Enrichment of Rumen Solid-Phase Bacteria for Production of Volatile Fatty Acids by Long-Term Subculturing In Vitro
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Experimental Design
2.3. Microbial Sequencing Analysis
2.4. Statistical Analysis
3. Results
3.1. pH and Gas Production During In Vitro Subculturing
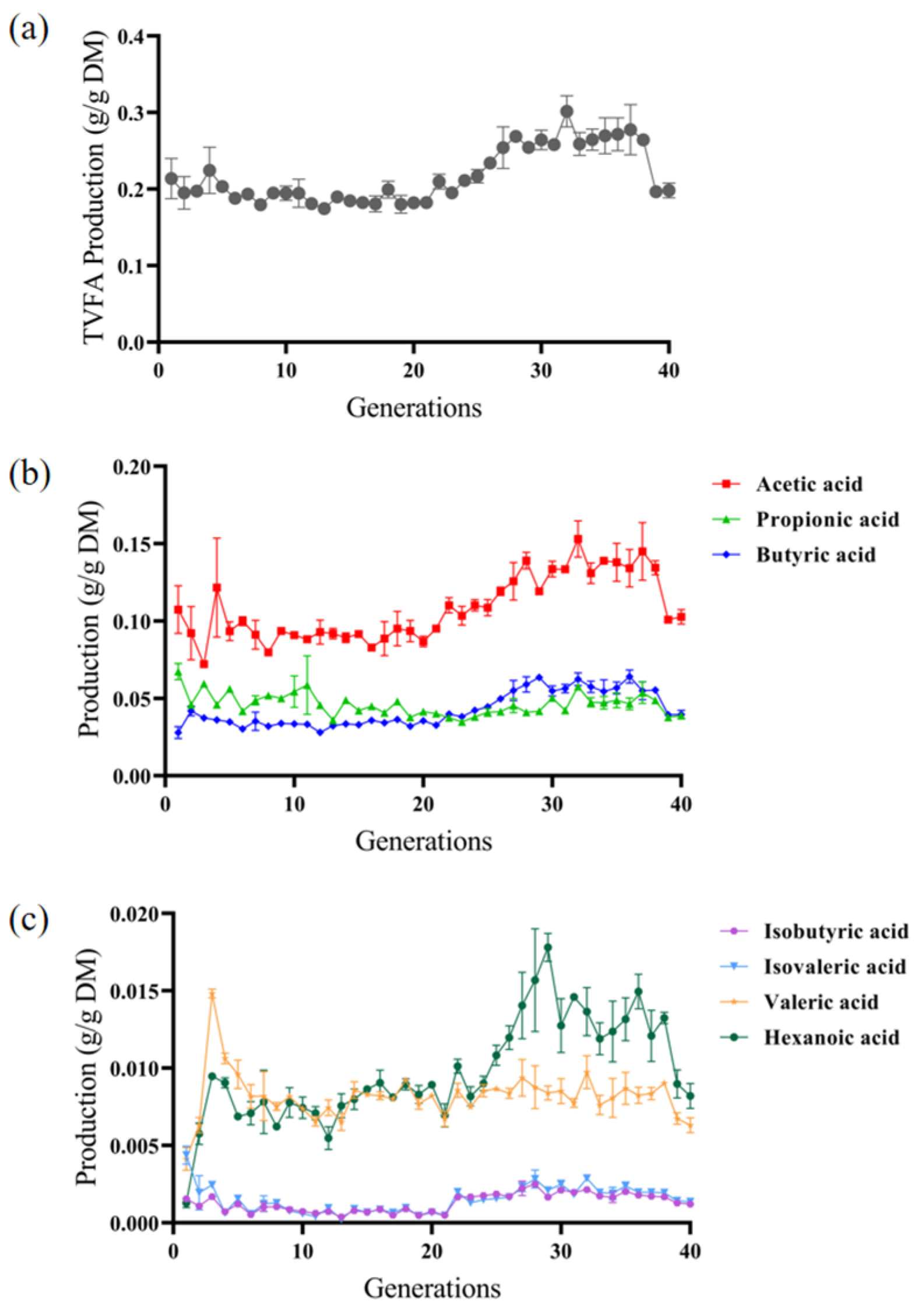
3.2. VFA Production During In Vitro Subculturing
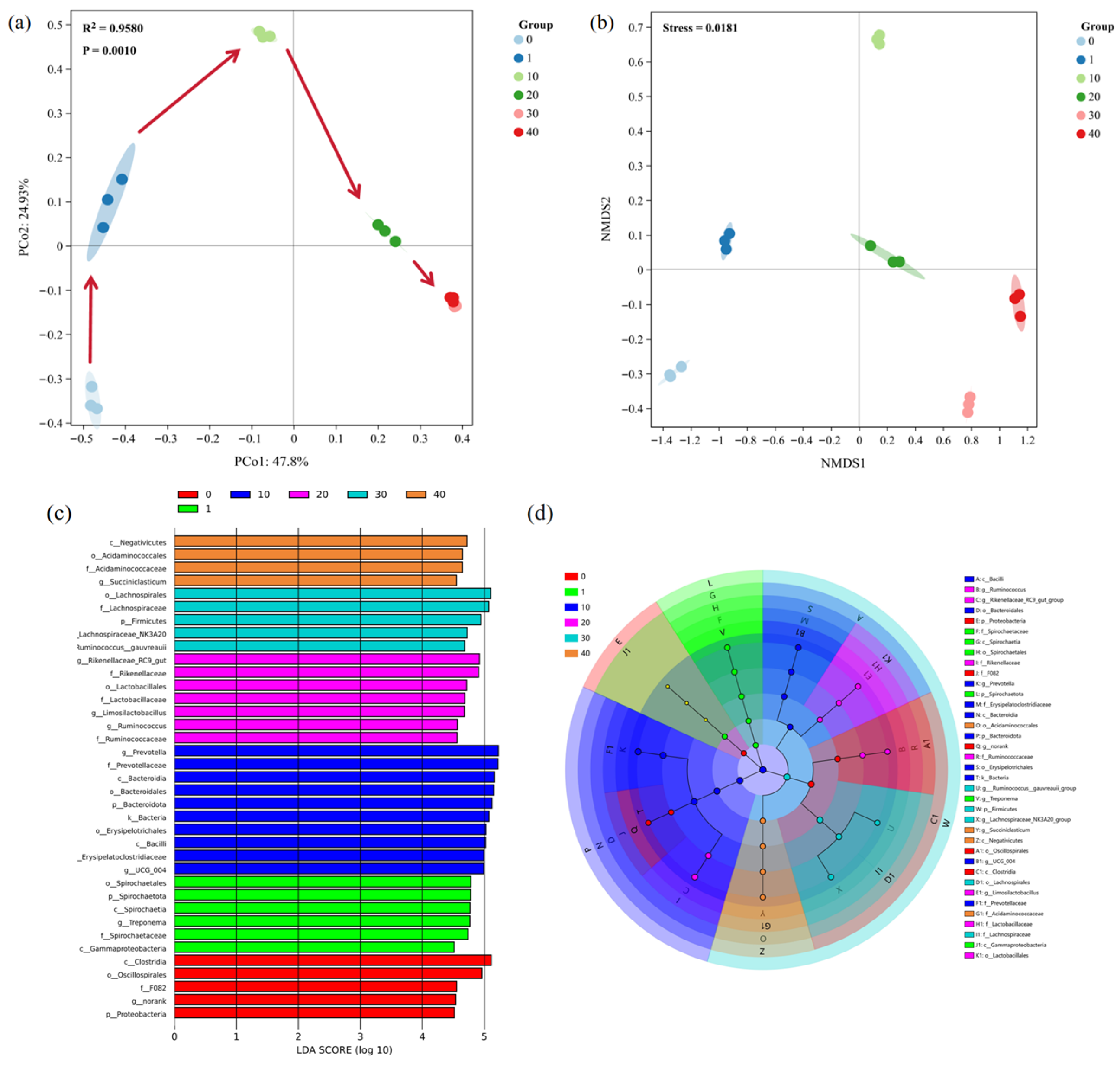
3.3. Analysis of Bacterial Communities During In Vitro Subculturing
3.3.1. Analysis of Alpha and Beta Diversity
3.3.2. Changes at the Phylum Level
3.3.3. Changes at the Genus Level
3.3.4. Effect of Bacterial Communities on VFA Production
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nguyen, L.N.; Nguyen, A.Q.; Johir, M.A.H.; Guo, W.; Ngo, H.H.; Chaves, A.V.; Nghiem, L.D. Application of Rumen and Anaerobic Sludge Microbes for Bio Harvesting from Lignocellulosic Biomass. Chemosphere 2019, 228, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, S.; Rapoport, A.; Singh, B. Biotechnological Potential of Lignocellulosic Biomass as Substrates for Fungal Xylanases and Its Bioconversion into Useful Products: A Review. Fermentation 2024, 10, 82. [Google Scholar] [CrossRef]
- Rashmi, R.; Tripathi, T.; Pandey, S.; Kumar, S. Transforming Lignocellulosic Biomass into Biofuels: Recent Innovations in Pretreatment and Bioconversion Techniques. Int. J. Res. Appl. Sci. Eng. Technol. 2024, 12, 1079–1089. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Y.; Sun, P.; Peng, C. A Review on Renewable Energy: Conversion and Utilization of Biomass. Smart Mol. 2024, 2, e20240019. [Google Scholar] [CrossRef]
- Dong, W.; Yang, Y.; Liu, C.; Zhang, J.; Pan, J.; Luo, L.; Wu, G.; Awasthi, M.K.; Yan, B. Caproic Acid Production from Anaerobic Fermentation of Organic Waste—Pathways and Microbial Perspective. Renew. Sustain. Energy Rev. 2023, 175, 113181. [Google Scholar] [CrossRef]
- Bhujbal, S.K.; Ghosh, P.; Vijay, V.K.; Rathour, R.; Kumar, M.; Singh, L.; Kapley, A. Biotechnological Potential of Rumen Microbiota for Sustainable Bioconversion of Lignocellulosic Waste to Biofuels and Value-Added Products. Sci. Total Environ. 2022, 814, 152773. [Google Scholar] [CrossRef]
- Liang, J.; Zhang, P.; Zhang, R.; Chang, J.; Chen, L.; Wang, G.; Tian, Y.; Zhang, G. Response of Rumen Microorganisms to pH during Anaerobic Hydrolysis and Acidogenesis of Lignocellulose Biomass. Waste Manag. 2024, 174, 476–486. [Google Scholar] [CrossRef]
- Zhao, W.; Abdelsattar, M.M.; Wang, X.; Zhang, N.; Chai, J. In Vitro Modulation of Rumen Fermentation by Microbiota from the Recombination of Rumen Fluid and Solid Phases. Microbiol. Spectr. 2023, 11, e03387-22. [Google Scholar] [CrossRef]
- Bowen, J.M.; McCabe, M.S.; Lister, S.J.; Cormican, P.; Dewhurst, R.J. Evaluation of Microbial Communities Associated with the Liquid and Solid Phases of the Rumen of Cattle Offered a Diet of Perennial Ryegrass or White Clover. Front. Microbiol. 2018, 9, 2389. [Google Scholar] [CrossRef]
- Su, M.; Hao, Z.; Shi, H.; Li, T.; Wang, H.; Li, Q.; Zhang, Y.; Ma, Y. Metagenomic Analysis Revealed Differences in Composition and Function Between Liquid-Associated and Solid-Associated Microorganisms of Sheep Rumen. Front. Microbiol. 2022, 13, 851567. [Google Scholar] [CrossRef]
- Wang, P.; Qi, M.; Barboza, P.; Leigh, M.B.; Ungerfeld, E.; Selinger, L.B.; McAllister, T.A.; Forster, R.J. Isolation of High-Quality Total RNA from Rumen Anaerobic Bacteria and Fungi, and Subsequent Detection of Glycoside Hydrolases. Can. J. Microbiol. 2011, 57, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, P.; Ye, J.; Wu, Y.; Fang, W.; Gou, X.; Zeng, G. Improvement of Methane Production from Rice Straw with Rumen Fluid Pretreatment: A Feasibility Study. Int. Biodeterior. Biodegrad. 2016, 113, 9–16. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, G.; Zhang, P.; Ma, X.; Li, F.; Zhang, H.; Tao, X.; Ye, J.; Nabi, M. Rumen Fluid Fermentation for Enhancement of Hydrolysis and Acidification of Grass Clipping. J. Environ. Manag. 2018, 220, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Fang, W.; Chang, J.; Zhang, G.; Ma, W.; Nabi, M.; Zubair, M.; Zhang, R.; Chen, L.; Huang, J.; et al. Long-Term Rumen Microorganism Fermentation of Corn Stover In Vitro for Volatile Fatty Acid Production. Bioresour. Technol. 2022, 358, 127447. [Google Scholar] [CrossRef]
- American Public Health Association (APHA). Standard Methods for the Examination of Water and Wastewater, 21st ed.; American Public Health Association: Washington, DC, USA, 2005; 1220p. [Google Scholar]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for Dietary Fiber, Neutral Detergent Fiber, and Nonstarch Polysaccharides in Relation to Animal Nutrition. J. Dairy. Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Lin, M.; Dai, X.; Weimer, P.J. Shifts in Fermentation End Products and Bacterial Community Composition in Long-Term, Sequentially Transferred In Vitro Ruminal Enrichment Cultures Fed Switchgrass with and without Ethanol as a Co-Substrate. Bioresour. Technol. 2019, 285, 121324. [Google Scholar] [CrossRef]
- Menke, K.H.; Raab, L.; Salewski, A.; Steingass, H.; Fritz, D.; Schneider, W. The Estimation of the Digestibility and Metabolizable Energy Content of Ruminant Feedingstuffs from the Gas Production When They Are Incubated with Rumen Liquor In Vitro. J. Agric. Sci. 1979, 93, 217–222. [Google Scholar] [CrossRef]
- Nguyen, A.Q.; Nguyen, L.N.; Johir, M.A.H.; Ngo, H.-H.; Chaves, A.V.; Nghiem, L.D. Derivation of Volatile Fatty Acid from Crop Residues Digestion Using a Rumen Membrane Bioreactor: A Feasibility Study. Bioresour. Technol. 2020, 312, 123571. [Google Scholar] [CrossRef]
- Jin, W.; Xu, X.; Yang, F. Application of Rumen Microorganisms for Enhancing Biogas Production of Corn Straw and Livestock Manure in a Pilot-Scale Anaerobic Digestion System: Performance and Microbial Community Analysis. Energies 2018, 11, 920. [Google Scholar] [CrossRef]
- Ozbayram, E.G.; Kleinsteuber, S.; Nikolausz, M.; Ince, B.; Ince, O. Effect of Bioaugmentation by Cellulolytic Bacteria Enriched from Sheep Rumen on Methane Production from Wheat Straw. Anaerobe 2017, 46, 122–130. [Google Scholar] [CrossRef]
- Moraïs, S.; Mizrahi, I. Islands in the Stream: From Individual to Communal Fiber Degradation in the Rumen Ecosystem. FEMS Microbiol. Rev. 2019, 43, 362–379. [Google Scholar] [CrossRef] [PubMed]
- Huws, S.A.; Edwards, J.E.; Lin, W.; Rubino, F.; Alston, M.; Swarbreck, D.; Caim, S.; Stevens, P.R.; Pachebat, J.; Won, M.-Y.; et al. Microbiomes Attached to Fresh Perennial Ryegrass Are Temporally Resilient and Adapt to Changing Ecological Niches. Microbiome 2021, 9, 143. [Google Scholar] [CrossRef]
- Accetto, T.; Avguštin, G. Non-Oral Prevotella Stepping into the Spotlight. Anaerobe 2021, 68, 102321. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.A.; Yang, C.; Zhang, J.; Kalwar, Q.; Liang, Z.; Li, C.; Du, M.; Yan, P.; Long, R.; Han, J.; et al. Effects of Dietary Energy Levels on Rumen Fermentation, Microbial Diversity, and Feed Efficiency of Yaks (Bos grunniens). Front. Microbiol. 2020, 11, 625. [Google Scholar] [CrossRef]
- Gharechahi, J.; Vahidi, M.F.; Bahram, M.; Han, J.-L.; Ding, X.-Z.; Salekdeh, G.H. Metagenomic Analysis Reveals a Dynamic Microbiome with Diversified Adaptive Functions to Utilize High Lignocellulosic Forages in the Cattle Rumen. ISME J. 2021, 15, 1108–1120. [Google Scholar] [CrossRef]
- Ozbayram, E.G.; Kleinsteuber, S.; Nikolausz, M. Biotechnological Utilization of Animal Gut Microbiota for Valorization of Lignocellulosic Biomass. Appl. Microbiol. Biotechnol. 2020, 104, 489–508. [Google Scholar] [CrossRef]
- Won, M.-Y.; Oyama, L.B.; Courtney, S.J.; Creevey, C.J.; Huws, S.A. Can Rumen Bacteria Communicate to Each Other? Microbiome 2020, 8, 23. [Google Scholar] [CrossRef]
- Sizova, M.V.; Muller, P.A.; Stancyk, D.; Panikov, N.S.; Mandalakis, M.; Hazen, A.; Hohmann, T.; Doerfert, S.N.; Fowle, W.; Earl, A.M.; et al. Oribacterium parvum sp. Nov. and Oribacterium asaccharolyticum sp. Nov., Obligately Anaerobic Bacteria from the Human Oral Cavity, and Emended Description of the Genus Oribacterium. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 8, 2642–2649. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Plugge, C.M.; Akkermans, A.D.L.; De Vos, W.M. Victivallis vadensis Gen. Nov., sp. Nov., a Sugar-Fermenting Anaerobe from Human Faeces. Int. J. Syst. Evol. Microbiol. 2003, 53, 211–215. [Google Scholar] [CrossRef]
- Zhang, L.; Chung, J.; Jiang, Q.; Sun, R.; Zhang, J.; Zhong, Y.; Ren, N. Characteristics of Rumen Microorganisms Involved in Anaerobic Degradation of Cellulose at Various pH Values. RSC Adv. 2017, 7, 40303–40310. [Google Scholar] [CrossRef]
- Liang, J.; Nabi, M.; Zhang, P.; Zhang, G.; Cai, Y.; Wang, Q.; Zhou, Z.; Ding, Y. Promising Biological Conversion of Lignocellulosic Biomass to Renewable Energy with Rumen Microorganisms: A Comprehensive Review. Renew. Sustain. Energy Rev. 2020, 134, 110335. [Google Scholar] [CrossRef]
- Meng, Y.; Mumme, J.; Xu, H.; Wang, K. A Biologically Inspired Variable-pH Strategy for Enhancing Short-Chain Fatty Acids (SCFAs) Accumulation in Maize Straw Fermentation. Bioresour. Technol. 2016, 201, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Dong, A.; Xu, Y.; Wu, Q.; Lambo, M.T.; Zhang, Y.; Dou, X.; Li, Y. Regulatory Effects of High Concentrate Diet Synergistically Fermented with Cellulase and Lactic Acid Bacteria: In Vitro Ruminal Fermentation, Methane Production, and Rumen Microbiome. Anim. Feed. Sci. Technol. 2025, 319, 116194. [Google Scholar] [CrossRef]
- Wainaina, S.; Lukitawesa; Kumar Awasthi, M.; Taherzadeh, M.J. Bioengineering of Anaerobic Digestion for Volatile Fatty Acids, Hydrogen or Methane Production: A Critical Review. Bioengineered 2019, 10, 437–458. [Google Scholar] [CrossRef]


| Items | Composition |
|---|---|
| Liquid A | Mix and dissolve CaCl2·2H2O 13.2 g, MnCl2·4H2O 10.0 g, CoCl2·6H2O 1.0 g, and FeCl3·6H2O 8.0 g in distilled water at constant volume to 100 mL |
| Solution B | Dissolve 4.0 g NH4HCO3 and 35 g NaHCO3 in 1000 mL distilled water |
| Liquid C | Dissolve 5.7 g NaH2PO4, 6.2 g KH2PO4, and 0.6 g MgSO4·7H2O in distilled water at constant volume to 1000 mL |
| Azurazine solution | 0.1% (w/v) |
| Mineral salt incubation buffer was configured from sterilized ultrapure water 400 mL + 0.1 mL solution A + 200 mL solution B + 200 mL solution C + 1 mL azurazine solution, and CO2 was passed continuously for 3 h. | |
| Items | 10 | 20 | 30 | 40 | SEM | p-Value |
|---|---|---|---|---|---|---|
| Proportions (%) | ||||||
| CO2 | 82.53 c | 90.40 a | 86.62 b | 91.40 a | 1.14 | <0.01 |
| CH4 | 9.81 a | 7.76 b | 9.80 a | 4.00 c | 0.73 | <0.01 |
| H2 | 0.4832 b | 0.0026 b | 0.8512 ab | 1.7413 a | 0.25 | <0.05 |
| Production (mL/g DM) | ||||||
| CO2 | 72.35 b | 74.54 b | 80.84 a | 82.79 a | 1.33 | <0.01 |
| CH4 | 8.59 a | 6.39 b | 9.15 a | 3.62 c | 0.67 | <0.01 |
| H2 | 0.4322 b | 0.0021 b | 0.7851 ab | 1.5762 a | 0.22 | <0.05 |
| Index | 0 | 1 | 10 | 20 | 30 | 40 | SEM | p-Value |
|---|---|---|---|---|---|---|---|---|
| Chao 1 | 3305.68 a | 3248.24 a | 1974.42 b | 2012.68 b | 1394.19 c | 999.01 d | 212.50 | <0.01 |
| ACE | 3310.05 a | 3289.62 a | 2079.14 b | 2159.06 b | 1472.94 c | 1020.70 d | 209.53 | <0.01 |
| Shannon | 6.38 a | 5.54 b | 3.87 d | 4.35 c | 4.29 c | 4.82 c | 0.21 | <0.01 |
| Simpson | 0.99 a | 0.96 b | 0.89 c | 0.96 b | 0.97 b | 0.97 b | 0.01 | <0.01 |
| Phylum | 0 | 1 | 10 | 20 | 30 | 40 | SEM | p-Value |
|---|---|---|---|---|---|---|---|---|
| Firmicutes | 47.47 bc | 36.46 d | 39.91 cd | 50.02 ab | 57.57 a | 55.71 ab | 2.07 | <0.01 |
| Bacteroidota | 32.51 bc | 39.19 b | 52.72 a | 38.83 b | 26.83 d | 28.96 d | 2.22 | <0.01 |
| Proteobacteria | 8.25 a | 8.00 a | 1.00 b | 1.06 b | 2.23 b | 1.80 b | 0.98 | <0.05 |
| Spirochaetota | 1.19 b | 13.60 a | 2.72 b | 2.10 b | 1.94 b | 1.96 b | 1.07 | <0.01 |
| Actinobacteriota | 1.44 b | 0.43 b | 1.23 b | 4.82 a | 5.00 a | 6.65 a | 0.60 | <0.01 |
| Desulfobacterota | 1.95 b | 0.29 c | 0.37 c | 0.98 bc | 3.77 a | 3.55 a | 0.36 | <0.01 |
| Verrucomicrobiota | 4.07 a | 1.15 bc | 0.07 e | 0.85 cd | 1.63 b | 0.54 de | 0.32 | <0.01 |
| Synergistota | 0.32 bc | 0.12 c | 0.67 b | 1.21 a | 0.76 b | 0.38 bc | 0.01 | <0.01 |
| Fibrobacterota | 0.13 bc | 0.25 bc | 1.19 a | 0.06 c | 0.21 bc | 0.31 b | 0.10 | <0.01 |
| Planctomycetota | 1.07 a | 0.08 b | 0.01 b | 0.02 b | 0.01 b | 0.01 b | 0.10 | <0.01 |
| Phylum | Family | Genus | 0 | 1 | 10 | 20 | 30 | 40 | SEM | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|
| Firmicutes | Ruminococcaceae | Ruminococcus | 6.55 b | 2.17 c | 0.16 d | 8.27 a | 0.08 d | 0.01 d | 0.82 | <0.01 |
| Christensenellaceae | Christensenellaceae R-7 group | 5.97 a | 3.20 b | 0.71 d | 1.63 c | 0.08 e | 0.09 e | 0.51 | <0.01 | |
| Oscillospiraceae | NK4A214 group | 2.15 a | 0.70 bc | 1.48 ab | 0.76 bc | 0.37 c | 0.58 c | 0.18 | <0.01 | |
| UCG-002 | 1.79 a | 0.27 b | 0.04 c | 0.06 c | 0.00 c | 0.00 c | 0.16 | <0.01 | ||
| Acidaminococcaceae | Succiniclasticum | 2.14 b | 2.06 b | 1.43 b | 7.08 a | 7.15 a | 9.75 a | 0.83 | <0.01 | |
| Acidaminococcus | 0.00 d | 0.02 d | 0.08 cd | 0.41 c | 1.47 b | 2.43 a | 0.26 | <0.01 | ||
| Hungateiclostridiaceae | Saccharofermentans | 1.21 b | 1.52 ab | 0.07 c | 1.79 a | 0.03 c | 0.00 c | 0.19 | <0.01 | |
| Lachnospiraceae | Eubacterium ruminantium group | 1.12 b | 2.35 a | 0.06 c | 0.26 c | 0.03 c | 0.00 c | 0.22 | <0.01 | |
| Acetitomaculum | 0.79 c | 0.10 d | 0.10 d | 0.58 cd | 3.18 b | 3.82 a | 0.37 | <0.01 | ||
| Lachnospiraceae NK3A20 group | 0.74 cd | 0.47 d | 0.43 d | 1.74 c | 12.22 a | 9.40 b | 0.17 | <0.01 | ||
| Butyrivibrio | 0.55 c | 1.28 a | 0.97 b | 0.30 cd | 0.04 d | 0.00 d | 0.12 | <0.01 | ||
| Lachnospiraceae NK4A136 group | 0.25 b | 1.25 a | 1.30 a | 0.16 b | 0.05 b | 0.01 b | 0.14 | <0.01 | ||
| Oribacterium | 0.24 c | 1.70 b | 0.34 c | 0.17 c | 3.33 a | 2.06 b | 0.29 | <0.01 | ||
| probable genus 10 | 0.19 c | 1.30 a | 0.04 c | 0.68 b | 0.02 c | 0.00 c | 0.12 | <0.01 | ||
| Lachnospiraceae FCS020 group | 0.17 c | 1.29 a | 0.05 c | 0.64 b | 0.04 c | 0.01 c | 0.12 | <0.01 | ||
| Ruminococcus gauvreauii group | 0.13 c | 0.11 c | 0.30 c | 0.62 c | 10.20 a | 7.53 b | 1.00 | <0.01 | ||
| Syntrophococcus | 0.06 c | 0.06 c | 0.30 c | 0.57 b | 2.71 a | 2.84 a | 0.29 | <0.01 | ||
| Streptococcaceae | Streptococcus | 0.36 b | 4.48 a | 0.13 b | 0.31 b | 0.04 b | 0.00 b | 0.39 | <0.01 | |
| Erysipelotrichaceae | Erysipelotrichaceae UCG-009 | 0.15 b | 0.08 b | 1.03 a | 1.50 a | 0.10 b | 0.11 b | 0.15 | <0.01 | |
| UCG-004 | 0.12 c | 0.29 c | 20.67 a | 0.85 c | 3.86 b | 3.60 b | 1.77 | <0.01 | ||
| Anaerovoracaceae | Eubacterium nodatum group | 0.09 d | 0.04 d | 0.12 d | 0.51 c | 0.92 b | 1.27 a | 0.11 | <0.01 | |
| Lactobacillaceae | Limosilactobacillus | 0.08 b | 0.05 b | 0.73 b | 10.40 a | 0.10 b | 0.04 b | 0.93 | <0.01 | |
| Erysipelotrichaceae | Solobacterium | 0.08 c | 0.03 c | 0.15 c | 1.74 b | 1.51 b | 2.16 a | 0.22 | <0.01 | |
| Veillonellaceae | Megasphaera | 0.03 d | 0.65 b | 0.03 d | 0.30 c | 0.71 b | 1.23 a | 0.11 | <0.01 | |
| Clostridiaceae | Clostridium sensu stricto 1 | 0.03 b | 0.07 b | 1.26 b | 1.59 b | 5.32 a | 4.83 a | 0.54 | <0.01 | |
| Bacteroidota | Rikenellaceae | Rikenellaceae RC9 gut group | 11.03 b | 6.00 d | 5.13 d | 22.83 a | 8.77 bc | 6.94 cd | 1.48 | <0.01 |
| Prevotellaceae | Prevotella | 5.60 c | 25.84 b | 39.55 a | 8.27 c | 4.27 c | 7.86 c | 3.25 | <0.01 | |
| Prevotella_7 | 0.03 d | 0.18 d | 4.03 a | 0.46 d | 2.42 b | 1.63 c | 0.35 | <0.01 | ||
| Bacteroidaceae | Bacteroides | 0.05 c | 0.12 c | 1.39 b | 1.27 b | 4.55 a | 4.59 a | 0.47 | <0.01 | |
| Proteobacteria | Moraxellaceae | Acinetobacter | 5.18 a | 0.14 b | 0.00 b | 0.04 b | 0.00 b | 0.01 b | 0.48 | <0.01 |
| Succinivibrionaceae | Succinivibrio | 0.52 b | 1.46 a | 0.06 b | 0.05 b | 0.01 b | 0.01 b | 0.15 | <0.01 | |
| Ruminobacter | 0.44 b | 5.46 a | 0.13 b | 0.16 b | 0.01 b | 0.00 b | 0.71 | 0.15 | ||
| Verrucomicrobiota | Victivallaceae | Victivallis | 0.04 b | 0.02 b | 0.02 b | 0.02 b | 1.55 a | 0.27 b | 0.14 | <0.01 |
| Desulfobacterota | Desulfovibrionaceae | Desulfovibrio | 1.82 b | 0.22 c | 0.36 c | 0.97 bc | 3.77 a | 3.55 a | 0.37 | <0.01 |
| Actinobacteriota | Atopobiaceae | Olsenella | 0.12 c | 0.13 c | 1.01 c | 3.24 b | 3.24 b | 4.98 a | 0.47 | <0.01 |
| Spirochaetota | Spirochaetaceae | Treponema | 1.02 c | 13.59 a | 2.68 b | 2.09 bc | 1.94 bc | 1.95 bc | 1.07 | <0.01 |
| Synergistota | Synergistaceae | Pyramidobacter | 0.08 c | 0.11 bc | 0.50 b | 1.04 a | 0.34 bc | 0.18 bc | 0.09 | <0.01 |
| Fibrobacterota | Fibrobacteraceae | Fibrobacter | 0.13 bc | 0.25 bc | 1.19 a | 0.06 c | 0.21 bc | 0.31 b | 0.10 | <0.01 |
| Tenericutes | Anaeroplasmataceae | Anaeroplasma | 0.04 c | 0.76 b | 1.91 a | 0.20 c | 0.18 c | 0.30 c | 0.16 | <0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Cheng, Z.; Zong, Y.; Shen, Y.; Jama, S.M.; Lin, M. Enrichment of Rumen Solid-Phase Bacteria for Production of Volatile Fatty Acids by Long-Term Subculturing In Vitro. Fermentation 2025, 11, 173. https://doi.org/10.3390/fermentation11040173
Liu W, Cheng Z, Zong Y, Shen Y, Jama SM, Lin M. Enrichment of Rumen Solid-Phase Bacteria for Production of Volatile Fatty Acids by Long-Term Subculturing In Vitro. Fermentation. 2025; 11(4):173. https://doi.org/10.3390/fermentation11040173
Chicago/Turabian StyleLiu, Wengboyang, Zhiqiang Cheng, Yujie Zong, Yue Shen, Shakib Mohamed Jama, and Miao Lin. 2025. "Enrichment of Rumen Solid-Phase Bacteria for Production of Volatile Fatty Acids by Long-Term Subculturing In Vitro" Fermentation 11, no. 4: 173. https://doi.org/10.3390/fermentation11040173
APA StyleLiu, W., Cheng, Z., Zong, Y., Shen, Y., Jama, S. M., & Lin, M. (2025). Enrichment of Rumen Solid-Phase Bacteria for Production of Volatile Fatty Acids by Long-Term Subculturing In Vitro. Fermentation, 11(4), 173. https://doi.org/10.3390/fermentation11040173






