The Production Optimization of a Thermostable Phytase from Bacillus subtilis SP11 Utilizing Mustard Meal as a Substrate
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Isolation of Phytase-Producing Bacteria
2.3. Quantitative Screening of Phytase-Producing Bacteria
2.4. Phytase Assay
2.5. Identification of SP11 Isolate
2.6. Detection of Full-Length Gene Encoding Phytase
2.7. Single-Factor Analysis
2.8. Plackett–Burman Design (PBD)
2.9. Central Composite Design (CCD)
2.10. Characterization of Crude Phytase
2.11. In Vitro Dephytinization
2.12. Statistical Analysis
3. Results
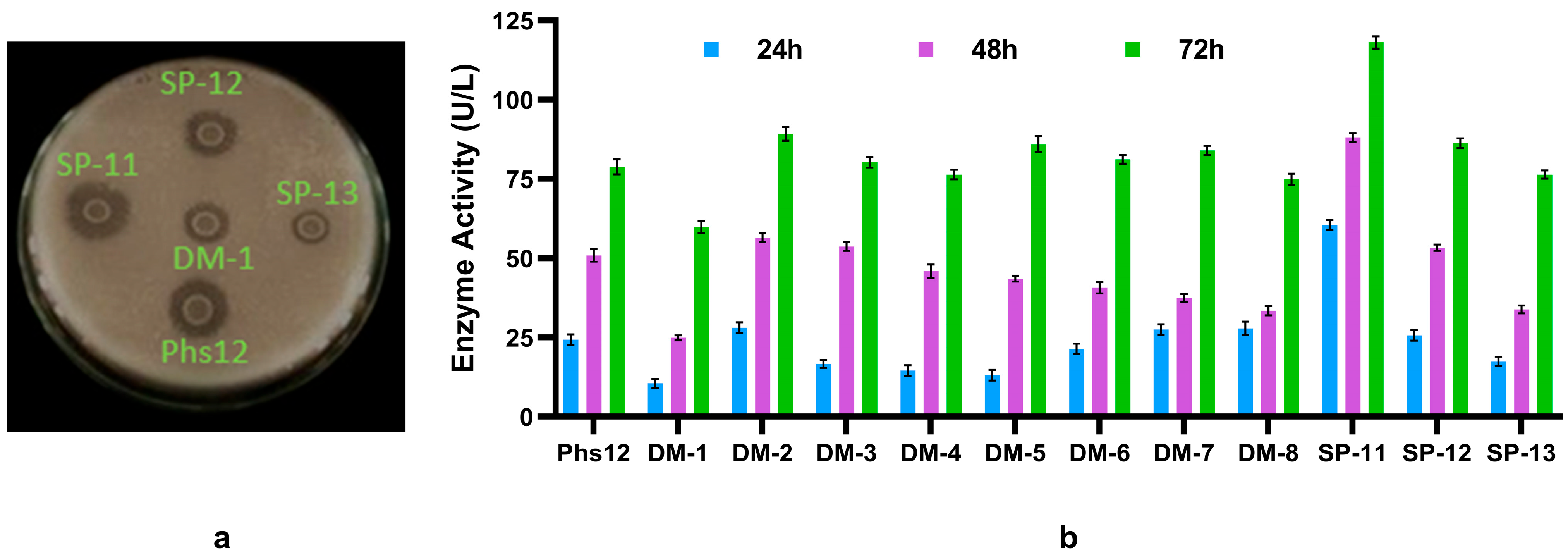
3.1. Isolation and Quantitative Screening of Phytase-Producing Bacteria
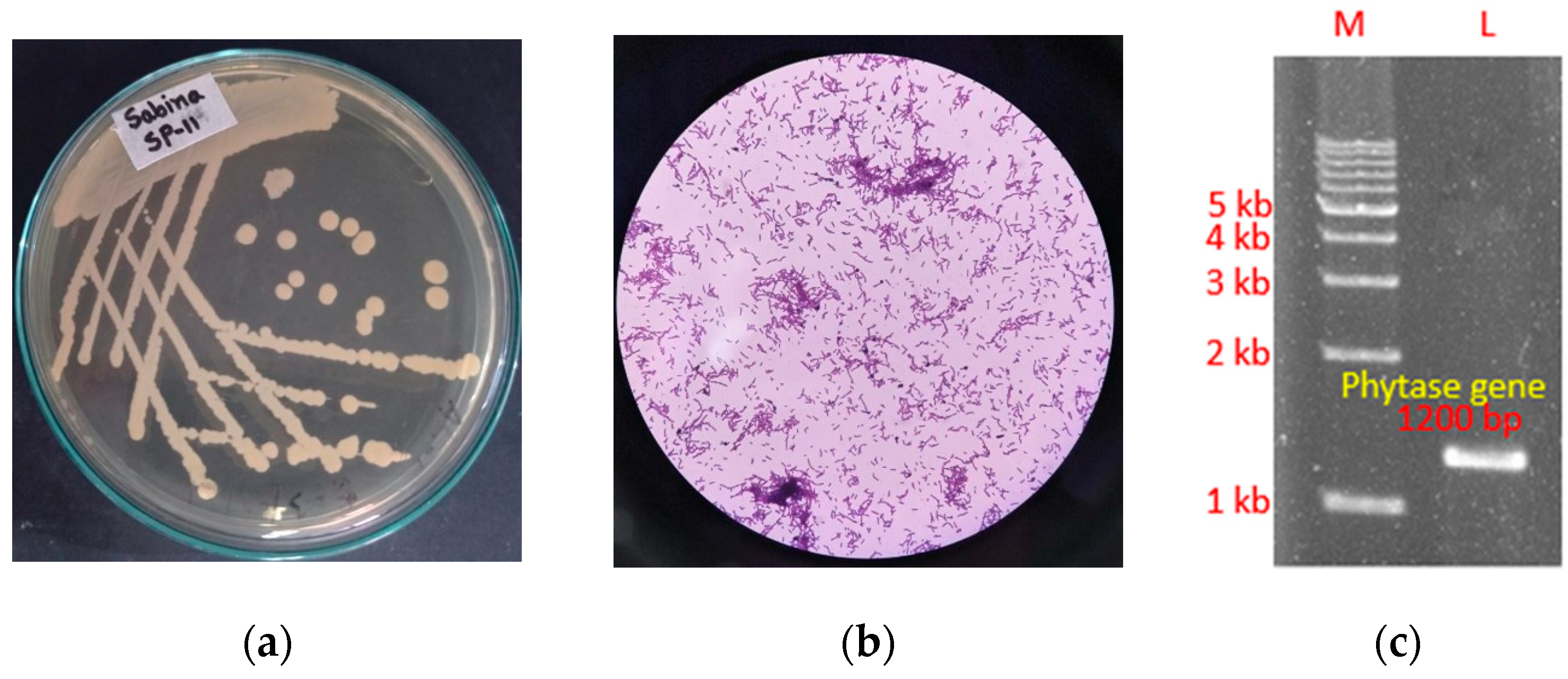
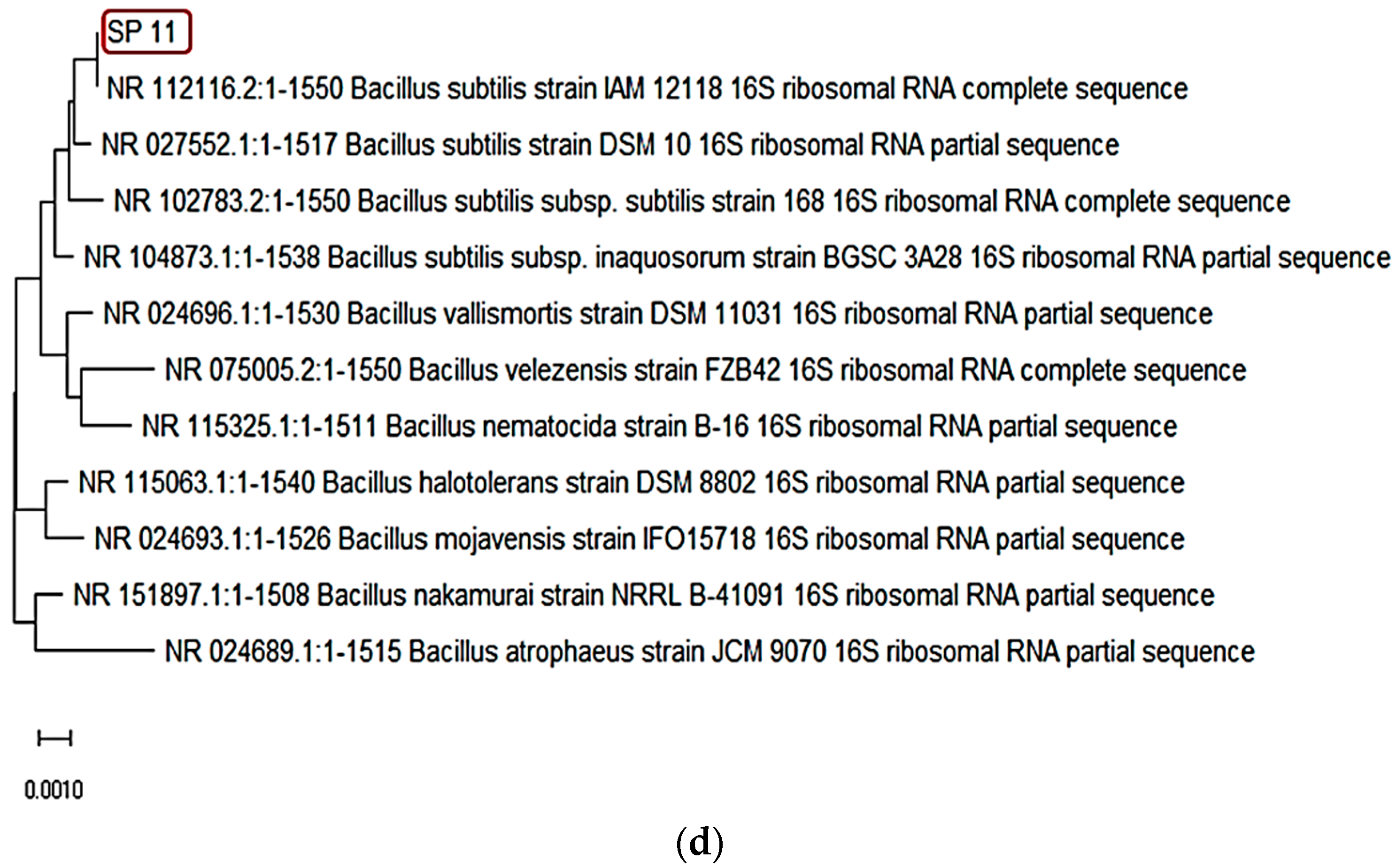
3.2. Identification of the SP11 Isolate and Detection of Full-Length Gene Encoding Phytase
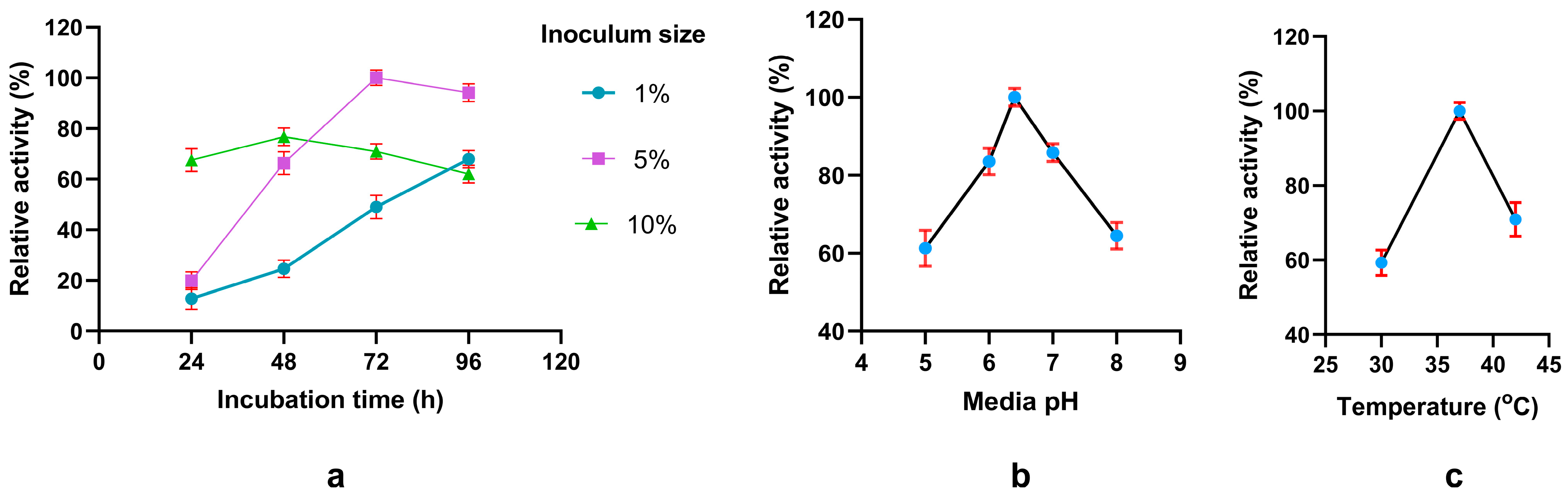
3.3. Single-Factor Analysis
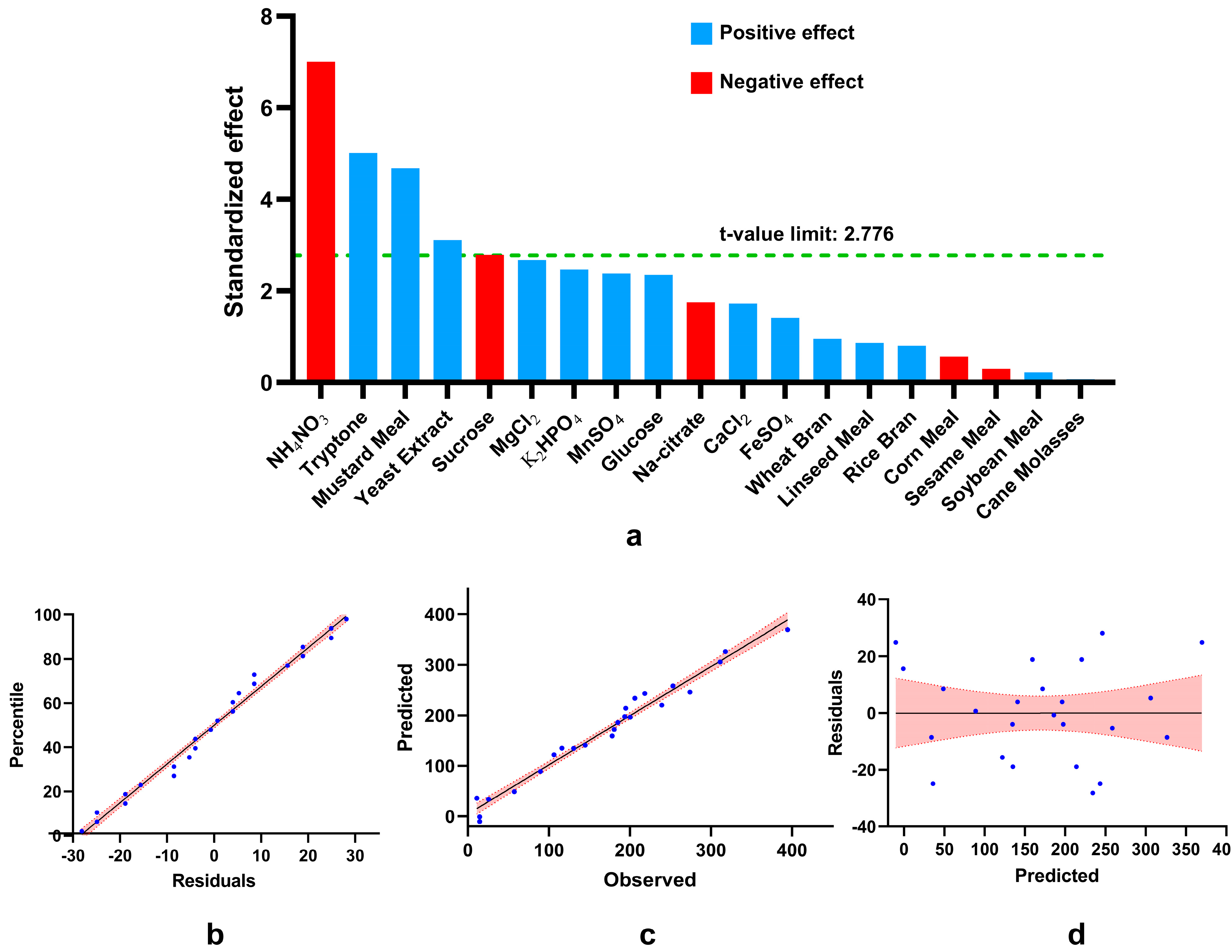
3.4. Determining Significant Variable for Phytase Production Using Plackett–Burman Design
+ 7.01 X7 + 0.57 X8 + 19.14 X9 − 57.08 X10 + 40.87 X11 + 25.40 X12 + 20.15 X13 −
22.77 X14 + 14.04 X15 + 21.80 X16 + 11.50 X17 + 19.41 X18 − 14.24 X19
3.5. Evaluation of the Model’s Fitness
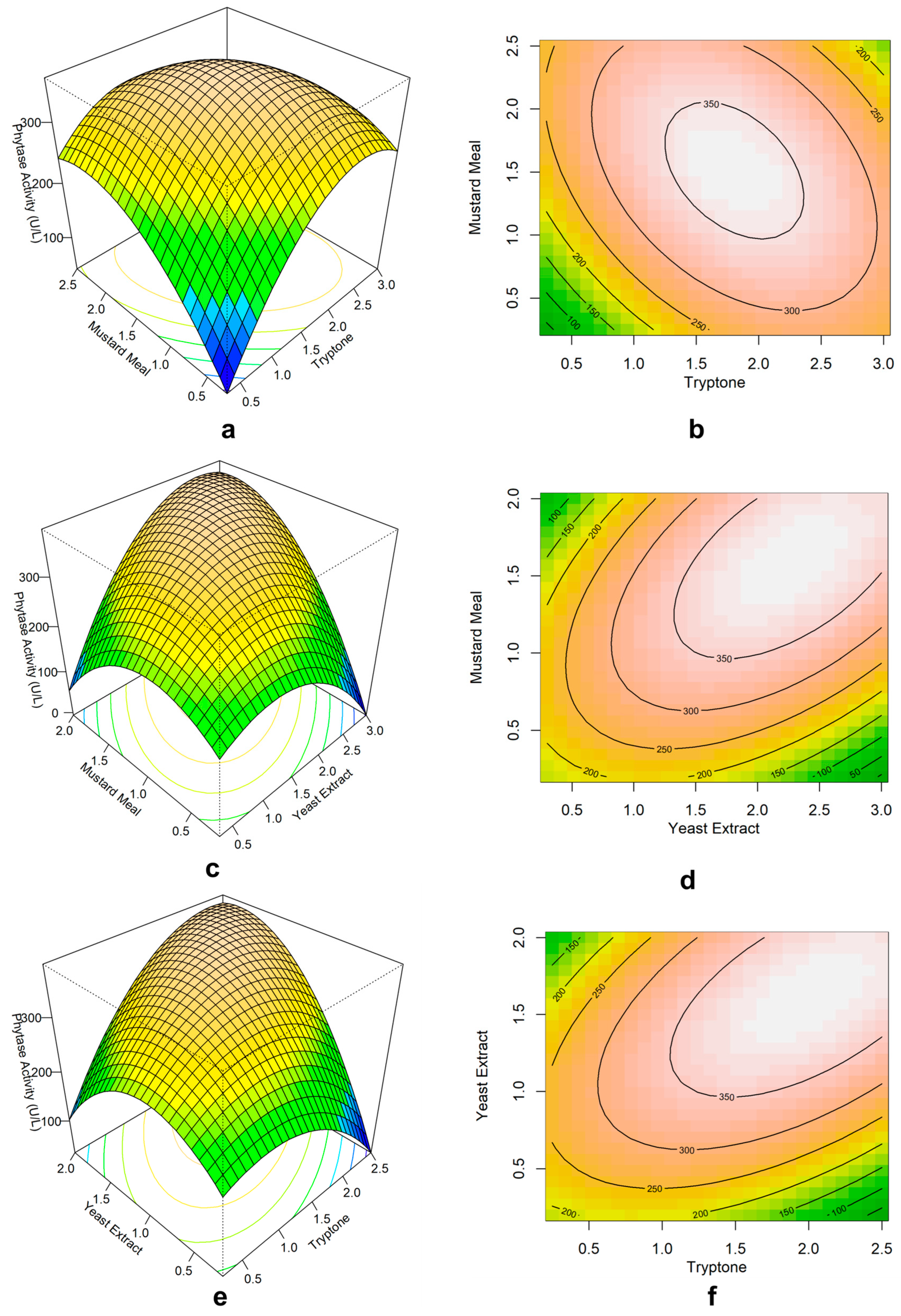
3.6. Central Composite Design to Determine the Optimum Amount
3.7. Statistical Analysis of the Central Composite Design
3.8. Response Surface Plots and Contour Plots
3.9. Verification of the Model
3.10. Time Course Profile
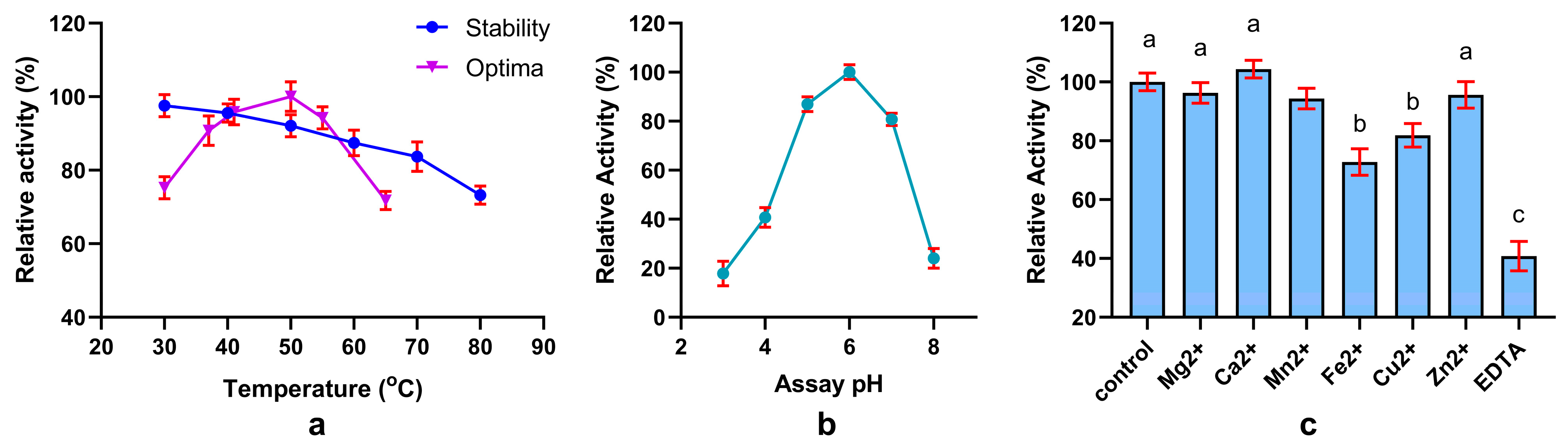
3.11. Characterization of Crude Phytase
3.12. In Vitro Dephytinization of Wheat Bran
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Haefner, S.; Knietsch, A.; Scholten, E.; Braun, J.; Lohscheidt, M.; Zelder, O. Biotechnological Production and Applications of Phytases. Appl. Microbiol. Biotechnol. 2005, 68, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Greiner, R.; Egli, I. Determination of the Activity of Acidic Phytate-Degrading Enzymes in Cereal Seeds. J. Agric. Food Chem. 2003, 51, 847–850. [Google Scholar] [CrossRef] [PubMed]
- Charles Bai, S.; Hardy, R.W.; Hamidoghli, A. Diet Analysis and Evaluation. In Fish Nutrition; Hardy, R.W., Kaushik, S.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 709–743. [Google Scholar]
- Kumar, A.; Singh, B.; Raigond, P.; Sahu, C.; Mishra, U.N.; Sharma, S.; Lal, M.K. Phytic Acid: Blessing in Disguise, a Prime Compound Required for Both Plant and Human Nutrition. Food Res. Int. 2021, 142, 110193. [Google Scholar] [CrossRef] [PubMed]
- Dersjant-Li, Y.; Awati, A.; Schulze, H.; Partridge, G. Phytase in Non-ruminant Animal Nutrition: A Critical Review on Phytase Activities in the Gastrointestinal Tract and Influencing Factors. J. Sci. Food Agric. 2015, 95, 878–896. [Google Scholar] [CrossRef]
- Joudaki, H.; Aria, N.; Moravej, R.; Rezaei Yazdi, M.; Emami-Karvani, Z.; Hamblin, M.R. Microbial Phytases: Properties and Applications in the Food Industry. Curr. Microbiol. 2023, 80, 374. [Google Scholar] [CrossRef]
- Valente Junior, D.T.; Genova, J.L.; Kim, S.W.; Saraiva, A.; Rocha, G.C. Carbohydrases and Phytase in Poultry and Pig Nutrition: A Review beyond the Nutrients and Energy Matrix. Animals 2024, 14, 226. [Google Scholar] [CrossRef]
- Mullaney, E.J.; Ullah, A.H.J. The Term Phytase Comprises Several Different Classes of Enzymes. Biochem. Biophys. Res. Commun. 2003, 312, 179–184. [Google Scholar] [CrossRef]
- Mullaney, E.J.; Daly, C.B.; Ullah, A.H.J. Advances in Phytase Research; Elsevier: Amsterdam, The Netherlands, 2000; pp. 157–199. [Google Scholar]
- Balaban, N.P.; Suleimanova, A.D.; Shakirov, E.V.; Sharipova, M.R. Histidine Acid Phytases of Microbial Origin. Microbiology 2018, 87, 745–756. [Google Scholar] [CrossRef]
- Kumar, V.; Yadav, A.N.; Verma, P.; Sangwan, P.; Saxena, A.; Kumar, K.; Singh, B. β-Propeller Phytases: Diversity, Catalytic Attributes, Current Developments and Potential Biotechnological Applications. Int. J. Biol. Macromol. 2017, 98, 595–609. [Google Scholar] [CrossRef]
- Hegeman, C.E.; Grabau, E.A. A Novel Phytase with Sequence Similarity to Purple Acid Phosphatases Is Expressed in Cotyledons of Germinating Soybean Seedlings. Plant Physiol. 2001, 126, 1598–1608. [Google Scholar] [CrossRef]
- Puhl, A.A.; Gruninger, R.J.; Greiner, R.; Janzen, T.W.; Mosimann, S.C.; Selinger, L.B. Kinetic and Structural Analysis of a Bacterial Protein Tyrosine Phosphatase-like Myo -inositol Polyphosphatase. Protein Sci. 2007, 16, 1368–1378. [Google Scholar] [CrossRef] [PubMed]
- Konietzny, U.; Greiner, R. Molecular and Catalytic Properties of Phytate-degrading Enzymes (Phytases). Int. J. Food Sci. Technol. 2002, 37, 791–812. [Google Scholar] [CrossRef]
- Chen, C.; Cheng, K.; Ko, T.; Guo, R. Current Progresses in Phytase Research: Three-Dimensional Structure and Protein Engineering. ChemBioEng Rev. 2015, 2, 76–86. [Google Scholar] [CrossRef]
- Kammoun, R.; Farhat, A.; Chouayekh, H.; Bouchaala, K.; Bejar, S. Phytase Production by Bacillus subtilis US417 in Submerged and Solid State Fermentations. Ann. Microbiol. 2012, 62, 155–164. [Google Scholar] [CrossRef]
- Animal Feed Phytase Market Size, Share & Trends Analysis Report by Form (Liquid, Granules or Powder), By Class (3-Phytase, 6-Phytase, Others), By Source (Plants, Microorganisms), By Livestock, By Region, And by Segment Forecasts, 2025–2034. Available online: https://www.insightaceanalytic.com/report/global-animal-feed-phytase-market/1460 (accessed on 28 May 2025).
- Phytase Imports In Bangladesh Market Size & Demand Based On Import Trade Data. Available online: https://www.volza.com/p/phytase/import/import-in-bangladesh/ (accessed on 28 May 2025).
- Howson, S.J.; Davis, R.P. Production of Phytate-Hydrolysing Enzyme by Some Fungi. Enzyme Microb. Technol. 1983, 5, 377–382. [Google Scholar] [CrossRef]
- Bae, H.D.; Yanke, L.J.; Cheng, K.-J.; Selinger, L.B. A Novel Staining Method for Detecting Phytase Activity. J. Microbiol. Methods 1999, 39, 17–22. [Google Scholar] [CrossRef]
- Li, G.-E.; Wu, X.-Q.; Ye, J.-R.; Hou, L.; Zhou, A.-D.; Zhao, L. Isolation and Identification of Phytate-Degrading Rhizobacteria with Activity of Improving Growth of Poplar and Masson Pine. World J. Microbiol. Biotechnol. 2013, 29, 2181–2193. [Google Scholar] [CrossRef]
- Bravo, A.; Sarabia, S.; Lopez, L.; Ontiveros, H.; Abarca, C.; Ortiz, A.; Ortiz, M.; Lina, L.; Villalobos, F.J.; Peña, G.; et al. Characterization of Cry Genes in a Mexican Bacillus Thuringiensis Strain Collection. Appl. Environ. Microbiol. 1998, 64, 4965–4972. [Google Scholar] [CrossRef]
- Suresh, S.; Radha, K. Effect of a Mixed Substrate on Phytase Production by Rhizopus oligosporus MTCC 556 Using Solid State Fermentation and Determination of Dephytinization Activities in Food Grains. Food Sci. Biotechnol. 2015, 24, 551–559. [Google Scholar] [CrossRef]
- Mussa, L.A.; Yadetie, D.M.; Temesgen, E.A.; Tefera, A.T.; Gemeda, M.T. Isolation and In-Vitro Characterization of Extracellular Phytase Producing Bacterial Isolates for Potential Application in Poultry Feed. BMC Microbiol. 2023, 23, 296. [Google Scholar] [CrossRef]
- Suliasih, S.; Widawati, S.; Ikhwani, A.; Suyadi, S.; Sudiana, I.M. Phytase Activity of Phytase-Producing Bacteria Isolated from Mangrove Sediment. IOP Conf. Ser. Earth Environ. Sci. 2022, 976, 12041. [Google Scholar] [CrossRef]
- Sardar, R.; Asad, M.; Ahmad, M.; Ahmad, T. Optimization of Phytase Production by Bacillus Sp. (HCYL03) under Solid-State Fermentation by Using Box-Behnken Design. Braz. Arch. Biol. Technol. 2022, 65, e22210307. [Google Scholar] [CrossRef]
- Rocky-Salimi, K.; Hashemi, M.; Safari, M.; Mousivand, M. Valorisation of Untreated Cane Molasses for Enhanced Phytase Production by Bacillus subtilis K46b and its Potential Role in Dephytinisation. J. Sci. Food Agric. 2017, 97, 222–229. [Google Scholar] [CrossRef]
- Cheva-Isarakul, B.; Tangtaweewipat, S.; Sangsrijun, P.; Yamauchi, K. Chemical Composition and Metabolizable Energy of Mustard Meal. J. Poult. Sci. 2003, 40, 221–225. [Google Scholar] [CrossRef]
- Sarker, A.K.; Saha, D.; Begum, H.; Zaman, A.; Rahman, M.M. Comparison of Cake Compositions, Pepsin Digestibility and Amino Acids Concentration of Proteins Isolated from Black Mustard and Yellow Mustard Cakes. AMB Express 2015, 5, 22. [Google Scholar] [CrossRef] [PubMed]
- El-Toukhy, N.M.K.; Youssef, A.S.; Mikhail, M. Isolation, Purification and Characterization of Phytase from Bacillus subtilis MJA. Afr. J. Biotechnol. 2013, 12, 2957–2967. [Google Scholar]
- Choi, Y.M.; Suh, H.J.; Kim, J.M. Purification and Properties of Extracellular Phytase from Bacillus Sp. KHU-10. J. Protein Chem. 2001, 20, 287–292. [Google Scholar] [CrossRef]
- McCapes, R.H.; Ekperigin, H.E.; Cameron, W.J.; Ritchie, W.L.; Slagter, J.; Stangeland, V.; Nagaraja, K.V. Effect of a New Pelleting Process on the Level of Contamination of Poultry Mash by Escherichia coli and Salmonella. Avian Dis. 1989, 33, 103–111. [Google Scholar] [CrossRef]
- Kerovuo, J.; Lauraeus, M.; Nurminen, P.; Kalkkinen, N.; Apajalahti, J. Isolation, Characterization, Molecular Gene Cloning, and Sequencing of a Novel Phytase from Bacillus subtilis. Appl. Environ. Microbiol. 1998, 64, 2079–2085. [Google Scholar] [CrossRef]
- Troxell, B.; Petri, N.; Daron, C.; Pereira, R.; Mendoza, M.; Hassan, H.M.; Koci, M.D. Poultry Body Temperature Contributes to Invasion Control through Reduced Expression of Salmonella Pathogenicity Island 1 Genes in Salmonella enterica Serovars Typhimurium and Enteritidis. Appl. Environ. Microbiol. 2015, 81, 8192–8201. [Google Scholar] [CrossRef]
- Rocky-Salimi, K.; Hashemi, M.; Safari, M.; Mousivand, M. A Novel Phytase Characterized by Thermostability and High PH Tolerance from Rice Phyllosphere Isolated Bacillus subtilis B.S.46. J. Adv. Res. 2016, 7, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Kierończyk, B.; Rawski, M.; Długosz, J.; Swiatkiewicz, S.; Józefiak, D. Avian Crop Function—A Review. Ann. Anim. Sci. 2016, 16, 653–678. [Google Scholar] [CrossRef]
- Farner, D.S. The Hydrogen Ion Concentration in Avian Digestive Tracts. Poult. Sci. 1942, 21, 445–450. [Google Scholar] [CrossRef]
- Shin, S.; Ha, N.-C.; Oh, B.-C.; Oh, T.-K.; Oh, B.-H. Enzyme Mechanism and Catalytic Property of β Propeller Phytase. Structure 2001, 9, 851–858. [Google Scholar] [CrossRef]
- Zhao, T.; Yong, X.; Zhao, Z.; Dolce, V.; Li, Y.; Curcio, R. Research Status of Bacillus Phytase. 3 Biotech. 2021, 11, 415. [Google Scholar] [CrossRef]
- Jain, J.; Singh, B. Phytase Production and Development of an Ideal Dephytinization Process for Amelioration of Food Nutrition Using Microbial Phytases. Appl. Biochem. Biotechnol. 2017, 181, 1485–1495. [Google Scholar] [CrossRef]


| Code | Variable | Low Value (−1) (%w/v) | High Value (+1) (% w/v) |
|---|---|---|---|
| X1 | Rice Bran | 0 | 1 |
| X2 | Wheat Bran | 0 | 1 |
| X3 | Corn Meal | 0 | 1 |
| X4 | Soybean Meal | 0 | 1 |
| X5 | Sesame Meal | 0 | 1 |
| X6 | Mustard Meal | 0 | 1 |
| X7 | Linseed Meal | 0 | 1 |
| X8 | Cane Molasses | 0 | 2 |
| X9 | Glucose | 0.4% | 1 |
| X10 | NH4NO3 | 0.2% | 0.5 |
| X11 | Tryptone | 0 | 1 |
| X12 | Yeast Extract | 0 | 0.5 |
| X13 | K2HPO4 | 0.02% | 0.1 |
| X14 | Sucrose | 0 | 1 |
| X15 | CaCl2 | 0.002% | 0.02 |
| X16 | MgCl2 | 0.002% | 0.02 |
| X17 | FeSO4 | 0.002% | 0.02 |
| X18 | MnSO4 | 0.002% | 0.02 |
| X19 | Sodium Citrate | 0 | 1 |
| Run | A | B | C | D | E | F | G | H | J | K | L | M | N | O | P | Q | R | S | T | Phytase Activity (U/L) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed | Predicted | Residuals | ||||||||||||||||||||
| 1 | 89.59 | 88.868 | 0.7225 | |||||||||||||||||||
| 2 | 194.99 | 213.874 | −18.8842 | |||||||||||||||||||
| 3 | 180.54 | 171.998 | 8.5425 | |||||||||||||||||||
| 4 | 394.57 | 369.679 | 24.8908 | |||||||||||||||||||
| 5 | 185.13 | 185.852 | −0.7225 | |||||||||||||||||||
| 6 | 57.29 | 48.748 | 8.5425 | |||||||||||||||||||
| 7 | 253.13 | 258.414 | −5.2842 | |||||||||||||||||||
| 8 | 317.90 | 326.443 | −8.5425 | |||||||||||||||||||
| 9 | 193.63 | 197.611 | −3.9808 | |||||||||||||||||||
| 10 | 11.05 | 35.941 | −24.8908 | |||||||||||||||||||
| 11 | 218.45 | 243.341 | −24.8908 | |||||||||||||||||||
| 12 | 144.84 | 140.859 | 3.9808 | |||||||||||||||||||
| 13 | 14.62 | −1.006 | 15.6258 | |||||||||||||||||||
| 14 | 130.56 | 134.541 | −3.9808 | |||||||||||||||||||
| 15 | 106.25 | 121.876 | −15.6258 | |||||||||||||||||||
| 16 | 178.16 | 159.276 | 18.8842 | |||||||||||||||||||
| 17 | 25.50 | 34.043 | −8.5425 | |||||||||||||||||||
| 18 | 116.11 | 134.994 | −18.8842 | |||||||||||||||||||
| 19 | 239.36 | 220.476 | 18.8842 | |||||||||||||||||||
| 20 | 206.04 | 234.189 | −28.1492 | |||||||||||||||||||
| 21 | 274.21 | 246.061 | 28.1492 | |||||||||||||||||||
| 22 | 311.44 | 306.156 | 5.2842 | |||||||||||||||||||
| 23 | 14.62 | −10.271 | 24.8908 | |||||||||||||||||||
| 24 | 200.26 | 196.279 | 3.9808 | |||||||||||||||||||
| Term | Coefficients | t-Statistics | F-Value | p-Value | Contribution |
|---|---|---|---|---|---|
| Constant | 169.09 | 20.73 | 0.000 | ||
| Rice Bran | 6.56 | 0.8 | 0.65 | 0.466 | 0.42% |
| Wheat Bran | 7.73 | 0.95 | 0.9 | 0.397 | 0.59% |
| Corn Meal | −4.55 | −0.56 | 0.31 | 0.607 | 0.20% |
| Soybean Meal | 1.83 | 0.22 | 0.05 | 0.834 | 0.03% |
| Sesame Meal | −2.45 | −0.3 | 0.09 | 0.779 | 0.06% |
| Mustard Meal | 38.22 | 4.68 | 21.95 | 0.009 | 14.38% |
| Linseed Meal | 7.01 | 0.86 | 0.74 | 0.439 | 0.48% |
| Cane Molasses | 0.57 | 0.07 | 0 | 0.948 | 0.00% |
| Glucose | 19.14 | 2.35 | 5.5 | 0.079 | 3.61% |
| NH4NO3 | −57.08 | −7 | 48.94 | 0.002 | 32.07% |
| Tryptone | 40.87 | 5.01 | 25.1 | 0.007 | 16.45% |
| Yeast Extract | 25.4 | 3.11 | 9.69 | 0.036 | 6.35% |
| K2HPO4 | 20.15 | 2.47 | 6.1 | 0.069 | 4.00% |
| Sucrose | −22.77 | −2.79 | 7.79 | 0.049 | 5.10% |
| CaCl2 | 14.04 | 1.72 | 2.96 | 0.16 | 1.94% |
| MgCl2 | 21.8 | 2.67 | 7.14 | 0.056 | 4.68% |
| FeSO4 | 11.5 | 1.41 | 1.99 | 0.231 | 1.30% |
| MnSO4 | 19.41 | 2.38 | 5.66 | 0.076 | 3.71% |
| Na-citrate | −14.24 | −1.75 | 3.05 | 0.156 | 2.00% |
| Analysis of Variance (ANOVA) | |||||
| Source | DF | Adj SS | Adj MS | F-Value | p-Value |
| Model | 19 | 237,392 | 12,494 | 7.82 | 0.030 |
| Residuals | 4 | 8840 | 2210 | ||
| R2 = 97.38% | |||||
| Run | Mustard Meal (X6) | Tryptone (X11) | Yeast Extract (X12) | Phytase Activity (U/L) | ||
|---|---|---|---|---|---|---|
| Observed | Predicted | Residual | ||||
| 1 | 0 | 0 | 0 | 388.8 | 361.9 | 26.8 |
| 2 | 1 | −1 | −1 | 257.2 | 244.1 | 13.1 |
| 3 | 1 | 1 | 1 | 427.0 | 386.5 | 40.5 |
| 4 | −1 | −1 | −1 | 219.2 | 236.4 | −17.2 |
| 5 | 0 | 0 | 0 | 374.7 | 361.9 | 12.8 |
| 6 | 0 | 0 | 0 | 378.7 | 361.9 | 16.8 |
| 7 | −1 | 1 | 1 | 323.6 | 313.4 | 10.2 |
| 8 | 0 | 0 | −1.68 | 199.4 | 189.3 | 10.1 |
| 9 | −1 | 1 | −1 | 231.8 | 246.4 | −14.6 |
| 10 | 0 | 0 | 1.68 | 274.6 | 317.7 | −43.1 |
| 11 | 0 | 0 | 0 | 353.8 | 361.9 | −8.2 |
| 12 | 0 | 0 | 0 | 349.7 | 361.9 | −12.3 |
| 13 | 1.68 | 0 | 0 | 232.0 | 287.4 | −55.4 |
| 14 | −1 | −1 | 1 | 153.9 | 151.2 | 2.7 |
| 15 | 1 | −1 | 1 | 367.7 | 329.8 | 37.9 |
| 16 | 1 | 1 | −1 | 169.2 | 148.6 | 20.6 |
| 17 | 0 | 1.68 | 0 | 287.7 | 310.2 | −22.5 |
| 18 | −1.68 | 0 | 0 | 242.0 | 219.5 | 22.5 |
| 19 | 0 | 0 | 0 | 331.6 | 361.9 | −30.3 |
| 20 | 0 | −1.68 | 0 | 243.7 | 254.1 | −10.5 |
| Level | Mustard meal (% w/v) | Tryptone (% w/v) | Yeast extract (% w/v) | |||
| −α | 0.3 | 0.25 | 0.2 | |||
| −1 | 0.85 | 0.71 | 0.57 | |||
| 0 | 1.65 | 1.38 | 1.1 | |||
| +1 | 2.45 | 2.04 | 1.64 | |||
| +α | 3 | 2.5 | 2 | |||
| Term | Coefficient | t-Statistics | p-Value |
| Constant | 361.9 | 24.86 | 0.000 |
| Mustard Meal | 20.19 | 2.09 | 0.063 |
| Tryptone | 16.66 | 1.72 | 0.115 |
| Yeast Extract | 38.16 | 3.95 | 0.003 |
| Mustard Meal*Mustard Meal | −38.35 | −4.08 | 0.002 |
| Tryptone*Tryptone | −28.2 | −3 | 0.013 |
| Yeast Extract*Yeast Extract | −38.33 | −4.08 | 0.002 |
| Mustard Meal*Tryptone | −26.4 | −2.09 | 0.063 |
| Mustard Meal*Yeast Extract | 42.7 | 3.38 | 0.007 |
| Tryptone*Yeast Extract | 38.1 | 3.02 | 0.013 |
| Source | DF | F-value | p-value |
| Model | 9 | 9.26 | 0.001 |
| Linear | 3 | 7.65 | 0.006 |
| Square | 3 | 11.82 | 0.001 |
| 2-Way Interaction | 3 | 8.3 | 0.005 |
| Lack-of-Fit | 5 | 4.55 | 0.061 |
| R2 = 89.28% | |||
| Mustard Meal | Tryptone | Yeast Extract | Predicted | 95% Cl | Desirability | Observed | Accuracy |
|---|---|---|---|---|---|---|---|
| 2.21% w/v | 1.95% w/v | 1.85% w/v | 401.121 (U/L) | (337 U/L, 465.3 U/L) | 0.91 | 436 U/L | 92% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, M.A.M.; Akhter, S.; Arif, T.; Mian, M.M.; Mamun, M.A.A.; Karim, M.M.; Khan, S.N. The Production Optimization of a Thermostable Phytase from Bacillus subtilis SP11 Utilizing Mustard Meal as a Substrate. Fermentation 2025, 11, 452. https://doi.org/10.3390/fermentation11080452
Khan MAM, Akhter S, Arif T, Mian MM, Mamun MAA, Karim MM, Khan SN. The Production Optimization of a Thermostable Phytase from Bacillus subtilis SP11 Utilizing Mustard Meal as a Substrate. Fermentation. 2025; 11(8):452. https://doi.org/10.3390/fermentation11080452
Chicago/Turabian StyleKhan, Md. Al Muid, Sabina Akhter, Tanjil Arif, Md. Mahmuduzzaman Mian, Md. Arafat Al Mamun, Muhammad Manjurul Karim, and Shakila Nargis Khan. 2025. "The Production Optimization of a Thermostable Phytase from Bacillus subtilis SP11 Utilizing Mustard Meal as a Substrate" Fermentation 11, no. 8: 452. https://doi.org/10.3390/fermentation11080452
APA StyleKhan, M. A. M., Akhter, S., Arif, T., Mian, M. M., Mamun, M. A. A., Karim, M. M., & Khan, S. N. (2025). The Production Optimization of a Thermostable Phytase from Bacillus subtilis SP11 Utilizing Mustard Meal as a Substrate. Fermentation, 11(8), 452. https://doi.org/10.3390/fermentation11080452






