Meta-Omics Analyses of Conventional and Regenerative Fermented Vegetables: Is There an Impact on Health-Boosting Potential?
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of Conventional and Regenerative Organic Produce
2.2. About the Farms
2.3. Fermentation Preparation and Sample Collection
2.4. Microbiome Analyses—16S rRNA and ITS2 Short Amplicon Sequencing
DNA Extraction, Sequencing, and Data Processing
2.5. Microbiome Analyses—16S rRNA Long Read Sequencing
2.6. Metabolomic Analyses
2.6.1. Sample Preparation for LC-MS Analysis
2.6.2. Chemical Derivatization
2.6.3. LC-MS Analysis
2.7. Analysis of Spectral Data
2.8. Statistical Analyses
2.8.1. Microbial Community Analysis
2.8.2. Metabolomic Analysis
2.8.3. Correlation and Network Analysis
3. Results
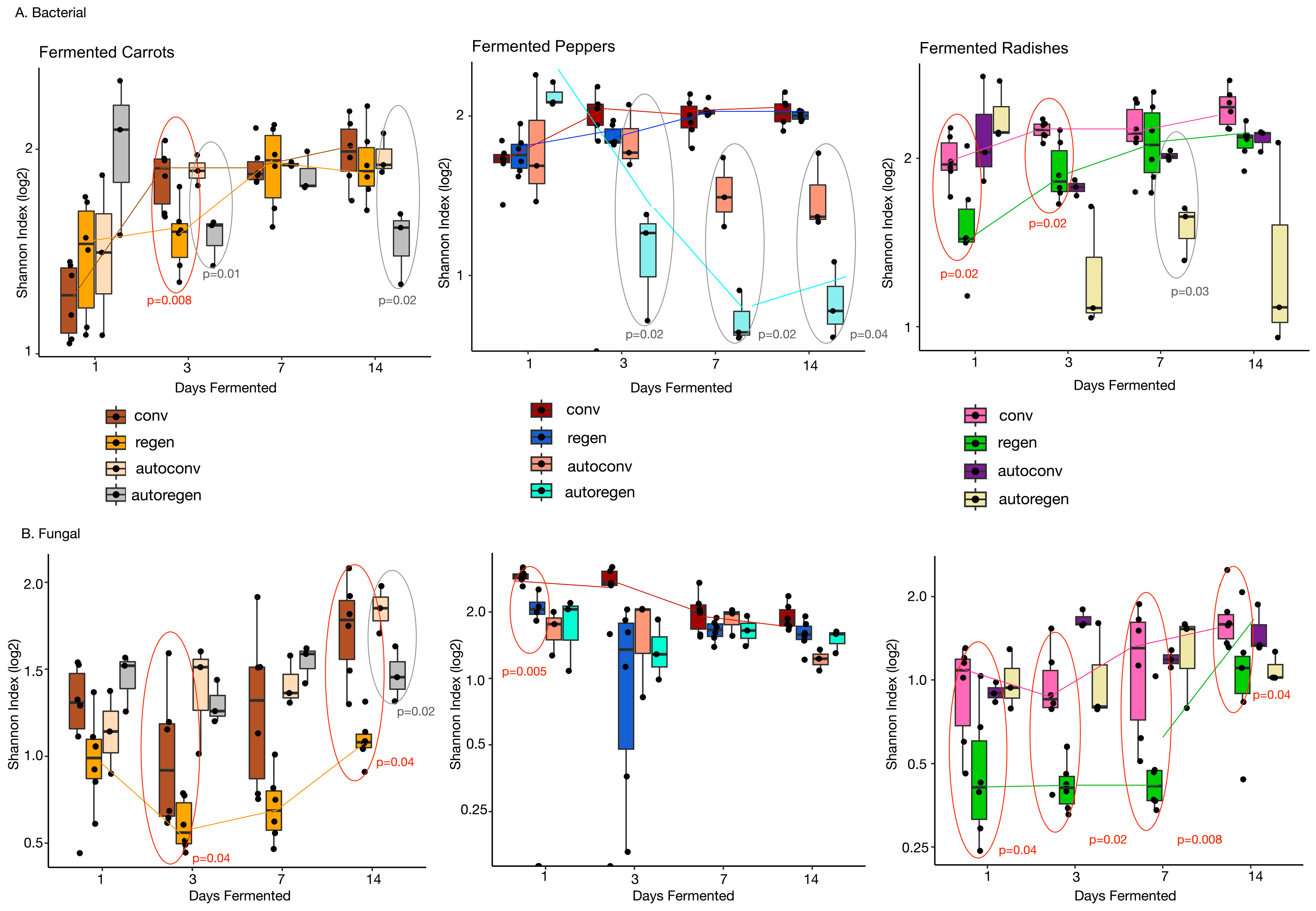
3.1. Microbial Alpha Diversity for Ferments Under Conventional or Regenerative Farming Practices
3.1.1. Bacteriome
3.1.2. Mycobiome
3.2. Alpha Diversity Differed Across Fermentation Time in Conventional and Regenerative Ferments
3.2.1. Bacteriome
3.2.2. Mycobiome
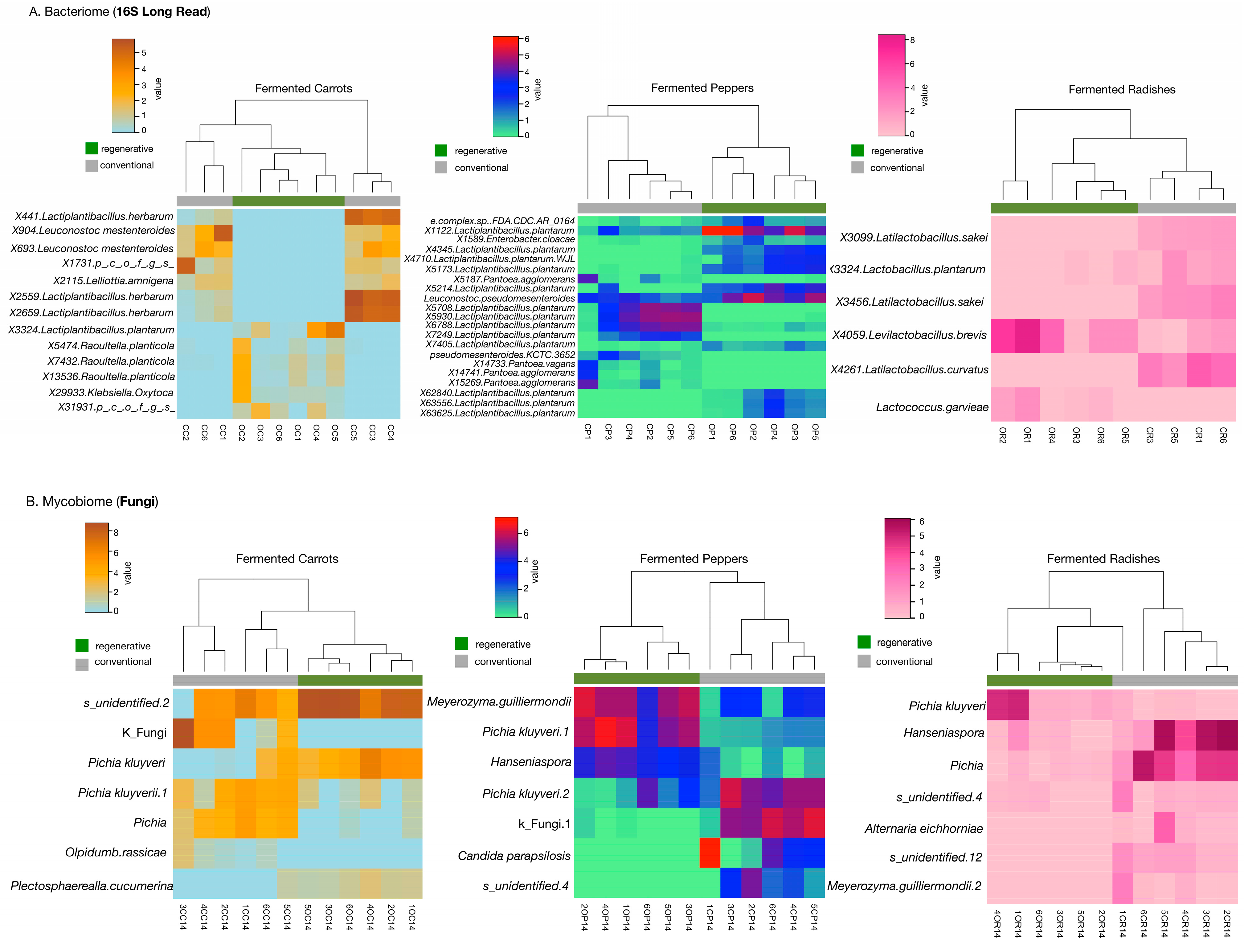
3.3. Conventional and Regenerative Fermented Vegetables Display Unique Bacteriome and Mycobiome Composition
3.4. Distinct Taxonomic Signatures Differentiate Conventional and Regenerative Fermented Vegetables
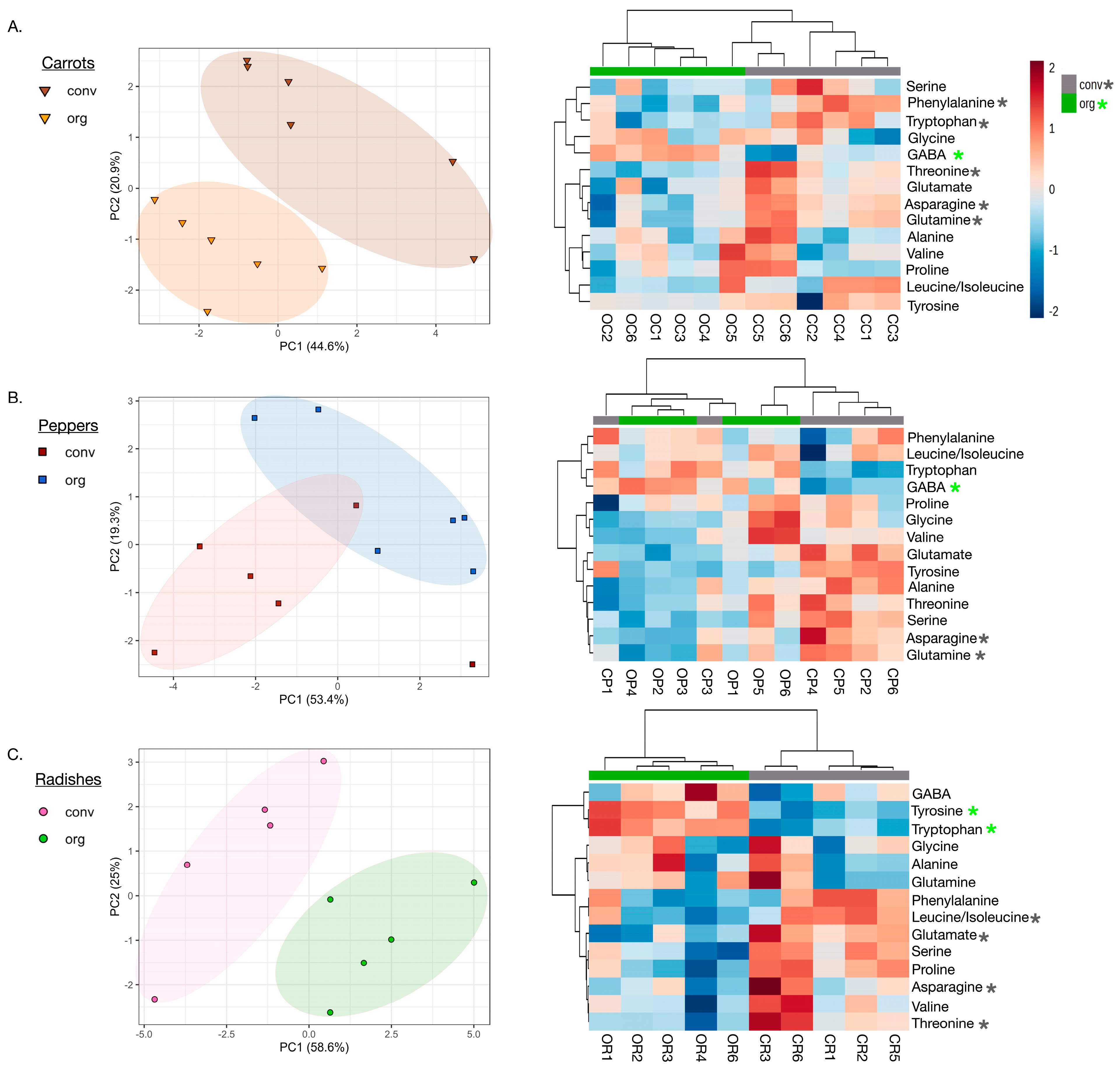
3.5. Metabolome Profiles for Conventional and Regenerative Fermented Vegetables
3.6. No Differences Observed in the Abundance of Organic Acids in Conventional vs. Regenerative Growing Systems
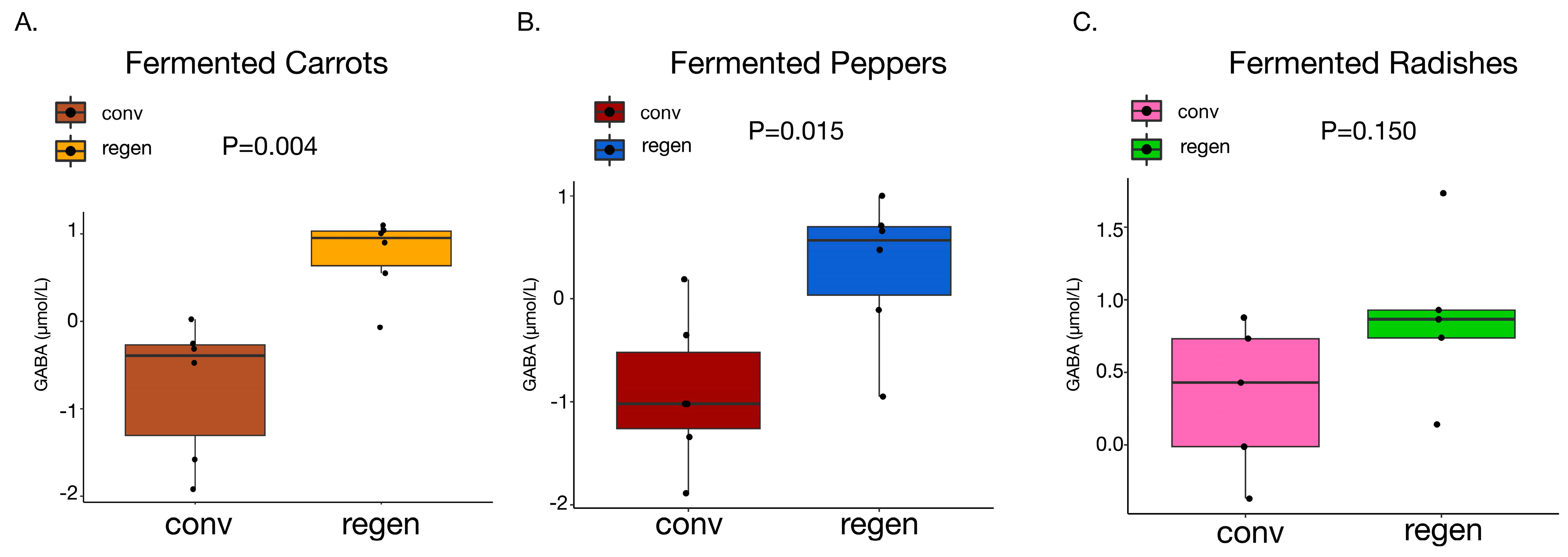
3.7. Higher Amounts of GABA Discovered in All Fermented Vegetables
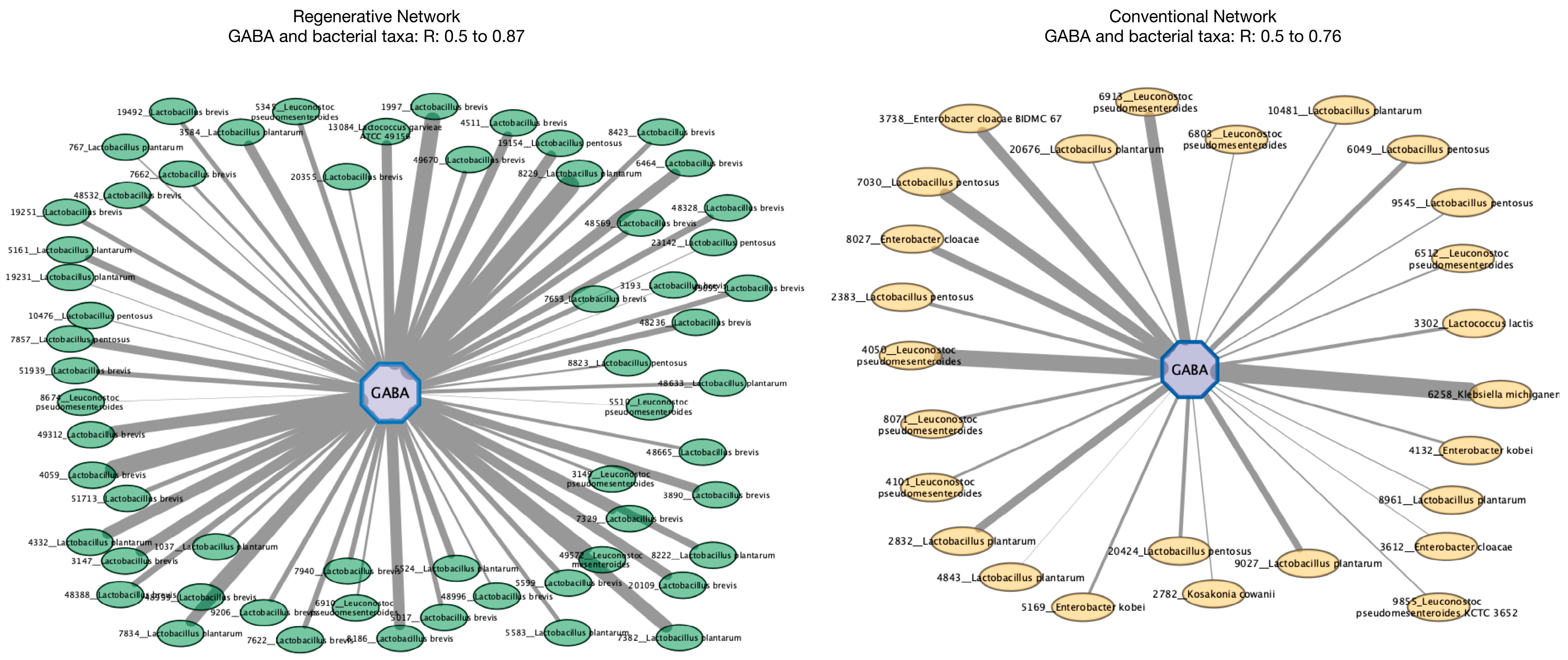
3.8. Network Dynamics Reveal GABA Is More Strongly Correlated to Probiotic Bacterial Taxa in Fermented Vegetables from the Regenerative Growing System
4. Discussion
4.1. Regenerative and Conventional Fermented Vegetables Display Minor Differences in Microbial Alpha Diversity but Reveal Unique Microbiome Compositions
4.2. Environmental Bacteria Found to Be More Characteristic of Fermented Carrots
4.3. Different Strains of Lactiplantibacillus plantarum Dominate Both Regenerative and Conventional Fermented Peppers
4.4. Regenerative and Conventional Radishes Display Distinct LAB
4.5. Typical Fungi Characteristic of Fermented Foods and Beverages Found in All Fermented Vegetables but a Unique Species, Plectosphaerealla cucumerina, Discovered in Regenerative Carrots
4.6. GABA Found in High Amounts in All Fermented Vegetables but Significantly Higher in Regenerative Ones
4.7. Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rasmussen, L.V.; Coolsaet, B.; Martin, A.; Mertz, O.; Pascual, U.; Corbera, E.; Dawson, N.; Fisher, J.A.; Franks, P.; Ryan, C.M. Social-Ecological Outcomes of Agricultural Intensification. Nat. Sustain. 2018, 1, 275–282. [Google Scholar] [CrossRef]
- Culman, S.W.; Young-Mathews, A.; Hollander, A.D.; Ferris, H.; Sánchez-Moreno, S.; O’Geen, A.T.; Jackson, L.E. Biodiversity Is Associated with Indicators of Soil Ecosystem Functions over a Landscape Gradient of Agricultural Intensification. Landsc. Ecol. 2010, 25, 1333–1348. [Google Scholar] [CrossRef]
- Young-Mathews, A.; Culman, S.W.; Sánchez-Moreno, S.; Toby O’Geen, A.; Ferris, H.; Hollander, A.D.; Jackson, L.E. Plant-Soil Biodiversity Relationships and Nutrient Retention in Agricultural Riparian Zones of the Sacramento Valley, California. Agrofor. Syst. 2010, 80, 41–60. [Google Scholar] [CrossRef]
- Raven, P.H.; Wagner, D.L. Agricultural Intensification and Climate Change Are Rapidly Decreasing Insect Biodiversity. Proc. Natl. Acad. Sci. USA 2021, 118, e2002548117. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.S.; Horwath, W.R.; Shennan, C.; Scow, K.M. Changes in Soil Chemical Properties Resulting from Organic and Low-Input Farming Practices. Agron. J. 1998, 90, 662–671. [Google Scholar] [CrossRef]
- Montgomery, D.R.; Biklé, A.; Archuleta, R.; Brown, P.; Jordan, J. Soil Health and Nutrient Density: Preliminary Comparison of Regenerative and Conventional Farming. PeerJ 2022, 10, e12848. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, D.R.; Biklé, A. The Hidden Half of Nature: The Microbial Roots of Life and Health; W. W. Norton & Company: New York, NY, USA, 2015; ISBN 9780393244410. [Google Scholar]
- Zhou, M.; Zhao, J. A Review on the Health Effects of Pesticides Based on Host Gut Microbiome and Metabolomics. Front. Mol. Biosci. 2021, 8, 632955. [Google Scholar] [CrossRef]
- Liu, Q.; Shao, W.; Zhang, C.; Xu, C.; Wang, Q.; Liu, H.; Sun, H.; Jiang, Z.; Gu, A. Organochloride Pesticides Modulated Gut Microbiota and Influenced Bile Acid Metabolism in Mice. Environ. Pollut. 2017, 226, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Rook, G.A.W. 99th Dahlem Conference on Infection, Inflammation and Chronic Inflammatory Disorders: Darwinian Medicine and the “hygiene” or “old Friends” Hypothesis. Clin. Exp. Immunol. 2010, 160, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Marco, M.L.; Hill, C.; Hutkins, R.; Slavin, J.; Tancredi, D.J.; Merenstein, D.; Sanders, M.E. Should There Be a Recommended Daily Intake of Microbes? J. Nutr. 2020, 150, 3061–3067. [Google Scholar] [CrossRef]
- Katz, S.E. The Art of Fermentation: An In-Depth Exploration of Essential Concepts and Processes from Around the World; Chelsea Green Publishing: White River Junction, VT, USA, 2012; ISBN 9781603582865. [Google Scholar]
- Klingenberg, P.G. Campbell-Platt: Fermented Foods of the World. A Dictionary and Guide. 291 Seiten. Butterworth, London, Boston, Durban U. A. 1987. Preis: 35,—£ (hardcover). Nahrung 1989, 33, 304. [Google Scholar] [CrossRef]
- Dunn, R.R.; Wilson, J.; Nichols, L.M.; Gavin, M.C. Toward a Global Ecology of Fermented Foods. Curr. Anthropol. 2021, 62, S220–S232. [Google Scholar] [CrossRef]
- Leeuwendaal, N.K.; Stanton, C.; O’Toole, P.W.; Beresford, T.P. Fermented Foods, Health and the Gut Microbiome. Nutrients 2022, 14, 1527. [Google Scholar] [CrossRef]
- Stanton, C.; Ross, R.P.; Fitzgerald, G.F.; Van Sinderen, D. Fermented Functional Foods Based on Probiotics and Their Biogenic Metabolites. Curr. Opin. Biotechnol. 2005, 16, 198–203. [Google Scholar] [CrossRef]
- Wastyk, H.C.; Fragiadakis, G.K.; Perelman, D.; Dahan, D.; Merrill, B.D.; Yu, F.B.; Topf, M.; Gonzalez, C.G.; Van Treuren, W.; Han, S.; et al. Gut-Microbiota-Targeted Diets Modulate Human Immune Status. Cell 2021, 184, 4137–4153.e14. [Google Scholar] [CrossRef]
- Taylor, B.C.; Lejzerowicz, F.; Poirel, M.; Shaffer, J.P.; Jiang, L.; Aksenov, A.; Litwin, N.; Humphrey, G.; Martino, C.; Miller-Montgomery, S.; et al. Consumption of Fermented Foods Is Associated with Systematic Differences in the Gut Microbiome and Metabolome. mSystems 2020, 5, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, E.S.; Garnås, E.; Jensen, K.J.; Hansen, L.H.; Olsen, P.S.; Ritz, C.; Krych, L.; Nielsen, D.S. Lacto-Fermented Sauerkraut Improves Symptoms in IBS Patients Independent of Product Pasteurisatio—A Pilot Study. Food Funct. 2018, 9, 5323–5335. [Google Scholar] [CrossRef] [PubMed]
- Bourrie, B.C.T.; Willing, B.P.; Cotter, P.D. The Microbiota and Health Promoting Characteristics of the Fermented Beverage Kefir. Front. Microbiol. 2016, 7, 647. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Pedersen, O. Gut Microbiota in Human Metabolic Health and Disease. Nat. Rev. Microbiol. 2020, 19, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Pokusaeva, K.; Fitzgerald, G.F.; van Sinderen, D. Carbohydrate Metabolism in Bifidobacteria. Genes Nutr. 2011, 6, 285–306. [Google Scholar] [CrossRef] [PubMed]
- SaeidiFard, N.; Djafarian, K.; Shab-Bidar, S. Fermented Foods and Inflammation: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Clin. Nutr. ESPEN 2020, 35, 30–39. [Google Scholar] [CrossRef]
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Foligné, B.; Gänzle, M.; Kort, R.; Pasin, G.; Pihlanto, A.; et al. Health Benefits of Fermented Foods: Microbiota and beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Devi, S.M.; Kurrey, N.K.; Halami, P.M. In Vitro Anti-Inflammatory Activity among Probiotic Lactobacillus Species Isolated from Fermented Foods. J. Funct. Foods 2018, 47, 19–27. [Google Scholar] [CrossRef]
- Schon, N.; Fraser, T.; Masters, N.; Stevenson, B.; Cavanagh, J.; Harmsworth, G.; Grelet, G.-A. Soil Health Research in the Context of Regenerative Agriculture; AgResearch: Hamilton, New Zealand, 2024. [Google Scholar]
- Schreefel, L.; Schulte, R.P.O.; de Boer, I.J.M.; Schrijver, A.P.; van Zanten, H.H.E. Regenerative Agriculture—The Soil Is the Base. Glob. Food Sec. 2020, 26, 100404. [Google Scholar] [CrossRef]
- LaCanne, C.E.; Lundgren, J.G. Regenerative Agriculture: Merging Farming and Natural Resource Conservation Profitably. PeerJ 2018, 6, e4428. [Google Scholar] [CrossRef] [PubMed]
- Guse, K. The Microbiome and Fermentation: From Human Evolution to Human Transformation. Ph.D. Thesis, University of Minnesota, Minneapolis, MN, USA, 2023. [Google Scholar]
- RumRaghuvanshi, R.; Grayson, A.G.; Schena, I.; Amanze, O.; Suwintono, K.; Quinn, R.A. Microbial Transformations of Organically Fermented Foods. Metabolites 2019, 9, 165. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; Grinevich, D.; Thakur, S.; Balamotis, M.A.; Yehezkel, T.B. Ultra-Accurate Microbial Amplicon Sequencing with Synthetic Long Reads. Microbiome 2021, 9, 130. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; Yun, K.; Mun, S.; Chung, W.-H.; Choi, S.-Y.; Nam, Y.; Lim, M.Y.; Hong, C.P.; Park, C.; Ahn, Y.J.; et al. The Effect of Taxonomic Classification by Full-Length 16S rRNA Sequencing with a Synthetic Long-Read Technology. Sci. Rep. 2021, 11, 1727. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Yao, D.; Chen, C. 2-Hydrazinoquinoline as a Derivatization Agent for LC-MS-Based Metabolomic Investigation of Diabetic Ketoacidosis. Metabolites 2013, 3, 993–1010. [Google Scholar] [CrossRef]
- Ma, Y.; Zhou, W.; Chen, P.; Urriola, P.E.; Shurson, G.C.; Ruan, R.; Chen, C. Metabolomic Evaluation of Scenedesmus Sp. as a Feed Ingredient Revealed Dose-Dependent Effects on Redox Balance, Intermediary and Microbial Metabolism in a Mouse Model. Nutrients 2019, 11, 1971. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2019, 2, 5–6. [Google Scholar]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of Phylogenetics and Evolution in R Language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.W.; Roberts, M.D.W. Package “labdsv”. Ordination Multivar. 2016, 775, 1–68. [Google Scholar]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H.; Chang, W.; Wickham, M.H. Package “ggplot2.” Create elegant data visualisations using the grammar of graphics. Version 2016, 2, 1–189. [Google Scholar]
- Sharma, A.K.; Davison, S.; Pafco, B.; Clayton, J.B.; Rothman, J.M.; McLennan, M.R.; Cibot, M.; Fuh, T.; Vodicka, R.; Robinson, C.J.; et al. The Primate Gut Mycobiome-Bacteriome Interface Is Impacted by Environmental and Subsistence Factors. NPJ Biofilms Microbiomes 2022, 8, 12. [Google Scholar] [CrossRef]
- Wheeler, M.L.; Limon, J.J.; Bar, A.S.; Leal, C.A.; Gargus, M.; Tang, J.; Brown, J.; Funari, V.A.; Wang, H.L.; Crother, T.R.; et al. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe 2016, 19, 865–873. [Google Scholar] [CrossRef]
- Sivamaruthi, B.S.; Kesika, P.; Chaiyasut, C. Toxins in Fermented Foods: Prevalence and Preventions-A Mini Review. Toxins 2018, 11, 4. [Google Scholar] [CrossRef] [PubMed]
- Skowron, K.; Budzyńska, A.; Grudlewska-Buda, K.; Wiktorczyk-Kapischke, N.; Andrzejewska, M.; Wałecka-Zacharska, E.; Gospodarek-Komkowska, E. Two Faces of Fermented Foods-The Benefits and Threats of Its Consumption. Front. Microbiol. 2022, 13, 845166. [Google Scholar] [CrossRef]
- Rembiałkowska, E. Quality of Plant Products from Organic Agriculture. J. Sci. Food Agric. 2007, 87, 2757–2762. [Google Scholar] [CrossRef]
- Gomiero, T. Food Quality Assessment in Organic vs. Conventional Agricultural Produce: Findings and Issues. Appl. Soil Ecol. 2018, 123, 714–728. [Google Scholar] [CrossRef]
- Sękowska, A. Raoultella Spp.-Clinical Significance, Infections and Susceptibility to Antibiotics. Folia Microbiol. 2017, 62, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Appel, T.M.; Quijano-Martínez, N.; De La Cadena, E.; Mojica, M.F.; Villegas, M.V. Microbiological and Clinical Aspects of Raoultella spp. Front. Public Health 2021, 9, 686789. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.P.; Pradhan, H.R.; Sharma, B.K.; Rijal, S.K. Histamine in Foods: Its Safety and Human Health Implications. J. Food. Sci. Technol. Nepal. 2013, 8, 1–11. [Google Scholar] [CrossRef]
- Kung, H.-F.; Lee, Y.-C.; Huang, Y.-L.; Huang, Y.-R.; Su, Y.-C.; Tsai, Y.-H. Degradation of Histamine by Lactobacillus Plantarum Isolated from Miso Products. J. Food Prot. 2017, 80, 1682–1688. [Google Scholar] [CrossRef] [PubMed]
- Todorov, S.D. Bacteriocins from Lactobacillus Plantarum—Production, Genetic Organization and Mode of Action: Produção, Organização Genética E Modo de Ação. Braz. J. Microbiol. 2009, 40, 209–221. [Google Scholar] [CrossRef]
- Butorac, K.; Banić, M.; Novak, J.; Leboš Pavunc, A.; Uroić, K.; Durgo, K.; Oršolić, N.; Kukolj, M.; Radović, S.; Scalabrin, S.; et al. The Functional Capacity of Plantaricin-Producing Lactobacillus Plantarum SF9C and S-Layer-Carrying Lactobacillus brevis SF9B to Withstand Gastrointestinal Transit. Microb. Cell Fact. 2020, 19, 106. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-C.; Weng, W.-L.; Lai, W.-L.; Tsai, H.-P.; Liu, W.-H.; Lee, M.-H.; Tsai, Y.-C. Effect of Lactobacillus Plantarum Strain K21 on High-Fat Diet-Fed Obese Mice. Evid. Based. Complement. Alternat. Med. 2015, 2015, 391767. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xu, X.; Bi, S.; Pan, X.; Lao, F.; Wu, J. Identification and Validation of Core Microbes Associated with Key Aroma Formation in Fermented Pepper Paste (Capsicum annuum L.). Food Res. Int. 2023, 163, 112194. [Google Scholar] [CrossRef]
- Zagorec, M.; Champomier-Vergès, M.-C. Lactobacillus Sakei: A Starter for Sausage Fermentation, a Protective Culture for Meat Products. Microorganisms 2017, 5, 56. [Google Scholar] [CrossRef]
- Lim, S.-M.; Jeong, J.-J.; Woo, K.H.; Han, M.J.; Kim, D.-H. Lactobacillus Sakei OK67 Ameliorates High-Fat Diet-Induced Blood Glucose Intolerance and Obesity in Mice by Inhibiting Gut Microbiota Lipopolysaccharide Production and Inducing Colon Tight Junction Protein Expression. Nutr. Res. 2016, 36, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.-M.; Han, S.-K.; Kim, J.-K.; Oh, S.-J.; Jang, H.-B.; Kim, D.-H. Lactobacillus Sakei Alleviates High-Fat-Diet-Induced Obesity and Anxiety in Mice by Inducing AMPK Activation and SIRT1 Expression and Inhibiting Gut Microbiota-Mediated NF-κB Activation. Mol. Nutr. Food Res. 2019, 63, e1800978. [Google Scholar] [CrossRef]
- Düz, M.; Doğan, Y.N.; Doğan, İ. Antioxidant Activitiy of Lactobacillus plantarum, Lactobacillus sake and Lactobacillus curvatus Strains Isolated from Fermented Turkish Sucuk. An. Acad. Bras. Cienc. 2020, 92, e20200105. [Google Scholar] [CrossRef]
- Park, D.-Y.; Ahn, Y.-T.; Park, S.-H.; Huh, C.-S.; Yoo, S.-R.; Yu, R.; Sung, M.-K.; McGregor, R.A.; Choi, M.-S. Supplementation of Lactobacillus Curvatus HY7601 and Lactobacillus Plantarum KY1032 in Diet-Induced Obese Mice Is Associated with Gut Microbial Changes and Reduction in Obesity. PLoS ONE 2013, 8, e59470. [Google Scholar] [CrossRef] [PubMed]
- Bock, H.-J.; Lee, N.-K.; Paik, H.-D. Neuroprotective Effects of Heat-Killed Levilactobacillus Brevis KU15152 on H2O2-Induced Oxidative Stress. J. Microbiol. Biotechnol. 2023, 33, 1189–1196. [Google Scholar] [CrossRef]
- Fan, X.; Zhang, Q.; Guo, W.; Wu, Q.; Hu, J.; Cheng, W.; Lü, X.; Rao, P.; Ni, L.; Chen, Y.; et al. The Protective Effects of Levilactobacillus Brevis FZU0713 on Lipid Metabolism and Intestinal Microbiota in Hyperlipidemic Rats. Food Sci. Hum. Wellness 2023, 12, 1646–1659. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, S.-E.; Kim, E.-J.; Seo, S.-H.; Whon, T.W.; Cho, K.-M.; Kwon, S.J.; Roh, S.W.; Son, H.-S. Long-Term Population Dynamics of Viable Microbes in a Closed Ecosystem of Fermented Vegetables. Food Res. Int. 2022, 154, 111044. [Google Scholar] [CrossRef]
- Englezos, V.; Di Gianvito, P.; Peyer, L.; Giacosa, S.; Segade, S.R.; Edwards, N.; Rolle, L.; Rantsiou, K.; Cocolin, L. Bioprotective Effect of Pichia Kluyveri and Lactiplantibacillus Plantarum in Winemaking Conditions. Am. J. Enol. Vitic. 2022, 73, 294–307. [Google Scholar] [CrossRef]
- Vicente, J.; Calderón, F.; Santos, A.; Marquina, D.; Benito, S. High Potential of Pichia Kluyveri and Other Pichia Species in Wine Technology. Int. J. Mol. Sci. 2021, 22, 1196. [Google Scholar] [CrossRef]
- Greppi, A.; Saubade, F.; Botta, C.; Humblot, C.; Guyot, J.-P.; Cocolin, L. Potential Probiotic Pichia Kudriavzevii Strains and Their Ability to Enhance Folate Content of Traditional Cereal-Based African Fermented Food. Food Microbiol. 2017, 62, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Martin, V.; Valera, M.J.; Medina, K.; Boido, E.; Carrau, F. Oenological Impact of the Hanseniaspora/Kloeckera Yeast Genus on Wines—A Review. Fermentation 2018, 4, 76. [Google Scholar] [CrossRef]
- Carlucci, A.; Raimondo, M.L.; Santos, J.; Phillips, A.J.L. Plectosphaerella Species Associated with Root and Collar Rots of Horticultural Crops in Southern Italy. Persoonia 2012, 28, 34–48. [Google Scholar] [CrossRef]
- Bailey, K.; Derby, J.-A.; Bourdôt, G.; Skipp, B.; Cripps, M.; Hurrell, G.; Saville, D.; Noble, A. Plectosphaerella Cucumerina as a Bioherbicide for Cirsium Arvense: Proof of Concept. Biocontrol 2017, 62, 693–704. [Google Scholar] [CrossRef]
- Zhou, J.; Bi, S.; Chen, H.; Chen, T.; Yang, R.; Li, M.; Fu, Y.; Jia, A.-Q. Anti-Biofilm and Antivirulence Activities of Metabolites from Plectosphaerella Cucumerina against Pseudomonas Aeruginosa. Front. Microbiol. 2017, 8, 769. [Google Scholar] [CrossRef] [PubMed]
- Ricucci, D.; Siqueira, J.F., Jr. Biofilms and Apical Periodontitis: Study of Prevalence and Association with Clinical and Histopathologic Findings. J. Endod. 2010, 36, 1277–1288. [Google Scholar] [CrossRef] [PubMed]
- Piraine, R.E.A.; Retzlaf, G.M.; Gonçalves, V.S.; Cunha, R.C.; Conrad, N.L.; Bochman, M.L.; Leite, F.P.L. Brewing and Probiotic Potential Activity of Wild Yeasts Hanseniaspora Uvarum PIT001, Pichia Kluyveri LAR001 and Candida Intermedia ORQ001. Eur. Food Res. Technol. 2023, 249, 133–148. [Google Scholar] [CrossRef]
- Gao, F.; Lv, Y.-W.; Long, J.; Chen, J.-M.; He, J.-M.; Ruan, X.-Z.; Zhu, H.-B. Butyrate Improves the Metabolic Disorder and Gut Microbiome Dysbiosis in Mice Induced by a High-Fat Diet. Front. Pharmacol. 2019, 10, 1040. [Google Scholar] [CrossRef]
- Yu, C.; Liu, S.; Chen, L.; Shen, J.; Niu, Y.; Wang, T.; Zhang, W.; Fu, L. Effect of Exercise and Butyrate Supplementation on Microbiota Composition and Lipid Metabolism. J. Endocrinol. 2019, 243, 125–135. [Google Scholar] [CrossRef]
- Vieira, E.L.M.; Leonel, A.J.; Sad, A.P.; Beltrão, N.R.M.; Costa, T.F.; Ferreira, T.M.R.; Gomes-Santos, A.C.; Faria, A.M.C.; Peluzio, M.C.G.; Cara, D.C.; et al. Oral Administration of Sodium Butyrate Attenuates Inflammation and Mucosal Lesion in Experimental Acute Ulcerative Colitis. J. Nutr. Biochem. 2012, 23, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.; Tang, J.; Feng, Q.; Niu, Z.; Shen, Q.; Wang, L.; Zhou, S. Gamma-Aminobutyric Acid (GABA): A Comprehensive Review of Dietary Sources, Enrichment Technologies, Processing Effects, Health Benefits, and Its Applications. Crit. Rev. Food Sci. Nutr. 2023, 64, 8852–8874. [Google Scholar] [CrossRef] [PubMed]
- Siucinska, E. Γ-Aminobutyric Acid in Adult Brain: An Update. Behav. Brain Res. 2019, 376, 112224. [Google Scholar] [CrossRef] [PubMed]
- Hepsomali, P.; Groeger, J.A.; Nishihira, J.; Scholey, A. Effects of Oral Gamma-Aminobutyric Acid (GABA) Administration on Stress and Sleep in Humans: A Systematic Review. Front. Neurosci. 2020, 14, 923. [Google Scholar] [CrossRef]
- Diez-Gutiérrez, L.; San Vicente, L.; Barrón, L.J.R.; Villarán, M.d.C.; Chávarri, M. Gamma-Aminobutyric Acid and Probiotics: Multiple Health Benefits and Their Future in the Global Functional Food and Nutraceuticals Market. J. Funct. Foods 2020, 64, 103669. [Google Scholar] [CrossRef]
- Bravo, J.A.; Forsythe, P.; Chew, M.V.; Escaravage, E.; Savignac, H.M.; Dinan, T.G.; Bienenstock, J.; Cryan, J.F. Ingestion of Lactobacillus Strain Regulates Emotional Behavior and Central GABA Receptor Expression in a Mouse via the Vagus Nerve. Proc. Natl. Acad. Sci. USA 2011, 108, 16050–16055. [Google Scholar] [CrossRef]
- Cui, Y.; Miao, K.; Niyaphorn, S.; Qu, X. Production of Gamma-Aminobutyric Acid from Lactic Acid Bacteria: A Systematic Review. Int. J. Mol. Sci. 2020, 21, 995. [Google Scholar] [CrossRef] [PubMed]
- Feehily, C.; Karatzas, K.A.G. Role of Glutamate Metabolism in Bacterial Responses towards Acid and Other Stresses. J. Appl. Microbiol. 2013, 114, 11–24. [Google Scholar] [CrossRef]
- Wu, Q.; Shah, N.P. High γ-Aminobutyric Acid Production from Lactic Acid Bacteria: Emphasis on Lactobacillus Brevis as a Functional Dairy Starter. Crit. Rev. Food Sci. Nutr. 2017, 57, 3661–3672. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; Dinan, T.G. Mind-Altering Microorganisms: The Impact of the Gut Microbiota on Brain and Behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef]
- Sudo, N.; Chida, Y.; Aiba, Y.; Sonoda, J.; Oyama, N.; Yu, X.-N.; Kubo, C.; Koga, Y. Postnatal Microbial Colonization Programs the Hypothalamic-Pituitary-Adrenal System for Stress Response in Mice. J. Physiol. 2004, 558, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Lyte, M.; Li, W.; Opitz, N.; Gaykema, R.P.A.; Goehler, L.E. Induction of Anxiety-like Behavior in Mice during the Initial Stages of Infection with the Agent of Murine Colonic Hyperplasia Citrobacter Rodentium. Physiol. Behav. 2006, 89, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Zorumski, C.F.; Paul, S.M.; Covey, D.F.; Mennerick, S. Neurosteroids as Novel Antidepressants and Anxiolytics: GABA-A Receptors and beyond. Neurobiol. Stress. 2019, 11, 100196. [Google Scholar] [CrossRef] [PubMed]
- Petty, F. GABA and Mood Disorders: A Brief Review and Hypothesis. J. Affect. Disord. 1995, 34, 275–281. [Google Scholar] [CrossRef]
- van de Wouw, M.; Walsh, A.M.; Crispie, F.; van Leuven, L.; Lyte, J.M.; Boehme, M.; Clarke, G.; Dinan, T.G.; Cotter, P.D.; Cryan, J.F. Distinct Actions of the Fermented Beverage Kefir on Host Behaviour, Immunity and Microbiome Gut-Brain Modules in the Mouse. Microbiome 2020, 8, 67. [Google Scholar] [CrossRef] [PubMed]
- Abildgaard, A.; Elfving, B.; Hokland, M.; Wegener, G.; Lund, S. Probiotic Treatment Reduces Depressive-like Behaviour in Rats Independently of Diet. Psychoneuroendocrinology 2017, 79, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Dhaliwal, J.; Singh, D.P.; Singh, S.; Pinnaka, A.K.; Boparai, R.K.; Bishnoi, M.; Kondepudi, K.K.; Chopra, K. Lactobacillus Plantarum MTCC 9510 Supplementation Protects from Chronic Unpredictable and Sleep Deprivation-Induced Behaviour, Biochemical and Selected Gut Microbial Aberrations in Mice. J. Appl. Microbiol. 2018, 125, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, D.; Nigam, P.S. Probiotics, Prebiotics, Synbiotics, and Fermented Foods as Potential Biotics in Nutrition Improving Health via Microbiome-Gut-Brain Axis. Fermentation 2022, 8, 303. [Google Scholar] [CrossRef]
- Huma, N.; Davison, S.; Guse, K.; Walker, G.; Rutschke, S.; Sackett, A.; Blanco, G.; Damian, J.P.; Faulk, C.; Gomez, A. Regular Consumption of Kombucha Alleviates Depression Symptoms and Modulates the Gut Microbiome in Mice. Curr. Dev. Nutr. 2024, 8, 102874. [Google Scholar] [CrossRef]
- Hornick, S.B. Factors Affecting the Nutritional Quality of Crops. Am. J. Altern. Agric. 1992, 7, 63–68. [Google Scholar] [CrossRef]
- Panthee, B.; Gyawali, S.; Panthee, P.; Techato, K. Environmental and Human Microbiome for Health. Life 2022, 12, 456. [Google Scholar] [CrossRef] [PubMed]
- Bodor, A.; Bounedjoum, N.; Vincze, G.E.; Erdeiné Kis, Á.; Laczi, K.; Bende, G.; Szilágyi, Á.; Kovács, T.; Perei, K.; Rákhely, G. Challenges of Unculturable Bacteria: Environmental Perspectives. Rev. Environ. Sci. Technol. 2020, 19, 1–22. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guse, K.; Mao, Q.; Chen, C.; Gomez, A. Meta-Omics Analyses of Conventional and Regenerative Fermented Vegetables: Is There an Impact on Health-Boosting Potential? Fermentation 2025, 11, 22. https://doi.org/10.3390/fermentation11010022
Guse K, Mao Q, Chen C, Gomez A. Meta-Omics Analyses of Conventional and Regenerative Fermented Vegetables: Is There an Impact on Health-Boosting Potential? Fermentation. 2025; 11(1):22. https://doi.org/10.3390/fermentation11010022
Chicago/Turabian StyleGuse, Kylene, Qingqing Mao, Chi Chen, and Andres Gomez. 2025. "Meta-Omics Analyses of Conventional and Regenerative Fermented Vegetables: Is There an Impact on Health-Boosting Potential?" Fermentation 11, no. 1: 22. https://doi.org/10.3390/fermentation11010022
APA StyleGuse, K., Mao, Q., Chen, C., & Gomez, A. (2025). Meta-Omics Analyses of Conventional and Regenerative Fermented Vegetables: Is There an Impact on Health-Boosting Potential? Fermentation, 11(1), 22. https://doi.org/10.3390/fermentation11010022








