scholar] [CrossRef]
- Cappello, M.S.; Poltronieri, P.; Blaiotta, G.; Zacheo, G. Molecular and physiological characteristics of a grape yeast strain containing atypical genetic material. Int. J. Food Microbiol. 2010, 144, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Bely, M.; Stoeckle, P.; Masneuf-Pomarède, I.; Dubourdieu, D. Impact of mixed Torulaspora delbrueckii-Saccharomyces cerevisiae culture on high-sugar fermentation. Int. J. Food Microbiol. 2008, 122, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Rainieri, S.; Pretorius, I.S. Selection and improvement of wine yeasts. Ann. Microbiol. 2000, 50, 15–32. [Google Scholar]
- Henschke, P.A. Wine yeast. In Yeast Sugar Metabolism; Zimmermann, F.K., Entian, K.D., Eds.; Technomic Publishing Co Inc.: Lancaster, PA, USA, 1997; pp. 527–560. [Google Scholar]
- Salmon, J.M.; Vincent, O.; Mauricio, J.C.; Bely, M.; Barre, P. Sugar transport inhibition and apparent loss of activity in Saccharomyces cerevisiae as a major limiting factor of enological fermentations. Am. J. Enol. Vitic. 1993, 44, 56–64. [Google Scholar]
- Blomberg, A.; Adler, L. Physiology of osmotolerance in fungi. Adv. Microb. Physiol. 1992, 33, 145–212. [Google Scholar] [PubMed]
- Romano, P. Lievito starter e qualità aromatica del vino. Inf. Agric. 2006, 62, 27–31. [Google Scholar]
- Csoma, H.; Zakany, N.; Capece, A.; Romano, P.; Sipiczki, M. Biological diversity of Saccharomyces yeasts of spontaneously fermenting wines in four wine regions: Comparative genotypic and phenotypic analysis. Int. J. Food Microbiol. 2010, 140, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Romano, P.; Fiore, C.; Paraggio, M.; Caruso, M.; Capece, A. Function of yeast species and strains in wine flavour. Int. J. Food Microbiol. 2003, 86, 169–180. [Google Scholar] [CrossRef]
- Manzanares, P.; Ramón, D.; Querol, A. Screening of non-Saccharomyces wine yeasts for the production of b-D-xylosidase activity. Int. J. Food Microbiol. 1999, 46, 105–112. [Google Scholar] [CrossRef]
- Beltran, G.; Torija, M.J.; Novo, M.; Ferrer, N.; Poblet, M.; Guillamón, J.M.; Rozès, N.; Mas, A. Analysis of yeast populations during alcoholic fermentation: A six year follow-up study. Syst. Appl. Microbiol. 2002, 25, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Cocolin, L.; Bisson, L.F.; Mills, D.A. Direct profiling of the yeast dynamics in wine fermentations. FEMS Microbiol. Lett. 2000, 189, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Santamaría, P.; Garijo, P.; López, R.; Tenorio, C.; Rosa Gutiérrez, A. Analysis of yeast population during spontaneous alcoholic fermentation: Effect of the age of the cellar and the practice of inoculation. Int. J. Food Microbiol. 2005, 103, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Borrull, A.; Poblet, M.; Rozès, N. New insights into the capacity of commercial wine yeasts to grow on sparkling wine media. Factor screening for improving wine yeast selection. Food Microbiol. 2015, 48, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Kemp, B.; Alexandre, H.; Robillard, B.; Marchal, R. Effect of production phase on bottle-fermented sparkling wine quality. J. Agric. Food Chem. 2015, 63, 19–38. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.R.; Goyashiki, R.; Ramakrishnan, V.; Karpel, J.E.; Bisson, L.F. Genes required for ethanol tolerance and utilization in Saccharomyces cerevisiae. Am. J. Enol. Vitic. 2008, 59, 401–411. [Google Scholar]
- Bisson, L.F.; Block, D.E. Ethanol tolerance in Saccharomyces. In Biodiversity and Biotechnology of Wine Yeasts; Ciani, M., Ed.; Research Signpost: Kerala, India, 2002. [Google Scholar]
- Martí-Raga, M.; Martín, V.; Gil, M.; Sancho, M.; Zamora, F.; Mas, A.; Beltran, G. Contribution of yeast and base wine supplementation to sparkling wine composition. J. Sci. Food Agric. 2016. [Google Scholar] [CrossRef] [PubMed]
- Laurent, M.; Valade, M. La propagation des levains de tirage. Vign. Champen. 1998, 3, 29–52. [Google Scholar]
- Monk, P.R.; Storer, R.J. The kinetics of yeast growth and sugar utilization in tirage: The influence of different methods of starter culture preparation and inoculation levels. Am. J. Enol. Vitic. 1986, 37, 72–76. [Google Scholar]
- Benucci, I.; Liburdi, K.; Cerreti, M.; Esti, M. Characterization of active dry wine yeast during starter culture (Pied de cuve). Preparation for sparkling wine production. J. Food Sci. 2016, 81, 2015–2020. [Google Scholar] [CrossRef] [PubMed]
- Pampulha, M.E.; Loureiro-Dias, M.C. Activity of glycolytic enzymes of Saccharomyces cerevisiae in the presence of acetic acid. Appl. Microbiol. Biotechnol. 1989, 34, 375–380. [Google Scholar] [CrossRef]
- Martínez-Rodríguez, A.J.; Polo, M.C. Characterization of the Nitrogen Compounds Released during Yeast Autolysis in a Model Wine System. J. Agric. Food Chem. 2000, 48, 1081–1085. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, C.; Feuillat, M. Yeast autolysis. In Wine Microbiology and Biotechnology; Fleet, G.H., Ed.; Harwood Academic Publishers: Chur, Switzerland, 1993; pp. 225–242. [Google Scholar]
- Martínez-Rodríguez, A.J.; Polo, M.C.; Carrascosa, A.V. Structural and ultrastructural changes in yeast cells during autolysis in a model wine system and in sparkling wines. Int. J. Food Microbiol. 2001, 71, 45–51. [Google Scholar] [CrossRef]
- Martínez-Rodriguez, A.J.; Carrascosa, A.V.; Polo, M.C. Release of nitrogen compounds to the extracellular medium by three strains of Saccharomyces cerevisiae during induced autolysis in a model wine system. Int. J. Food Microbiol. 2001, 68, 155–160. [Google Scholar] [CrossRef]
- Molnar, I.; Oura, E.; Suomalainen, H. Study of volatile substrates produced during the autolysis of champagne yeast. Acta Aliment. 1981, 10, 27–36. [Google Scholar]
- Torresi, S.; Frangipane, M.T.; Garzillo, A.M.V.; Massantini, R.; Contini, M. Effects of a β-glucanase enzymatic preparation on yeast lysis during aging of traditional sparkling wines. Food Res. Int. 2014, 55, 83–92. [Google Scholar] [CrossRef]
- Moreno-Arribas, M.V.; Polo, M.C. Amino Acids and Biogenic Amines. In Wine Chemistry and Biochemistry; Moreno-Arribas, M.V., Polo, M.C., Eds.; Springer: New York, NY, USA, 2009; pp. 163–189. [Google Scholar]
- Todd, B.E.N.; Fleet, G.H.; Henschke, P.A. Promotion of autolysis through the interaction of killer and sensitive yeasts: Potential application in sparkling wine production. Am. J. Enol. Vitic. 2000, 51, 65–72. [Google Scholar]
- Jolly, J.; Augustyn, O.P.H.; Pretorius, I.S. The role and use of non-Saccharomyces yeasts in wine production. S. Afr. J. Enol. Vitic. 2006, 27, 15–39. [Google Scholar]
- Suárez-Lepe, J.A.; Morata, A. New trends in yeast selection for winemaking. Trends Food Sci. Technol. 2012, 23, 39–50. [Google Scholar] [CrossRef]
- Benito, S.; Gálvez, L.; Palomero, F.; Calderón, F.; Morata, A.; Palmero, D.; Suárez-Lepe, J.A. Schizosaccharomyces selective differential media. Afr. J. Microbiol. Res. 2013, 7, 3026–3036. [Google Scholar]
- Lasik, M. The application of malolactic fermentation process to create good-quality grape wine produced in cool-climate countries: A review. Eur. Food Res. Technol. 2013, 237, 843–850. [Google Scholar] [CrossRef]
- Bartowsky, E.J.; Borneman, A.R. Genomic variations of Oenococcus oeni strains and the potential to impact on malolactic fermentation and aroma compounds in wine. Appl. Microbiol. Biotechnol. 2011, 92, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Auge, D.; Valade, M.; Moncomble, D. Acidity of Champagne wines: Use of malolactic fermentation? Vign. Champen. Epernay 2000, 121, 44–56. [Google Scholar]
- Girbau-Solà, T.; López-Barajas, M.; López-Tamames, E.; Buxaderas, S. Foam aptitude of Trepat and Monastrell red varieties in cava elaboration. 2. Second fermentation and aging. J. Agric. Food Chem. 2002, 50, 5600–5604. [Google Scholar] [CrossRef] [PubMed]
- Girbau-Solà, T.; López-Tamames, E.; Buján, J.; Buxaderas, S. Foam aptitude of Trepat and Monastrell red varieties in cava elaboration. 1. Base wine characteristics. J. Agric. Food Chem. 2002, 50, 5596–5599. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Barajas, M.; Lopez-Tamames, E.; Buxaderas, S.; de la Torre-Boronat, M.C. Effect of vinification and variety on foam capacity of wine. Am. J. Enol. Vitic. 1998, 49, 397–402. [Google Scholar]
- Pozo-Bayón, M.Á.; Monagas, M.; Bartolomé, B.; Moreno-Arribas, M.V. Wine features related to safety and consumer health: An integrated perspective. Crit. Rev. Food Sci. Nutr. 2012, 52, 31–54. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, I.M.; Pinho, O. Biogenic amines in Portuguese traditional foods and wines. J. Food Prot. 2006, 69, 2293–2303. [Google Scholar] [CrossRef] [PubMed]
- Ancín-Azpilicueta, C.; González-Marco, A.; Jiménez-Moreno, N. Current knowledge about the presence of amines in wine. Crit. Rev. Food Sci. Nutr. 2008, 48, 257–275. [Google Scholar] [CrossRef] [PubMed]
- Maynard, L.S.; Schenker, V.J. Monoamine-oxidase inhibition by ethanol in vitro. Nature 1996, 196, 575. [Google Scholar] [CrossRef]
- Coton, M.; Romano, A.; Spano, G.; Ziegler, K.; Vetrana, C.; Desmarais, C.; Lonvaud-Funel, A.; Lucas, P.; Coton, E. Occurrence of biogenic amine-forming lactic acid bacteria in wine and cider. Food Microbiol. 2010, 27, 1078–1085. [Google Scholar] [CrossRef] [PubMed]
- Landete, J.M.; de Las Rivas, B.; Marcobal, A.; Muñoz, R. Molecular methods for the detection of biogenic amine-producing bacteria on foods. Int. J. Food Microbiol. 2007, 117, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Spano, G.; Russo, P.; Lonvaud-Funel, A.; Lucas, P.; Alexandre, H.; Grandvalet, C.; Coton, E.; Coton, M.; Barnavon, L.; Bach, B.; et al. Biogenic amines in fermented foods. Eur. J. Clin. Nutr. 2010, 64, S95–S100. [Google Scholar] [CrossRef] [PubMed]
- Beneduce, L.; Romano, A.; Capozzi, V.; Lucas, P.; Barnavon, L.; Bach, B.; Vuchot, P.; Grieco, F.; Spano, G. Biogenic amine in wines. Ann. Microbiol. 2010, 60, 573–578. [Google Scholar] [CrossRef]
- Landete, J.M.; Ferrer, S.; Polo, L.; Pardo, I. Biogenic Amines in Wines from Three Spanish Regions. J. Agric. Food Chem. 2005, 53, 1119–1124. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.; Moses, S.G.; Pretorius, I.S.; Cordero Otero, R.R. The Thr505 and Ser557 residues of the AGT1-encoded alpha-glucoside transporter are critical for maltotriose transport in Saccharomyces cerevisiae. J. Appl. Microbiol. 2008, 104, 1103–1111. [Google Scholar] [CrossRef] [PubMed]
- Marcobal, A.; Martín-Alvarez, P.J.; Polo, M.C.; Muñoz, R.; Moreno-Arribas, M.V. Formation of biogenic amines throughout the industrial manufacture of red wine. J. Food Prot. 2006, 69, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Torrea, D.; Ancín, C. Content of biogenic amines in a Chardonnay wine obtained through spontaneous and inoculated fermentations. J. Agric. Food Chem. 2002, 50, 4895–4899. [Google Scholar] [CrossRef] [PubMed]
- Caruso, M.; Capece, A.; Salzano, G.; Romano, P. Typing of Saccharomyces cerevisiae and Kloeckera apiculata strains from Aglianico wine. Lett. Appl. Microbiol. 2002, 34, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Granchi, L.; Romano, P.; Mangani, S.; Guerrini, S.; Vincenzini, M. Production of biogenic amines by wine microorganisms. Bull. l’OIV Off. Int. Vigne Vin 2005, 78, 595–610. [Google Scholar]
- Delage, N.; d’Harlingue, A.; Colonna Ceccaldi, B.; Bompeix, G. Occurrence of mycotoxins in fruit juices and wine. Food Control 2003, 14, 225–227. [Google Scholar] [CrossRef]
- Esti, M.; Benucci, I.; Liburdi, K.; Acciaro, G. Monitoring of ochratoxin A fate during alcoholic fermentation of wine-must. Food Control 2012, 27, 53–56. [Google Scholar] [CrossRef]
- Anli, E.; Bayram, M. Ochratoxin A in Wines. Food Rev. Int. 2009, 25, 214–232. [Google Scholar] [CrossRef]
- Angioni, A.; Caboni, P.; Garau, A.; Farris, A.; Orro, D.; Budroni, M.; Cabras, P. In vitro interaction between ochratoxin A and different strains of Saccharomyces cerevisiae and Kloeckera apiculata. J. Agric. Food Chem. 2007, 55, 2043–2048. [Google Scholar] [CrossRef] [PubMed]
- Caridi, A. Enological functions of parietal yeast mannoproteins. Antonie Van Leeuwenhoek 2006, 89, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Caridi, A.; Cufari, J.A.; Ramondino, D. Isolation and clonal pre-selection of enological Saccharomyces. J. Gen. Appl. Microbiol. 2002, 48, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Csutorás, C.; Rácz, L.; Rácz, K.; Fűtő, P.; Forgó, P.; Kiss, A. Monitoring of ochratoxin A during the fermentation of different wines by applying high toxin concentrations. Microchem. J. 2013, 107, 182–184. [Google Scholar] [CrossRef]
- Meca, G.; Blaiotta, G.; Ritieni, A. Reduction of ochratoxin A during the fermentation of Italian red wine Moscato. Food Control 2010, 21, 579–583. [Google Scholar] [CrossRef]
- Piotrowska, M.; Nowak, A.; Czyzowska, A. Removal of ochratoxin A by wine Saccharomyces cerevisiae strains. Eur. Food Res. Technol. 2013, 236, 441–447. [Google Scholar] [CrossRef]
- Petruzzi, L.; Bevilacqua, A.; Corbo, M.R.; Garofalo, C.; Baiano, A.; Sinigaglia, M. Selection of autochthonous Saccharomyces cerevisiae strains as wine starters using a polyphasic approach and ochratoxin A Removal. J. Food Prot. 2014, 77, 1168–1177. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; Capozzi, V.; Spano, G.; Corbo, M.R.; Sinigaglia, M.; Bevilacqua, A. Metabolites of microbial origin with an impact on health: Ochratoxin A and biogenic amines. Front. Microbiol. 2016, 7, 482. [Google Scholar] [CrossRef] [PubMed]
- Penacho, V.; Valero, E.; Gonzalez, R. Transcription profiling of sparkling wine second fermentation. Int. J. Food Microbiol. 2012, 153, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Tini, V.; Zambonelli, C.; Benevelli, M.; Castellari, L. The autolysogenic Saccharomyces cerevisiae strains for the sparkling wines production. Ind. Bevande 1995, 24, 113–118. [Google Scholar]
- Peppler, H.J. Yeast extracts. In Fermented Foods. Economic Microbiology; Rose, A.H., Ed.; Academic Press: London, UK, 1982; Volume 7, pp. 293–311. [Google Scholar]
- Romano, P.; Soli, M.G.; Suzzi, G.; Grazia, L.; Zambonelli, C. Improvement of a Wine Saccharomyces cerevisiae Strain by a Breeding Program. Appl. Environ. Microbiol. 1985, 50, 1064–1067. [Google Scholar] [PubMed]
- Giudici, P.; Solieri, L.; Pulvirenti, A.M.; Cassanelli, S. Strategies and perspectives for genetic improvement of wine yeasts. Appl. Microbiol. Biotechnol. 2005, 66, 622–628. [Google Scholar] [CrossRef] [PubMed]
- Coloretti, F.; Zambonelli, C.; Tini, V. Characterization of flocculent Saccharomyces interspecific hybrids for the production of sparkling wines. Food Microbiol. 2006, 23, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Winge, O.; Lausten, O. Artificial species hybridisation in yeast. Comp. Rend. Trav. Lab. Carslberg. Sér. Physiol. 1938, 22, 235–244. [Google Scholar]
- Castellari, M.; Ferruzzi, A.; Magrini, P.; Giudici, P.; Passarelli, C. Zambonelli Unbalanced wine fermentation by cryotolerant vs. non-cryotolerant. Saccharomyces strains. Vitis 1994, 33, 49–52. [Google Scholar]
- Massoutier, C.; Alexandre, H.; Feuillat, M.; Charpentier, C. Isolation and characterization of cryotolerant Saccharomyces strains. Vitis 1998, 37, 55–59. [Google Scholar]
- Suzzi, G.; Romano, P.; Zambonelli, C. Flocculation of wine yeasts: Frequency, differences, and stability of the character. Can. J. Microbiol. 1984, 30, 36–39. [Google Scholar] [CrossRef]

| Molecular Method | Reference |
|---|---|
| Random amplified polymorphic DNA (RAPD) PCR | [39] |
| Interdelta sequences analysis | [40] |
| Pulse field electrophoresis (PFGE) electrophoretic karyotypes | [38] |
| Mitochondrial DNA (mtDNA) restriction analysis | [33,34,36,41] |
| Polymorphic microsatellite loci (SSRs, simple sequence repeats) | [40,42] |
| Multilocus sequence typing (MLST) | [43,44] |
| Technological and Qualitative Characteristics | Reference |
|---|---|
| Resistance to low pH, sugars, ethanol, and sulphur dioxide contents | [26,46,51,55,56,57] |
| Low volatile acidity production | [52,58,59,60] |
| Low production of sulphur compounds (H2S, SO2) | [26,46,47,51,52,53,61] |
| Fermentation vigour | [51] |
| Desired enzymatic activities (e.g., β-glucosidase, β-xylosidase, protease, polygalacturonase, pectinase, glucanase, xylanase, and decarboxylase activities) | [46,47,57,62,63] |
| Desired fermentation-associated metabolites (glycerol, succinic acid, acetic acid, acetaldehyde, n-propanol, iso-butanol, isoamyl alcohol, and β-phenylethanol) | [46,47,57,62,63,64] |
| Implantation aptitude | [65,66,67] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garofalo, C.; Arena, M.P.; Laddomada, B.; Cappello, M.S.; Bleve, G.; Grieco, F.; Beneduce, L.; Berbegal, C.; Spano, G.; Capozzi, V. Starter Cultures for Sparkling Wine. Fermentation 2016, 2, 21. https://doi.org/10.3390/fermentation2040021
Garofalo C, Arena MP, Laddomada B, Cappello MS, Bleve G, Grieco F, Beneduce L, Berbegal C, Spano G, Capozzi V. Starter Cultures for Sparkling Wine. Fermentation. 2016; 2(4):21. https://doi.org/10.3390/fermentation2040021
Chicago/Turabian StyleGarofalo, Carmela, Mattia Pia Arena, Barbara Laddomada, Maria Stella Cappello, Gianluca Bleve, Francesco Grieco, Luciano Beneduce, Carmen Berbegal, Giuseppe Spano, and Vittorio Capozzi. 2016. "Starter Cultures for Sparkling Wine" Fermentation 2, no. 4: 21. https://doi.org/10.3390/fermentation2040021
APA StyleGarofalo, C., Arena, M. P., Laddomada, B., Cappello, M. S., Bleve, G., Grieco, F., Beneduce, L., Berbegal, C., Spano, G., & Capozzi, V. (2016). Starter Cultures for Sparkling Wine. Fermentation, 2(4), 21. https://doi.org/10.3390/fermentation2040021