Abstract
The unique microbial communities present on fruit surfaces significantly influence the fermentation process and product quality of artisanal cider production, constituting a microbial terroir analogous to that recognized in viticulture. In this study, we investigated the microbial composition and diversity associated with the apple varietals (Empire, Golden Delicious, and Idared) cultivated by two different orchard producers in the Hudson River Valley of New York. Using 16S rRNA and ITS amplicon sequencing, we identified distinct bacterial and fungal communities that varied significantly according to the apple varietal and orchard location. Notably, the orchard was the dominant factor shaping both the bacterial and fungal communities on the apples’ surfaces, with the varietal differences also playing a significant, albeit secondary, role. For example, we found that the bacterial genera Acidophilim sp. and 1174-901-12 sp., as well as the fungus Sporobolmyces patagonicus, were important markers of the orchard in which the apples were cultivated. These microbial signatures persisted into the early stages of cider fermentation, suggesting their potential influence on the cider quality and flavor profile. Our findings underscore the critical importance of the microbial terroir in cider production, and suggest that targeted management practices can leverage regional microbial diversity to enhance the distinctiveness and marketability of artisanal cider products.
1. Introduction
The microbial communities inhabiting plant surfaces and tissues significantly influence plant health by facilitating nutrient cycling, enhancing disease resistance, and modulating stress responses [1,2]. These microbiomes also profoundly shape the sensory and biochemical attributes of agricultural products, underpinning the concept of terroir—the unique regional characteristics derived from local environmental factors, such as climate, soil composition, and management practices [3].
While the role of the microbial terroir has been extensively studied in viticulture, particularly in wine production [4,5,6], where the microbial composition directly influences the fermentation dynamics, sensory properties, and regional wine identity, its significance remains relatively underexplored in orchard fruit production [3,7,8]. This knowledge gap is noteworthy given the growing consumer interest in artisanal and regionally distinct agricultural products. Apples (Malus domestica), a culturally and economically significant fruit crop in the northeastern United States (US), represent an ideal system for investigating microbial terroirs, due to their extensive varietal diversity and central role in fermented products such as cider [9].
The Hudson Valley region of New York (NY) is a major contributor to the state’s status as the second-largest apple producer in the US. The region provides an exceptional context for studying the microbial terroirs of apples. This region is characterized by varied microclimates; distinct soil profiles; diverse orchard management practices; and a rich, long-standing tradition of apple cultivation [8,10]. Such environmental and agricultural diversity has likely contributed to the significant variability in the microbial communities associated with apple surfaces and internal tissues, thereby influencing both apple quality and subsequent fermentation processes.
Natural fermentation processes, such as cidermaking, rely substantially on the indigenous microbial populations present on and within apples, which significantly affect the flavor, aroma, and overall quality of the resulting cider. These microbial signatures can thus underpin distinct regional identities, enabling producers to market their ciders based on their unique, terroir-driven characteristics, enhancing their economic and cultural value [8,11]. However, few studies have empirically evaluated how orchard-specific factors—including apple varietal selection and producer management practices—impact the apple microbiome and its persistence throughout fermentation processes.
This study systematically examined the microbial community composition and diversity across different apple varietals cultivated in two distinct orchards in the Hudson Valley region of New York. Using high-throughput sequencing approaches targeting bacterial and fungal genetic markers, we investigated whether specific microbial signatures could be identified and consistently linked to apple varietals and orchard practices, thereby extending the concept of terroir to a microbiological dimension. Understanding these microbial dynamics could have significant implications for orchard management, consumer perceptions of apple quality, and the branding of apples as regional specialty products. Our findings highlight the complex interactions between cultivar selection, production practices, and environmental factors that collectively shape the microbial terroir of apples.
2. Materials and Methods
2.1. Field Sites and Sampling Processing
To investigate the effect of varietals and producers on cider apples’ microbiomes, we collected apples of the same three varietals grown in two different orchards located in the mid-Hudson River Valley of New York (NY). More specifically, three Empire apples, three Golden Delicious apples, and three Idared apples, all late-season varietals, were collected from both Montgomery Place Orchard, Annandale-On-Hudson, NY, on 31 October 2017 and 1 November 2017, and from Vosburgh Orchards, Elizaville, NY, on 2 November 2017. The apple varietals Empire, Golden Delicious, and Idared each possess distinct characteristics influencing their suitability for fresh consumption and cidermaking [12,13]. Empire apples, developed by crossing McIntosh and Red Delicious, are known for their sweet–tart flavor and crisp texture [14]. Golden Delicious apples offer a sweet, mellow flavor and tender flesh, appreciated both fresh and in blended ciders, where they contribute aromatic complexity and balanced sweetness [15]. Idared apples, developed from a Jonathan and Wagener cross, possess a tangy, tart flavor and firm texture, characteristics valued in cidermaking for producing ciders with vibrant acidity and structure [16]. The apples were sampled on a schedule that minimized environmental differences due to seasonality, such as temperature and rain. Similarly, at each orchard, three trees were randomly chosen within the southeast part of the orchard to minimize the impact of the prevailing winds and, thus, the surrounding landscape on the apples’ microbiomes. From each tree, three apples, all located on the east side of the tree, with one from the bottom, one from the middle, and one from the top section, were sampled using sterile swabs that were moistened in individual 15 mL falcon tubes prepped with a sterile 0.15 M saline solution and rubbed onto each apple while still attached to the tree [17]. The swabs were then placed back into the falcon tubes and packed on ice and then stored at −80 °C until further processing.
2.2. Fermentation Conditions
In addition to the swabs that were taken to determine the surface microbiome composition, apples from each orchard and for each varietal were randomly collected into approximately two-gallon plastic bags for sampling. During the sampling process, gloves sterilized with a 70% ethanol solution were used consistently to minimize contamination of the apples’ microbiomes. The harvested apples were transported to the laboratory and processed into cider within 24 h of collection. The apples were manually diced into approximately half-inch cubes and homogenized into a coarse pulp using a food processor. The seeds and stems were included, and the apples were intentionally left unwashed to preserve their natural microbial communities. Juice extraction was performed manually by pressing the pulp through sterilized cheesecloth. All the equipment and surfaces utilized, including the cheesecloth (sterilized via boiling); cutting boards; knives; gloved hands (cleaned with 70% ethanol); and processing containers, such as food processors, jars, and airlocks (sanitized using a Star San solution), were thoroughly sterilized or disinfected to maintain aseptic conditions throughout processing.
Each processing batch yielded approximately 72 ounces (approximately 2.1 L) of apple juice, subsequently divided into three aliquots of approximately 24 ounces each, stored in sterilized 32-ounce mason jars fitted with airlocks (Figure 1). Immediately following pressing and after one week, 7 mL aliquots were aseptically withdrawn and frozen promptly at −18 °C in a domestic freezer before transfer to −80 °C storage until subsequent analyses. After the primary fermentation period concluded, the cider remained undisturbed in the original jars on the lees for secondary fermentation.
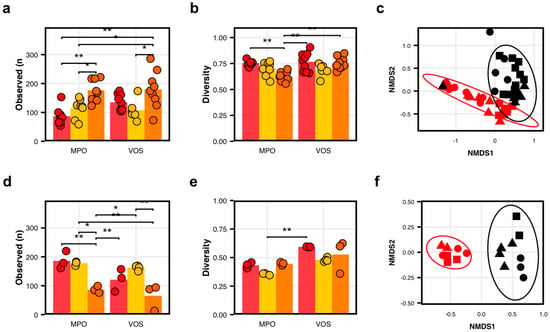
Figure 1.
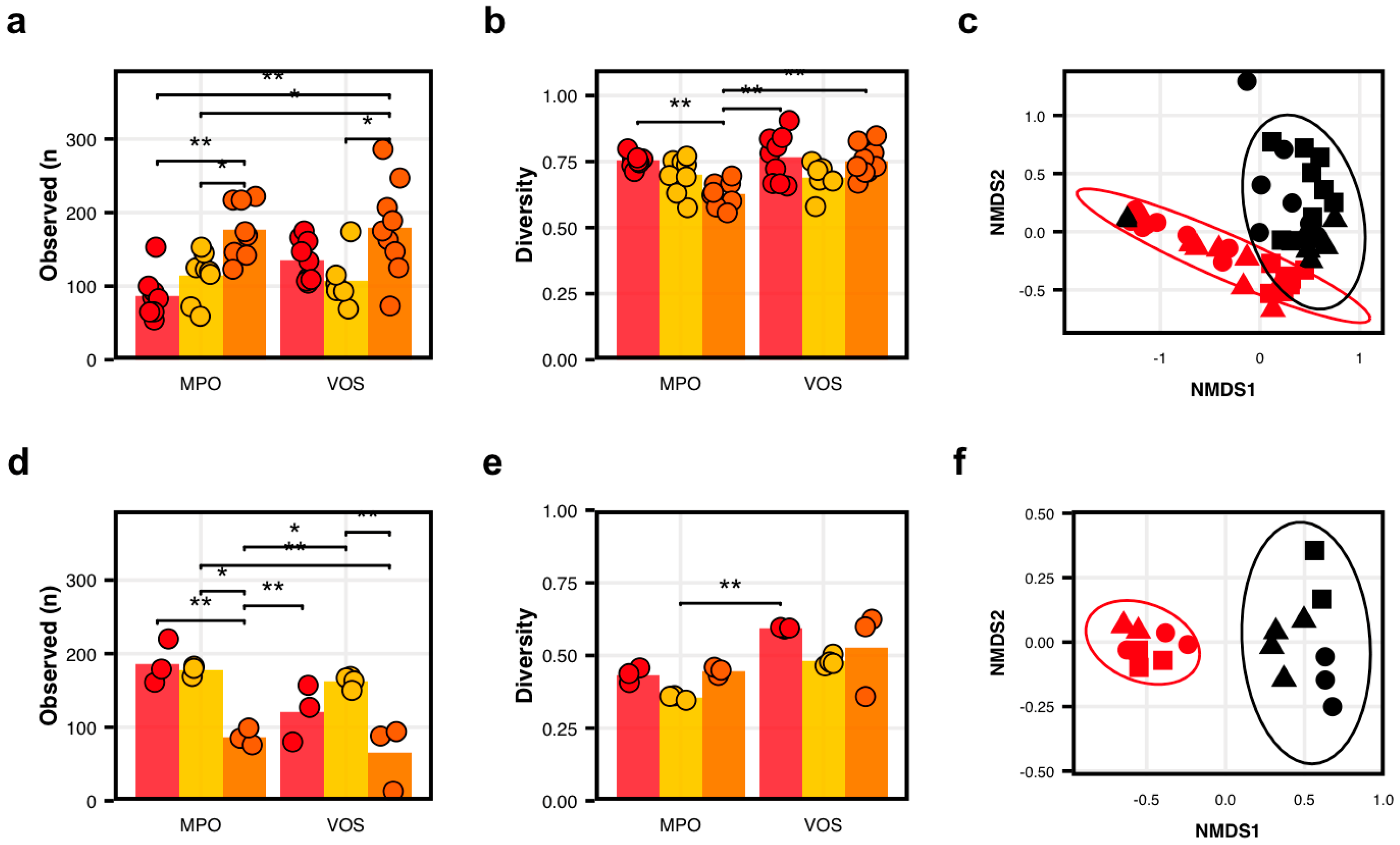
Microbial diversity in cider apples. (a–c) Bacterial communities differed among apple varietals (Empire, Golden, and Idared) and between orchards with respect to (a) observed ASVs number (p < 0.0001), (b) Pielou’s Evenness (p = 0.007), and (c) overall community structure (PERMANOVA, adj-p = 0.006). (d–f) Fungal communities also differed significantly among varietals and between orchards with respect to (d) observed ASVs number (p = 0.03), (e) Pielou’s Evenness (p < 0.0001), and (f) overall community structure (PERMANOVA, adj-p = 0.001). In panels (a,b,d,e), colors represent varietals, Empire (red), Golden (yellow), and Idared (orange), with dots representing individual samples. Statistical comparisons were made using linear models with Tukey tests; significance indicated as p < 0.05 (*) and p < 0.01 (**). In panels (c,f), shapes represent varietals—Empire (circles), Golden (triangles), and Idared (squares)—and colors represent orchards—Montgomery Place (red) and Vosburg (black).
2.3. DNA Extraction and Sequencing
The swabs of the preharvest apples were thawed at room temperature and then incubated at 60 °C at 220 rpm for 10 min. The liquid was then transferred to 2 mL centrifuge tubes with the cut swab tip and vortexed horizontally at max speed for five minutes. The liquid was transferred and then the DNA was isolated under a flame as described in the Qiagen (formerly MO BIO) PowerFood Microbial Kit protocol, with the exception of adding an additional minute to the first centrifuge step (Qiagen, Hilden, Germany). Purified genomic DNA (gDNA) was stored in nuclease-free H2O at −20 °C until further analysis.
We then used 16S rRNA and internal transcribed spacer (ITS) amplicon sequencing to characterize the bacterial and fungal populations, respectively. As per Earth Microbiome Project [18], we used the 515F and 806R primers to amplify and sequence the V4 region of the 16S rRNA gene. For the ITS amplicon sequencing, we used the ITS1f and ITS2 primers [19]. Then, the 16S rRNA and ITS amplicons were pooled separately and purified on a 2% agarose gel using a Qiagen gel extraction kit. The amplicon libraries were sequenced at Wright Labs, Huntington, Pennsylvania (PA), using Illumina MiSeq v2 paired-end sequencing (2 × 250 bp reads) with 20% PhiX spike-in. All the raw reads are available from the Sequence Reads Archive of the NCBI (BioProject ID: PRJNA1262505).
2.4. Quantifying Microbial Diversity
To characterize the bacterial diversity of the 16S amplicons and the fungal diversity of the ITS amplicons for each ferment, we identified and tabulated the different amplicon sequence variants (i.e., ASVs). More specifically, we processed the 16S rRNA and ITS reads using the DADA2 pipeline version 1.26 [20] using standard parameters unless specified, and implemented them in R version 4.4.2. In total, we obtained a total of 3,819,707 pairs of forward and reverse reads for the 16S rRNA, with an average sequencing depth of 56,026 reads per sample, and we obtained a total of 16,321,566 reads for the ITS, with an average sequencing depth per sample of 859,029 reads. Each sequence read was further processed, quality-checked, trimmed, assessed for chimeric contaminants, and de-noised for possible sequencing errors. While the 16S rRNA forward and reverse reads were trimmed at 240 bp and 225 bp, respectively, we only considered forward reads for the ITS for which we removed any trace of the primers within the read using Cutadapt v4.8.
We then assigned each ASV to a taxonomic rank, usually at the genus level, providing additional information about the biology of each microbiome community. Taxonomy was assigned using the DADA2 native taxa identifier function trained on the SILVA ribosomal RNA gene database version 138.1 [21,22] for the 16S rRNA and the UNITE version 10.0 for the ITS identification [23]. A complete list of all the ASVs and their abundance in each sample can be found in Table S1 for the 16S rRNA and Table S2 for the ITS, and the complete taxonomic assignment can be found in Table S3 and Table S4 for the 16S and ITS, respectively. In addition, mapping files linking the sample names and the different treatments are provided in Table S5 and Table S6 for the 16S and ITS analyses, respectively.
2.5. Microbial Community Analysis
The microbiome diversity was analyzed using phyloseq version 1.30.0 [24]. For both the bacterial and the fungal datasets, we estimated the number of observed ASVs and Pielou’s Evenness on samples rarefied to the lowest sampling depth. The latter included evenness measures among the different ASVs present in a sample, providing an overall measure of diversity based on the relative abundance [25]. We tested for differences between treatments using linear modeling and comparing the different statistical models with Akaike’s Information Criterion (AIC).
To test whether there were statistical differences in the population structure among the different varietals and environmental factors, we performed a Principal Coordinate Analysis (PCoA) of the distance matrices estimated from the Bray–Curtis dissimilarity index and the phylogenetic signal calculated as weighted UniFrac distance scores [26]. To test for statistical differences between groups, we used a permutational multivariate analysis of variance (PERMANOVA) implemented via the adonis function of vegan version 2.5.6 [27]. The latter is a non-parametric method that estimates F-values from distance matrices and relies on permutations to determine the statistical significance between group means. We also conducted pairwise PERMANOVA tests using the pairwiseAdonis package (version 0.4.1) [28]. Pairwise comparisons were conducted for different orchard–variety combinations to assess differences in microbial community structure based on the Bray–Curtis distance matrices that were previously estimated. Statistical significance was determined based on 999 permutations, and the p-values were adjusted for multiple comparisons using the false discovery rate (FDR) method.
Finally, a random forest analysis was conducted using the randomForest package (version 4.7-1) [29] implemented in R. Random forest models were built to determine orchard identities based on the microbial community composition. Models were generated using 500 trees (ntree = 500), and the importance of each predictor (taxon) was evaluated by calculating the mean decrease in accuracy. The models’ accuracy was assessed using the out-of-bag (OOB) error estimate. The confidence intervals for the OOB error rates were determined via bootstrapping (1000 iterations) by resampling the dataset with replacement and recalculating the model accuracy for each bootstrap iteration. A random seed (123) was set to ensure the reproducibility of the results. All computations were performed in R version 4.4.2 and data visualization was performed using ggplot2 [30]. We used ChatGPT 4.1 to assist with code improvement and data visualization.
3. Results
3.1. Bacterial Communities Differ Among Apple Varietals and Between Orchards
We first investigated the bacterial communities found to be associated with three apple varietals cultivated by two different producers at the time of harvest in late fall. Using 16S rRNA amplicon sequencing, we identified a total 21 different phyla, 107 orders, and 432 genera among the bacterial ASVs. While a full list of the taxonomic groups is available in Table S3, the most common bacteria phyla were Proteobacteria (86.9% ± 0.03), followed by Bacteroidota (4.2 ± 0.005%), Myxococcota (4.0 ± 0.01%), and Acinobacteria (3.3% ± 0.004%). At the order level, the bacteria ASVs were distributed within Rhizobiales (44.0 ± 0.04%), Sphingomonadales (14.4 ± 0.01%), and Burkholderiales (10.9 ± 0.02%) (Figure S1).
Overall, we found that the varietal and producer significantly impacted the microbial composition of the apples, both in terms of the diversity and community composition. More specifically, we found that the apple varietal impacted the number of observed bacterial types (Anova: F(2,42) = 14.0; p < 0.0001; Figure 1a), with Idared hosting the highest number of observed ASVs. We also found that the interaction between the varietal and producer had an impact on the bacterial diversity (Anova: F(2,42) = 5.6; p = 0.007; Figure 1b). Interestingly, we found that the Idared apples showed the highest diversity for both producers and that the Empire apples were statistically lower in diversity than the other varietals only for at Montgomery Place Orchards. Crucially, when considering the overall community compositions, accounting for the bacterial identity and relative abundance, we found striking differences when integrating the various varietal and orchard combinations (Adonis2: F(3,44) = 4.42; adj-p = 0.001). The location where the apples were grown explained the variance in our model (Adonis2: F(1,46) = 9.57; adj-p = 0.001; Figure 1c). In addition to finding a significant difference in the overall community structures, we also found 30 ASVs that differed significantly in abundance between the two orchards, including 10 that were in higher abundance at Montgomery Place Orchards, such as Pseudomonas spp. and Polaromonas sp., and 20 that were in higher abundance at Vosburg Orchards, including Methylobacterium sp. and 1174-90-12 sp., a known Rhizobiales (adj-ps < 0.0001; Figure S2. Moreover, a machine-learning-based random forest modelization of the bacterial communities was able to predict the source of the apples’ microbial communities with a 96.5% accuracy (error rate: 3.5% ± 2.4; 95% CI: 0–8.3%). The same model suggested that two genera were particularly important for identifying the signature of each orchard, i.e., Acidophilim sp. and 1174-901-12 sp., which were both in higher abundance at Vosburg Orchards.
The varietal also played an important role in shaping the apples’ microbiomes (Adonis2: F(2,45) = 2.7185; adj-p = 0.001; Figure 1c). When considering both producers at once, we detected significant differences between the Empire and Idared apples (Pairwise.Adonis2: F(1,32) = 4.28; adj-p = 0.001) as well as between the Golden and Idared (F(1,29) = 2.66; adj-p = 0.004), but no difference between the Empire and Golden apples (F(1,29) = 1.16; adj-p = 0.27). Moreover, when considering each varietal in the producer context, we found that all but one varietal–producer comparison harbored different bacterial communities (adj-ps < 0.05; Table S7). Interestingly, we did not detect a difference in the bacteria signature between the Empire and Golden varietals from Vosburg Orchards (adj-p = 0.45). These results, taken together, strongly suggest that there exists a measurable bacterial terroir on apple surfaces that is shaped by the apple varietal and orchard producer.
3.2. Fungal Communities Differ Among Apple Varietals and Between Orchards
We then investigated the fungal communities found in the same apples using ITS amplicon sequencing. Here, we identified a total of seven phyla, 191 families, and 345 genera among the fungal ASVs (Table S4). The most common fungal phyla were Ascomycota (57.0% ± 0.03), followed by Basidiomycota (42.8 ± 0.003%). When it came to order, the fungal ASVs were mostly distributed within Sporidiobolales (32.7 ± 0.08%), Eurotiales (24.1 ± 0.07%), and Dothideales (11.7 ± 0.07%) (Figure S3).
Overall, we found that, unlike the bacterial communities, the main factor significantly shaping the fungal communities was the apple producer. While we found that the varietal had an effect on the number of observed fungal types (Anova: F(2,13) = 22.0; p < 0.0001; Figure 1d), it did not have a particularly strong effect on fungal diversity (Anova: F(2,13) = 4.56; p = 0.03; Figure 1e). On the other hand, the producer had a limited impact on the number of fungal ASVs (Anova: F(1,13) = 5.6; p = 0.03; Figure 1d) and fungal diversity (Anova: F(1,13) = 18,20; p < 0.0001; Figure 1e). Interestingly, we found that the number of observed fungal ASVs and diversity were higher for the Empire apples, suggesting a negative correlation with the bacterial diversity of the same apples.
Crucially, the impact of the producer stood out compared to the varietal when considering the overall fungal communities on the apples’ surfaces. The strongest signal was obtained for the producer who grew the apples (Adonis2: F(1,17) = 16.1; adj-p = 0.001; Figure 1f). In addition to finding a significant difference in the overall community structure, we also found 49 ASVs that differed significantly in abundance between the two orchards, including 26 that were in higher abundance at Montgomery Place Orchards, including Sporobolomyces sp. and Candida sp., and 23 in higher abundance at Vosburg Orchards, such as Filobasicium sp. and Nigrograna sp. (adj-ps < 0.0001; Figure S4). Moreover, a machine-learning-based random forest modelization of the fungal communities was able to predict the source of the apples’ microbial communities with a 96.0% (error rate: 4% ± 4.1; 95% CI: 0–15.8%) accuracy. The same model suggested that one species was particularly important for identifying the signature of each orchard, i.e., Sporobolmyces patagonicus, which was in higher abundance at Montgomery Place Orchards.
Interestingly, we found that the varietal did not play as important a role in shaping the apples’ fungal microbiomes (Adonis2: F(2,16) = 1.04; adj-p = 0.36; Figure 1f). Yet, when using random forest modelizations, we found that the varietal could be used to make an accurate prediction when it came to comparing Empire and Idared and Golden and Idared (error rate: 4.8% ± 6.3; 95% CI: 0–23.1%). Each time, one species, i.e., Stereum complicatum, seemed to play an important role in identifying the Idared apples. When considering each varietal within the producer context, however, we found that only five varietal–producer comparisons showed marginal differences between the fungal communities, but such signatures disappeared when adjusting the p-values for multiple comparisons (adj-ps > 0.05; Table S8). These results, taken together, suggest that the orchard produces a detectable fungal terroir signature on the apple surfaces and that the varietal does, too, but to a lesser extent.
3.3. Bacterial Communities in Fermented Cider Differ Among Varietals and Between Orchards
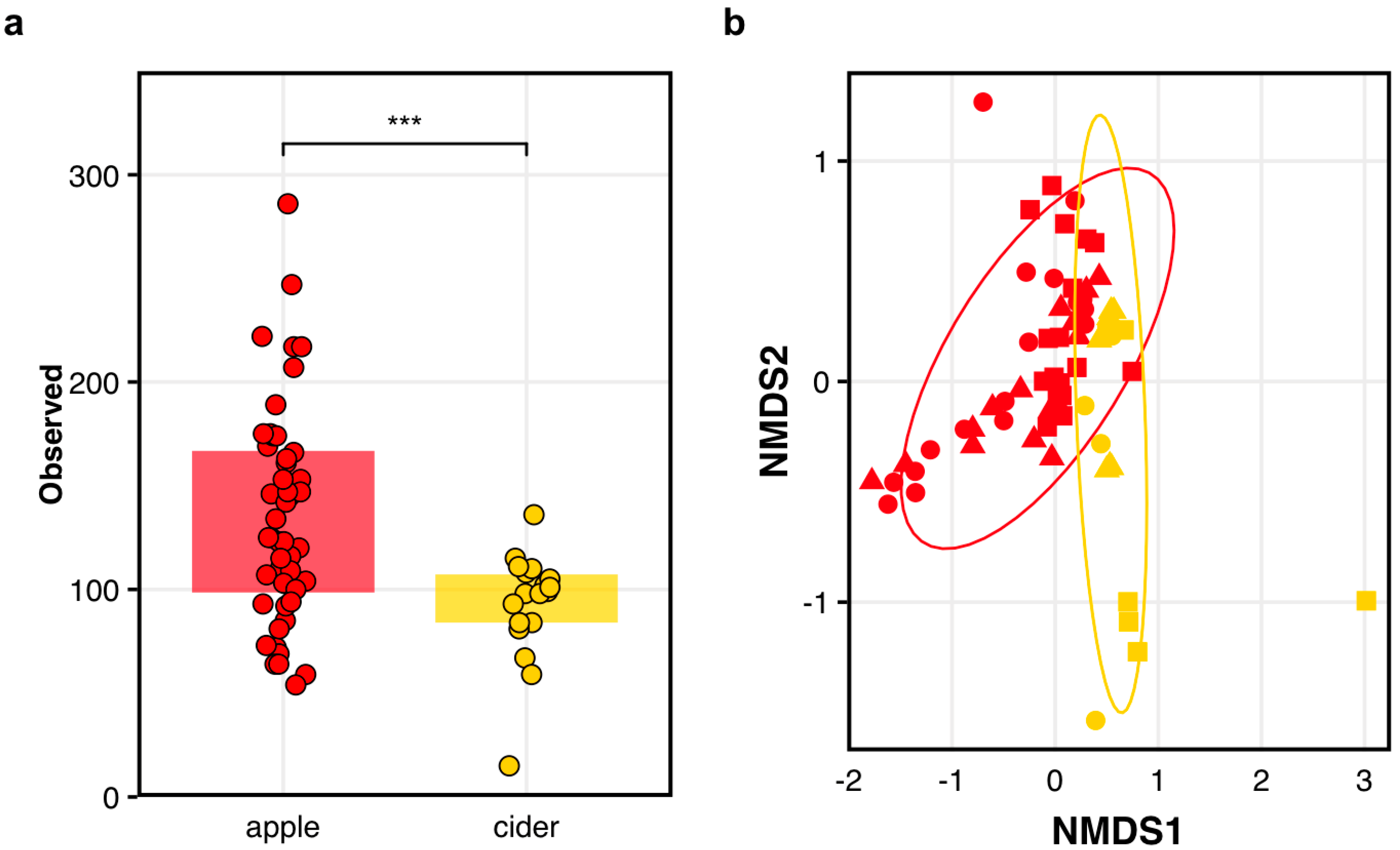
Finally, to investigate whether the apples’ microbial terroirs at harvest impacted cidermaking, we characterized the bacterial communities in the fermented cider one week after harvest. In short, we found some evidence suggesting that there is indeed a microbial terroir signature in fermented cider. As expected, given the important differences in the growing conditions, we found a significant reduction in the observed bacterial ASVs (Anova: F(1,54) = 13.81; adj-p = 0.005; Figure 2a) and bacteria community structures (Adonis2: F(1,64) = 8.52; adj-p = 0.001; Figure 2b) in the cider compared to the apples. But again, we found that the orchard was responsible for the greatest differences in the bacteria communities found in 1-week-old cider (Adonis2: F(1,64) = 10.0; adj-p = 0.001), and there were also significant differences among the ciders made from different varietals (Adonis2: F(2,63) = 2.55; adj-p = 0.002). Crucially, these differences were also found in many of the varietal and orchard contexts (Adonis2: F(1,54) = 6.51; adj-p = 0.001; Figure 2b; Table S9).
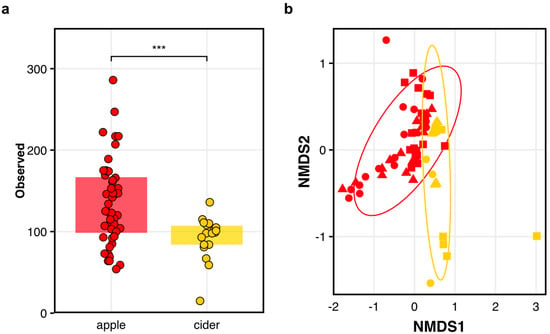
Figure 2.
Comparing microbial diversity of apples and fermented ciders. We observed significant differences in (a) the number of observed ASVs between the apples (red) and fermented ciders (gold) where each circle represents an independent data point (*** p = 0.0004), and in (b) the overall population structures between the apples and fermented ciders (adj-p = 0.001). The varietals presented are Empire (circles), Golden (triangles), and Idared (squares) where each symbol is an independent data point.
4. Discussion
Our findings indicate that a microbial terroir is evident in cider apple production, aligning closely with previous research conducted in viticulture. In wine production, it has been well documented that indigenous microbial communities on grape surfaces are strongly influenced by local environmental conditions and vineyard management practices, significantly shaping fermentation dynamics and regional identities [3,7,8]. Here, we extend this understanding to cider apples, demonstrating that similar ecological factors shape apple microbial communities, emphasizing their potential to influence cider quality and identity. Specifically, our analyses revealed significant differences in the bacterial communities between orchards, with the producer identity exhibiting a particularly pronounced effect. Varietal differences also contributed significantly, albeit to a lesser degree. These findings suggest that both orchard-specific management and apple varietal characteristics are the critical factors structuring the microbial assemblages found on apple surfaces, closely mirroring the mechanisms identified in grape microbial ecology [1,2,9].
Importantly, these microbial signatures persisted through the initial stages of cider fermentation. The microbial community patterns identified on the harvested apples remained detectable in the cider after one week of fermentation, suggesting that the initial microbial composition likely impacted the subsequent fermentation processes and potentially the cider flavor and quality [8,11]. While direct links between these microbial profiles and sensory or metabolic outcomes were beyond the scope of the current study, the persistence of orchard-specific microbial signatures into fermentation highlights their possible role in differentiating ciders and supporting regional branding strategies.
In addition to the age of the orchards and the agricultural practices, geographical and environmental factors, including the local climate and orchard-specific conditions, likely drove the observed microbial community differences [31]. For instance, Montgomery Place Orchard’s proximity to the Hudson River and Saw Kill tributary exposes it to increased moisture, distinct airflow patterns, and unique microclimatic conditions relative to Vosburg Orchard, located further inland in the mid-Hudson Valley. These environmental variations have likely fostered unique microbial assemblages, as evidenced by our data [7,10]. One organism of particular interest identified at Montgomery Place was Sporobolomyces patagonicus. This yeast species is known to thrive on plant surfaces in moist environments, making its prevalence at Montgomery Place consistent with the orchard’s environmental characteristics [32,33]. The presence and relative abundance of S. patagonicus underscores the potential of using specific microbial taxa as indicators of orchard microclimates and microbial terroirs in cider production.
While our study provides robust evidence supporting a microbial terroir in apple and cider production, several limitations should be acknowledged. For example, the temporal scope was restricted to harvest and after one week of fermentation. The longer-term dynamics of the entire fermentation process were not assessed, limiting our understanding of how microbial communities evolve throughout fermentation and their ultimate impact on cider quality and sensory properties. Additionally, the study’s limited geographic range and number of orchards may restrict the generalizability of the findings to broader contexts. Future research should aim to explicitly link microbial composition and diversity to cider fermentation outcomes by a qualitative determination of the metabolites and fungal communities in cider [34]. Connecting the genetic variations in the bacterial and fungal populations to changes in the metabolic pathways would deepen our understanding of microbial terroirs and could inform targeted orchard management practices aimed at enhancing cider quality and regional distinctiveness [3,12,13,35].
In conclusion, our study provides clear evidence supporting the concept of a microbial terroir in cider apple production. Both the varietal and orchard-specific environmental factors shape the microbial communities, with implications for fermentation dynamics, product differentiation, and regional identity. Recognizing and leveraging this microbial diversity could offer substantial qualitative and economic benefits for artisanal cider producers.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation11070369/s1, Figure S1: Bacterial diversity in apples and cider; Figure S2 Change in relative abundance of specific bacterial ASVs; Figure S3: Fungal diversity in apples; Figure S4: Change in relative abundance of specific fungal ASVs; Table S1: List of bacterial ASVs; Table S2: List of fungal ASVs; Table S3: Bacterial taxonomy; Table S4: Fungal taxonomy; Table S5: Metadata for bacterial data; Table S6: Metadata for fungal data; Table S7: Statistical analysis of pairwise comparisons for bacterial community analyses; Table S8: Statistical analysis of pairwise comparisons for fungal community analyses; Table S9: Statistical analysis of pairwise comparisons for bacterial community analyses between apples and ciders.
Author Contributions
Conceptualization, G.G.P. and S.S.J.; methodology, G.G.P. and L.C.M.; validation, P.L. and L.D.-S.; formal analysis, G.G.P. and L.C.M.; data curation, G.G.P.; writing—original draft preparation, G.G.P. and L.C.M.; writing—review and editing, G.G.P., L.C.M., P.L., L.D.-S. and S.S.J.; visualization, L.C.M. and G.G.P.; supervision, G.G.P.; project administration, G.G.P.; funding acquisition, G.G.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Bard College (Annandale-On-Hudson, NY, USA). This work was also supported in part through the NYU IT High Performance Computing resources, services, and staff expertise (New York, NY, USA).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the raw reads are available from the Sequence Reads Archive of the NCBI (BioProject ID: PRJNA1262505).
Acknowledgments
The authors thank Mary Verrelli for conducting the original laboratory work in 2017 as part of their senior thesis at Bard College, as well as Cathy Collins and Doug Taylor for their assistance in the field and the lab, respectively. During the preparation of this manuscript/study, the author(s) used ChatGPT 4.1 for the purposes of improving the R code for the statistical analysis and data visualization. The authors have reviewed and edited the output and take full responsibility for the content of this publication.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going Back to the Roots: The Microbial Ecology of the Rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The Rhizosphere Microbiome and Plant Health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; van der Lelie, D.; Zarraonaindia, I. Microbial Terroir for Wine Grapes. Proc. Natl. Acad. Sci. USA 2014, 111, 5–6. [Google Scholar] [CrossRef]
- Mas, A.; Portillo, M.C. Strategies for Microbiological Control of the Alcoholic Fermentation in Wines by Exploiting the Microbial Terroir Complexity: A Mini-Review. Int. J. Food Microbiol. 2022, 367, 109592. [Google Scholar] [CrossRef]
- Franco, G.C.; Leiva, J.; Nand, S.; Lee, D.M.; Hajkowski, M.; Dick, K.; Withers, B.; Soto, L.; Mingoa, B.-R.; Acholonu, M.; et al. Soil Microbial Communities and Wine Terroir: Research Gaps and Data Needs. Foods 2024, 13, 2475. [Google Scholar] [CrossRef]
- Silva, V.; Brito, I.; Alexandre, A. The Vineyard Microbiome: How Climate and the Main Edaphic Factors Shape Microbial Communities. Microorganisms 2025, 13, 1092. [Google Scholar] [CrossRef]
- Van Leeuwen, C.; Seguin, G. The Concept of Terroir in Viticulture. J. Wine Res. 2006, 17, 1–10. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Collins, T.S.; Masarweh, C.; Allen, G.; Heymann, H.; Ebeler, S.E.; Mills, D.A. Associations among Wine Grape Microbiome, Metabolome, and Fermentation Behavior Suggest Microbial Contribution to Regional Wine Characteristics. mBio 2016, 7, 10–1128. [Google Scholar] [CrossRef]
- Leff, J.W.; Fierer, N.; Angelo, T. Apple Orchard Microbiomes: Composition, Diversity, and Management. Appl. Environ. Microbiol. 2013, 79, 2632–2640. [Google Scholar]
- Lfattah, A.; Wisniewski, M.; Droby, S.; Schena, L. Spatial and Compositional Variation in the Fungal Communities of Organic and Conventionally Grown Apple Fruit at the Consumer Point-of-Purchase. Hortic. Res. 2016, 3, 16047. [Google Scholar]
- Knight, S.; Goddard, M.R. Quantifying Separation and Similarity in a Saccharomyces Cerevisiae Metapopulation. ISME J. 2015, 9, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Lea, A.G.H.; Drilleau, J.F. Cider-Making. In Fermented Beverage Production; Springer: Berlin/Heidelberg, Germany, 2003; pp. 59–87. [Google Scholar]
- Merwin, I.A.; Valois, S.; Padilla-Zakour, O.I. Cider Apples and Cider-Making Techniques in Europe and North America. In Horticultural Reviews; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2008; pp. 365–415. ISBN 9780470380147. [Google Scholar]
- Empire. Available online: https://www.applesfromny.com/varieties/empire/ (accessed on 5 May 2025).
- Golden Delicious. Available online: https://www.applesfromny.com/varieties/golden-delicious/ (accessed on 5 May 2025).
- Idared. Available online: https://www.applesfromny.com/varieties/idared/ (accessed on 5 May 2025).
- Lax, S.; Hampton-Marcell, J.T.; Gibbons, S.M.; Colares, G.B.; Smith, D.; Eisen, J.A.; Gilbert, J.A. Forensic Analysis of the Microbiome of Phones and Shoes. Microbiome 2015, 3, 21. [Google Scholar] [CrossRef] [PubMed]
- Walters, W.; Hyde, E.R.; Berg-Lyons, D.; Ackermann, G.; Humphrey, G.; Parada, A.; Gilbert, J.A.; Jansson, J.K.; Caporaso, J.G.; Fuhrman, J.A.; et al. Improved Bacterial 16S RRNA Gene (V4 and V4-5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems 2016, 1, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Mills, D.A. Improved Selection of Internal Transcribed Spacer-Specific Primers Enables Quantitative, Ultra-High-Throughput Profiling of Fungal Communities. Appl. Environ. Microbiol. 2013, 79, 2519–2526. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Glöckner, F.O.; Yilmaz, P.; Quast, C.; Gerken, J.; Beccati, A.; Ciuprina, A.; Bruns, G.; Yarza, P.; Peplies, J.; Westram, R.; et al. 25 Years of Serving the Community with Ribosomal RNA Gene Reference Databases and Tools. J. Biotechnol. 2017, 261, 169–176. [Google Scholar] [CrossRef]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; May, T.W.; Frøslev, T.G.; Pawlowska, J.; Lindahl, B.; Põldmaa, K.; Truong, C.; et al. The UNITE database for molecular identification and taxonomic communication of fungi and other eukaryotes: Sequences, taxa and classifications reconsidered. Nucleic Acids Res. 2024, 52, D791–D797. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Pielou, E.C. The Measurement of Diversity in Different Types of Biological Collections. J. Theor. Biol. 1966, 13, 131–144. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package. In CRAN: Contributed Packages; CRAN: Windhoek, Namibia, 2001. [Google Scholar]
- Martinez Arbizu, P. PairwiseAdonis: Pairwise Multilevel Comparison Using Adonis. 2020. Available online: https://github.com/pmartinezarbizu/pairwiseAdonis (accessed on 5 May 2025).
- Liaw, A.; Wiener, M. Classification and Regression by RandomForest. Available online: https://journal.r-project.org/articles/RN-2002-022/RN-2002-022.pdf (accessed on 5 May 2025).
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D.; van den Brand, T. Ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. In CRAN: Contributed Packages; CRAN: Windhoek, Namibia, 2007. [Google Scholar]
- Miura, T.; Sánchez, R.; Castañeda, L.E.; Godoy, K.; Barbosa, O. Is Microbial Terroir Related to Geographic Distance between Vineyards? Environ. Microbiol. Rep. 2017, 9, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Fatemi, S.; Haelewaters, D.; Urbina, H.; Brown, S.; Houston, M.L.; Aime, M.C. Sporobolomyces lactucae sp. nov. (Pucciniomycotina, Microbotryomycetes, Sporidiobolales): An Abundant Component of Romaine Lettuce Phylloplanes. J. Fungi 2022, 8, 302. [Google Scholar] [CrossRef] [PubMed]
- Libkind, D.; Gadanho, M.; van Broock, M.; Sampaio, J.P. Sporidiobolus longiusculus sp. nov. and Sporobolomyces patagonicus sp. nov., Novel Yeasts of the Sporidiobolales Isolated from Aquatic Environments in Patagonia, Argentina. Int. J. Syst. Evol. Microbiol. 2005, 55, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Fazio, N.A.; Albertin, W.; Masneuf-Pomarede, I.; Randazzo, C.L.; Caggia, C. Structure of Culturable Indigenous Yeast Population and Genetic Diversity of Saccharomyces cerevisiae and Non-Saccharomyces Yeasts during Spontaneous Fermentation of Etna Vineyards Grapes. Int. J. Food Microbiol. 2025, 440, 111282. [Google Scholar] [CrossRef]
- Liu, Q.; Hao, N.; Mi, L.; Peng, S.; Marie-Colette, A.K.; Zhao, X.; Wang, J. From Microbial Communities to Aroma Profiles: A Comparative Study of Spontaneous Fermentation in Merlot and Cabernet Sauvignon Wines. Food Chem. X 2025, 26, 102317. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).