Metabolic Engineering of Zymomonas mobilis for Xylonic Acid Production from Lignocellulosic Hydrolysate
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Growth Conditions
2.2. Construction of Plasmid and Transformation
2.3. Construction of Stable Xylonic Acid Producer
2.4. Batch Fermentation for Xylonic Production
2.5. Analytical Procedures
2.6. Statistical Analysis
3. Results and Discussion
3.1. Expression of Xylose Dehydrogenase Using Plasmid for Xylonic Acid Production in Z. mobilis
3.2. Construction of Stable Heterologous Xylonic Acid Producing Strains by CRISPR-Cas System
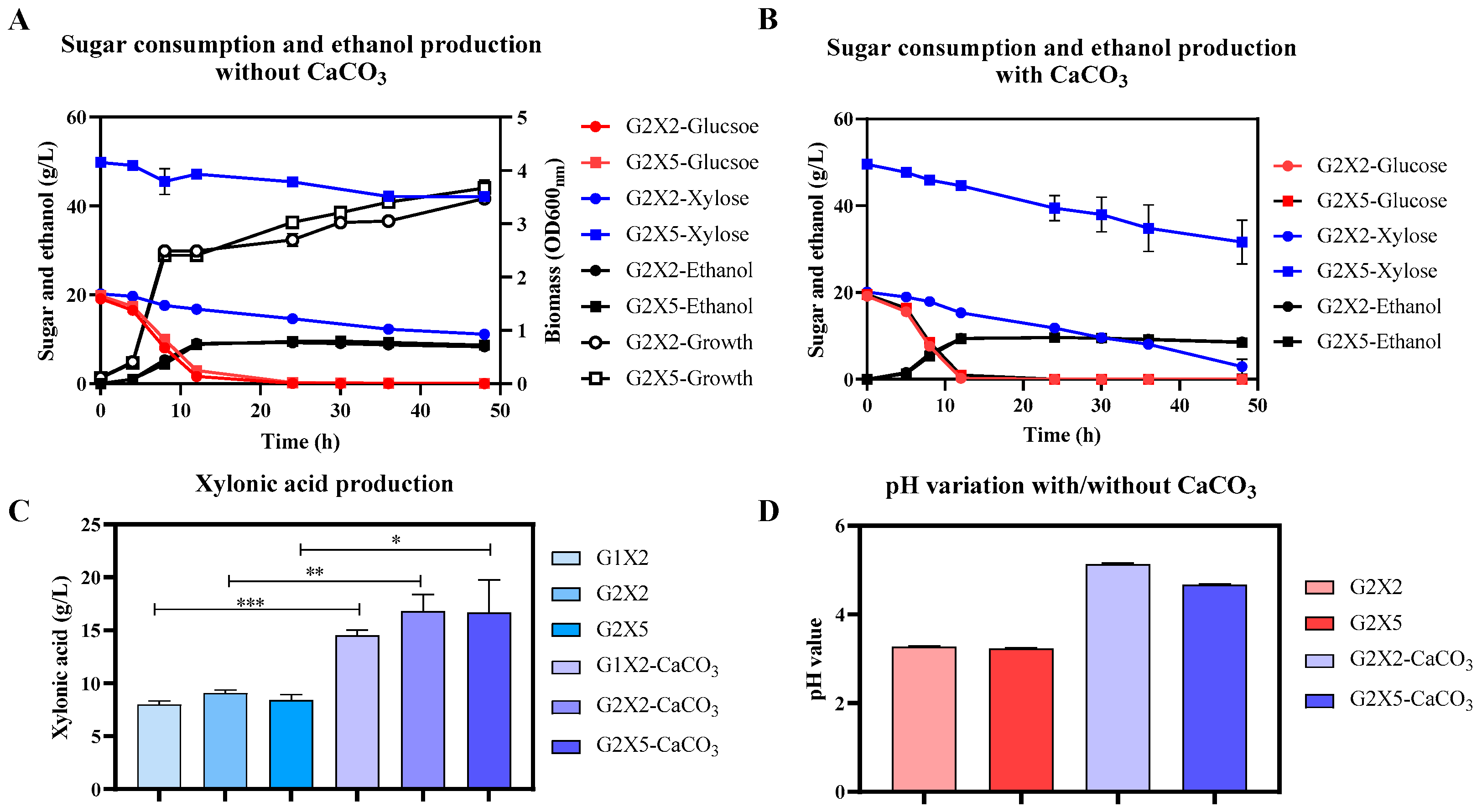
3.3. The Effect of Oxygen and High Cell Density on Xylonic Acid and Ethanol Production
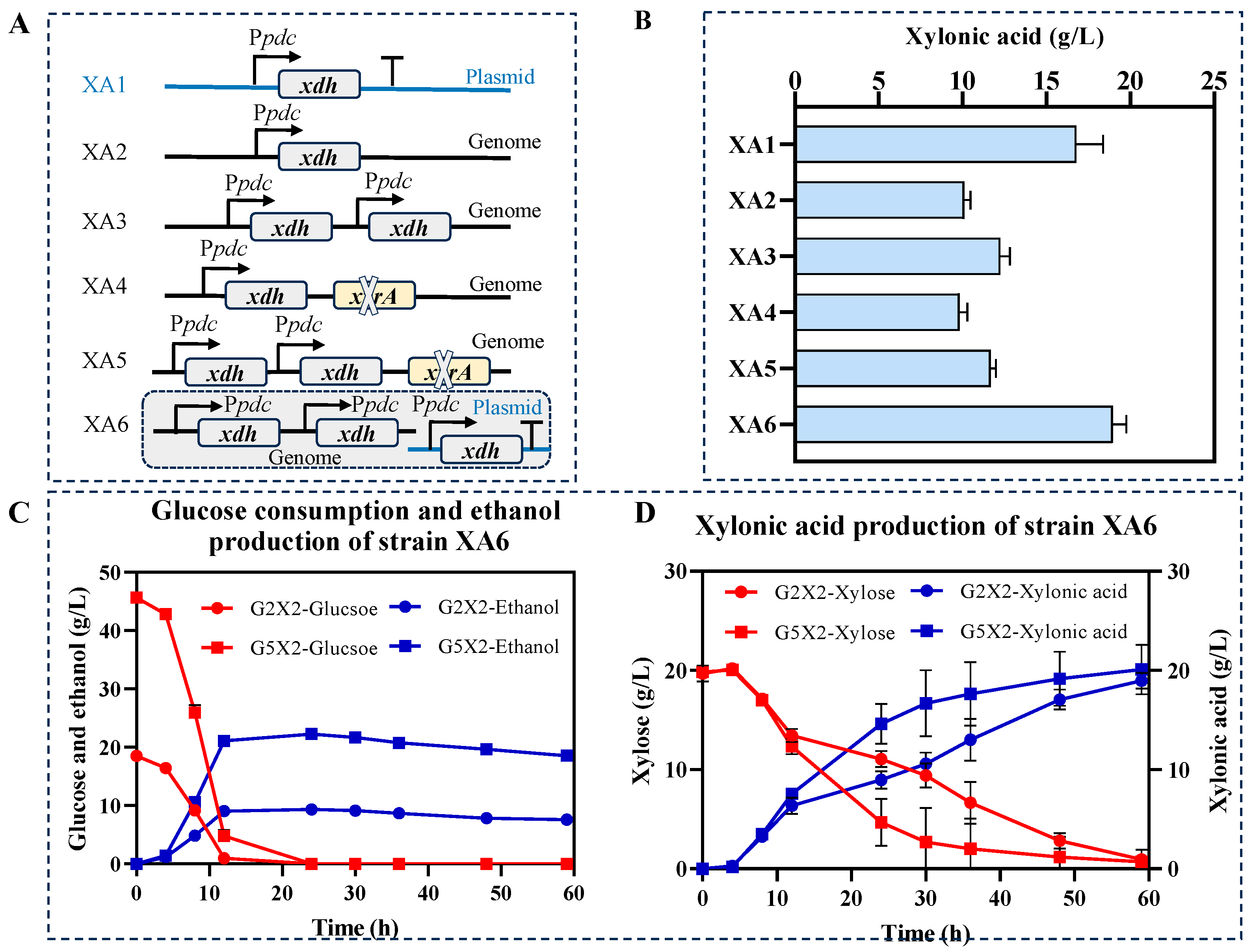
3.4. Increase in xdh Copy Numbers to Enhance Xylonic Acid Production
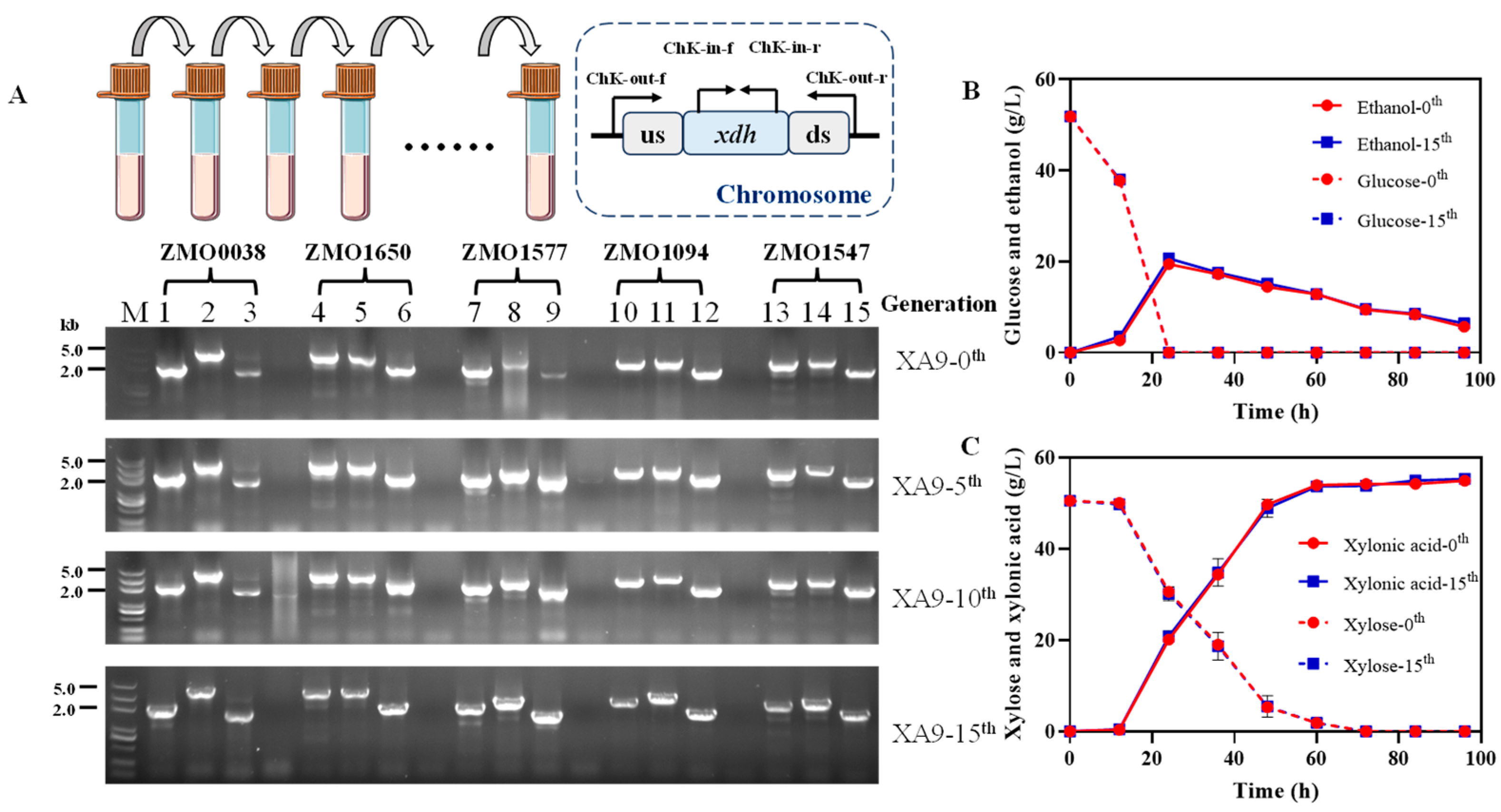
3.5. Genetic and Physiologic Stability of Strain XA9
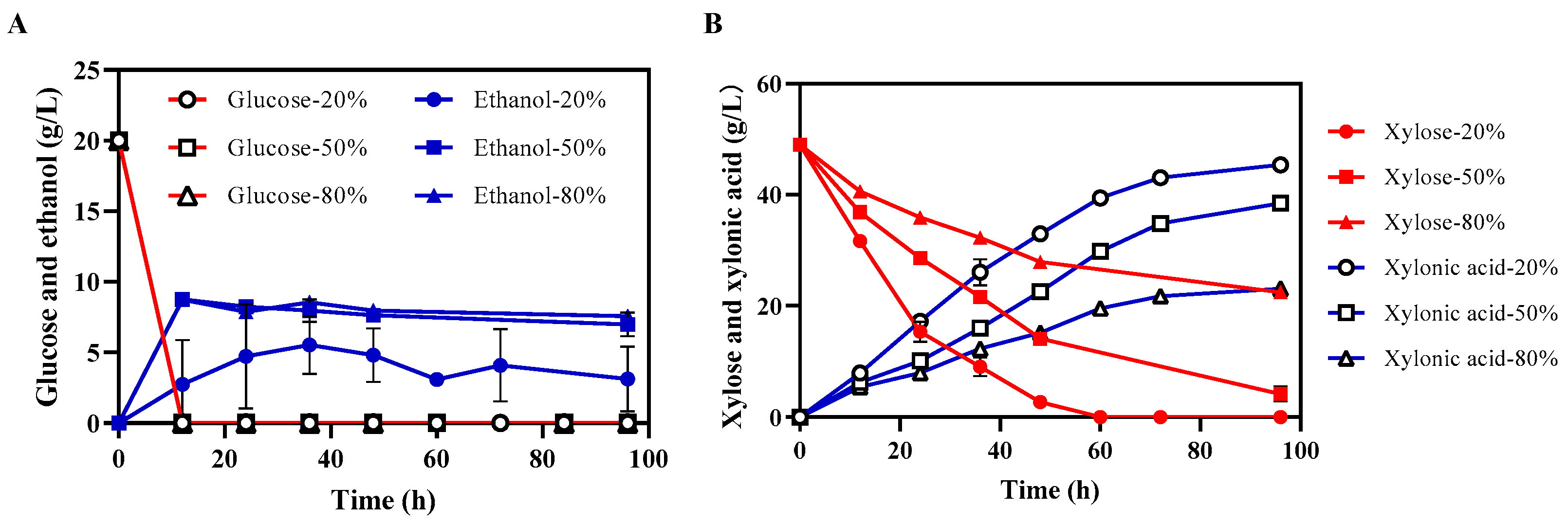
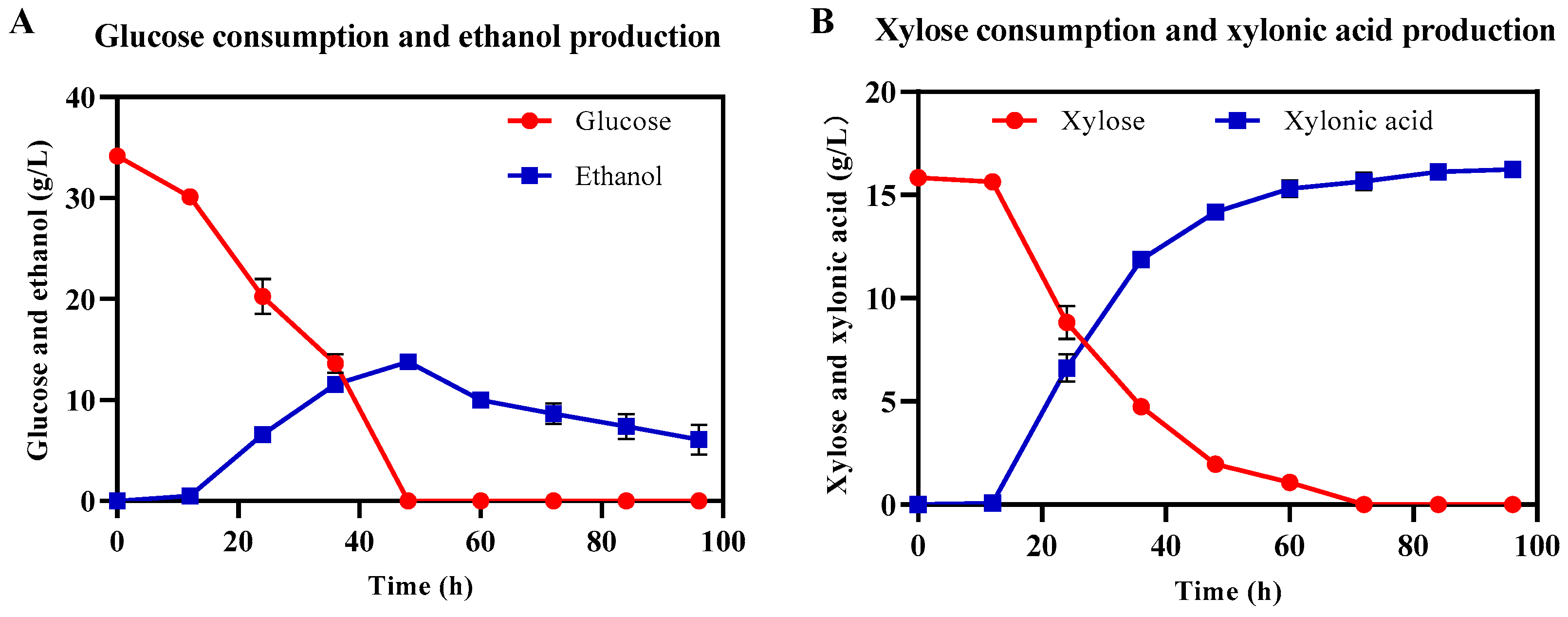
3.6. Xylonic Acid and Ethanol Co-Production Using Lignocellulosic Hydrolysate
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cao, Y.; Xian, M.; Zou, H.; Zhang, H. Metabolic engineering of Escherichia coli for the production of xylonate. PLoS ONE 2013, 8, e67305. [Google Scholar] [CrossRef] [PubMed]
- Trichez, D.; Carneiro, C.; Braga, M.; Almeida, J.R.M. Recent progress in the microbial production of xylonic acid. World J. Microbiol. Biotechnol. 2022, 38, 127. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, S.; Wang, Y.; Liang, X.; Xu, P.; Gao, C.; Ma, C. Production of d-xylonate from corn cob hydrolysate by a metabolically engineered Escherichia coli strain. ACS Sustain. Chem. Eng. 2018, 7, 2160–2168. [Google Scholar] [CrossRef]
- Banares, A.B.; Nisola, G.M.; Valdehuesa, K.N.G.; Lee, W.K.; Chung, W.J. Understanding D-xylonic acid accumulation: A cornerstone for better metabolic engineering approaches. Appl. Microbiol. Biotechnol. 2021, 105, 5309–5324. [Google Scholar] [CrossRef]
- Bondar, M.; da Fonseca, M.M.R.; Cesario, M.T. Xylonic acid production from xylose by Paraburkholderia sacchari. Biochem. Eng. J. 2021, 170, 107982. [Google Scholar] [CrossRef]
- Han, J.; Hamza, F.; Guo, J.; Sayed, M.; Pyo, S.-H.; Xu, Y. Advanced technological approaches and market status analysis of xylose bioconversion and utilization: Xylooligosacharides and xylonic acid as emerging products. Biotechnol. Adv. 2025, 79, 108509. [Google Scholar] [CrossRef]
- Su, B.; Song, D.; Zhu, H. Metabolic Engineering of Saccharomyces cerevisiae for Enhanced Carotenoid Production From Xylose-Glucose Mixtures. Front. Bioeng. Biotechnol. 2020, 8, 435. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Zhang, B.; Zhao, C.; Lin, J.; Wei, D. Overexpression of mGDH in Gluconobacter oxydans to improve D-xylonic acid production from corn stover hydrolysate. Microb. Cell Fact. 2022, 21, 35. [Google Scholar] [CrossRef]
- Zhang, M.; Wei, L.; Zhou, Y.; Du, L.; Imanaka, T.; Hua, Q. Genetic analysis of D-xylose metabolism pathways in Gluconobacter oxydans 621H. J. Ind. Microbiol. Biotechnol. 2013, 40, 379–388. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, G.; Zhang, J.; Bao, J. Fermentative production of high titer gluconic and xylonic acids from corn stover feedstock by Gluconobacter oxydans and techno-economic analysis. Bioresour. Technol. 2016, 219, 123–131. [Google Scholar] [CrossRef]
- Buchert, J.; Viikari, L.; Linko, M.; Markkanen, P. Production of xylonic acid by Pseudomonas fragi. Biotechnol. Lett. 1986, 8, 541–546. [Google Scholar] [CrossRef]
- Ishizaki, H.; Ihara, T.; Yoshitake, J.; Shimamura, M.; Imai, T.P.D.-I. D-Xylonic acid production by Enterobacter cloacae. Bull. Agric. Chem. Soc. Jpn. 1973, 47, 755–761. [Google Scholar]
- Wang, C.; Wei, D.; Zhang, Z.; Wang, D.; Shi, J.; Kim, C.H.; Jiang, B.; Han, Z.; Hao, J. Production of xylonic acid by Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 2016, 100, 10055–10063. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Lü, S.; Xu, Y.; Mo, Y.; Yu, S. Improving the performance of cell biocatalysis and the productivity of xylonic acid using a compressed oxygen supply. Biochem. Eng. J. 2015, 93, 196–199. [Google Scholar] [CrossRef]
- Liu, H.; Valdehuesa, K.N.; Nisola, G.M.; Ramos, K.R.; Chung, W.J. High yield production of D-xylonic acid from D-xylose using engineered Escherichia coli. Bioresour. Technol. 2012, 115, 244–248. [Google Scholar] [CrossRef]
- Toivari, M.; Nygard, Y.; Kumpula, E.P.; Vehkomaki, M.L.; Bencina, M.; Valkonen, M.; Maaheimo, H.; Andberg, M.; Koivula, A.; Ruohonen, L.; et al. Metabolic engineering of Saccharomyces cerevisiae for bioconversion of D-xylose to D-xylonate. Metab. Eng. 2012, 14, 427–436. [Google Scholar] [CrossRef]
- Toivari, M.; Vehkomaki, M.L.; Nygard, Y.; Penttila, M.; Ruohonen, L.; Wiebe, M.G. Low pH D-xylonate production with Pichia kudriavzevii. Bioresour. Technol. 2013, 133, 555–562. [Google Scholar] [CrossRef]
- Sundar, M.S.L.; Susmitha, A.; Rajan, D.; Hannibal, S.; Sasikumar, K.; Wendisch, V.F.; Nampoothiri, K.M. Heterologous expression of genes for bioconversion of xylose to xylonic acid in Corynebacterium glutamicum and optimization of the bioprocess. AMB Express 2020, 10, 68. [Google Scholar] [CrossRef]
- Yim, S.S.; Choi, J.W.; Lee, S.H.; Jeon, E.J.; Chung, W.J.; Jeong, K.J. Engineering of Corynebacterium glutamicum for consolidated conversion of hemicellulosic biomass into xylonic acid. Biotechnol. J. 2017, 12, 1700040. [Google Scholar] [CrossRef]
- Herrera, C.R.J.; Vieira, V.R.; Benoliel, T.; Carneiro, C.; De Marco, J.L.; de Moraes, L.M.P.; de Almeida, J.R.M.; Torres, F.A.G. Engineering Zymomonas mobilis for the production of xylonic acid from sugarcane bagasse hydrolysate. Microorganisms 2021, 9, 1372. [Google Scholar] [CrossRef]
- Panesar, P.S.; Marwaha, S.S.; Kennedy, J.F. Zymomonas mobilis: An alternative ethanol producer. J. Chem. Technol. Biotechnol. 2006, 81, 623–635. [Google Scholar] [CrossRef]
- He, M.; Wu, B.; Qin, H.; Ruan, Z.Y.; Tan, F.R.; Wang, J.L.; Shui, Z.X.; Dai, L.C.; Zhu, Q.L.; Pan, K.; et al. Zymomonas mobilis: A novel platform for future biorefineries. Biotechnol. Biofuels 2014, 7, 101. [Google Scholar] [CrossRef]
- Rogers, P.L.; Lee, K.J.; Skotnicki, M.L.; Tribe, D.E. Ethanol production by Zymomonas mobilis. In Microbial Reactions; Springer: Berlin/Heidelberg, Germany, 1982; pp. 37–84. [Google Scholar]
- Ohta, K.; Beall, D.S.; Mejia, J.P.; Shanmugam, K.T.; Ingram, L.O. Genetic improvement of Escherichia coli for ethanol production: Chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl. Environ. Microbiol. 1991, 57, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Zhang, J.; Geng, B.; Qiu, M.; Hu, M.; Yang, Q.; Bao, W.; Xiao, Y.; Zheng, Y.; Peng, W.; et al. Establishment and application of a CRISPR-Cas12a assisted genome-editing system in Zymomonas mobilis. Microb. Cell Fact. 2019, 18, 162. [Google Scholar] [CrossRef]
- Zheng, Y.; Han, J.; Wang, B.; Hu, X.; Li, R.; Shen, W.; Ma, X.; Ma, L.; Yi, L.; Yang, S.; et al. Characterization and repurposing of the endogenous type I-F CRISPR-Cas system of Zymomonas mobilis for genome engineering. Nucleic Acids Res. 2019, 47, 11461–11475. [Google Scholar] [CrossRef] [PubMed]
- Binan, G.; Yalun, W.; Xinyan, W.; Yongfu, Y.; Peng, Z.; Yunhaon, C.; Xuan, Z.; Chenguang, L.; Fengwu, B.; Ping, X.; et al. Efficient genome-editing tools to engineer the recalcitrant non-model industrial microorganism Zymomonas mobilis. Trends Biotechnol. 2024, 42, 1551–1575. [Google Scholar] [CrossRef] [PubMed]
- Nouri, H.; Fouladiha, H.; Moghimi, H.; Marashi, S.-A. A reconciliation of genome-scale metabolic network model of Zymomonas mobilis ZM4. Sci. Rep. 2020, 10, 7782. [Google Scholar] [CrossRef]
- Yan, X.; Bao, W.; Wu, Y.; Zhang, C.; Mao, Z.; Yuan, Q.; Hu, Z.; He, P.; Peng, Q.; Hu, M.; et al. Paradigm of engineering recalcitrant non-model microorganism with dominant metabolic pathway as a biorefinery chassis. Nat. Commun. 2024, 15, 10441. [Google Scholar] [CrossRef]
- Vriesekoop, F.; Pamment, N.B. Acetaldehyde stimulation of the growth of Zymomonas mobilis subjected to ethanol and other environmental stresses: Effect of other metabolic electron acceptors and evidence for a mechanism. Fermentation 2021, 7, 80. [Google Scholar] [CrossRef]
- Liu, Y.; Ghosh, I.N.; Martien, J.; Zhang, Y.; Amador-Noguez, D.; Landick, R. Regulated redirection of central carbon flux enhances anaerobic production of bioproducts in Zymomonas mobilis. Metab. Eng. 2020, 61, 261–274. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, C.; Bai, F. Making the biochemical conversion of lignocellulose more robust. Trends Biotechnol. 2024, 42, 418–430. [Google Scholar] [CrossRef]
- Shui, Z.X.; Qin, H.; Wu, B.; Ruan, Z.Y.; Wang, L.S.; Tan, F.R.; Wang, J.L.; Tang, X.Y.; Dai, L.C.; Hu, G.Q.; et al. Adaptive laboratory evolution of ethanologenic Zymomonas mobilis strain tolerant to furfural and acetic acid inhibitors. Appl. Microbiol. Biotechnol. 2015, 99, 5739–5748. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, T.; Nakamura, K. Simple and highly efficient transformation method for Zymomonas mobilis: Electroporation. Biosci. Biotechnol. Biochem. 1992, 56, 833. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zheng, Y.; Fu, H.; Chen, J.; Li, J.; Bian, Y.; Hu, P.; Lei, L.; Liu, Y.; Yang, J.; Peng, W. Development of a counterselectable system for rapid and efficient CRISPR-based genome engineering in Zymomonas mobilis. Microb. Cell Fact. 2023, 22, 208. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Shen, W.; Yan, X.; He, Q.; Cai, D.; Chen, S.; Wei, H.; Knoshaug, E.P.; Zhang, M.; Himmel, M.E.; et al. Metabolic engineering of Zymomonas mobilis for anaerobic isobutanol production. Biotechnol. Biofuels 2020, 13, 15. [Google Scholar] [CrossRef]
- Yang, S.; Tschaplinski, T.J.; Engle, N.L.; Carroll, S.L.; Martin, S.L.; Davison, B.H.; Palumbo, A.V.; Rodriguez, M., Jr.; Brown, S.D. Transcriptomic and metabolomic profiling of Zymomonas mobilis during aerobic and anaerobic fermentations. BMC Genomics 2009, 10, 34. [Google Scholar] [CrossRef]
- Silveira, M.M.; Wisbeck, E.; Lemmel, C.; Erzinger, G.; Lopes da Costa, J.P.; Bertasso, M.; Jonas, R. Bioconversion of glucose and fructose to sorbitol and gluconic acid by untreated cells of Zymomonas mobilis. J. Biotechnol. 1999, 75, 99–103. [Google Scholar] [CrossRef]
- Sarks, C.; Jin, M.; Sato, T.K.; Balan, V.; Dale, B.E. Studying the rapid bioconversion of lignocellulosic sugars into ethanol using high cell density fermentations with cell recycle. Biotechnol. Biofuels 2014, 7, 73. [Google Scholar] [CrossRef]
- Li, Z.; Waghmare, P.R.; Dijkhuizen, L.; Meng, X.; Liu, W. Research advances on the consolidated bioprocessing of lignocellulosic biomass. Eng. Microbiol. 2024, 4, 100139. [Google Scholar] [CrossRef]
- He, Y.; Li, H.; Chen, L.; Zheng, L.; Ye, C.; Hou, J.; Bao, X.; Liu, W.; Shen, Y. Production of xylitol by Saccharomyces cerevisiae using waste xylose mother liquor and corncob residues. Microb. Biotechnol. 2021, 14, 2059–2071. [Google Scholar] [CrossRef]
- Kurgan, G.; Onyeabor, M.; Holland, S.C.; Taylor, E.; Schneider, A.; Kurgan, L.; Billings, T.; Wang, X. Directed evolution of Zymomonas mobilis sugar facilitator Glf to overcome glucose inhibition. J. Ind. Microbiol. Biotechnol. 2022, 49, kuab066. [Google Scholar] [CrossRef] [PubMed]
- Kwak, S.; Jin, Y.-S. Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: A review and perspective. Microb. Cell Fact. 2017, 16, 82. [Google Scholar] [CrossRef] [PubMed]
- An, N.; Chen, X.; Sheng, H.; Wang, J.; Sun, X.; Yan, Y.; Shen, X.; Yuan, Q. Rewiring the microbial metabolic network for efficient utilization of mixed carbon sources. J. Ind. Microbiol. Biotechnol. 2021, 48, kuab040. [Google Scholar] [CrossRef] [PubMed]
- Menegon, Y.A.; Gross, J.; Jacobus, A.P. How adaptive laboratory evolution can boost yeast tolerance to lignocellulosic hydrolyses. Curr. Genet. 2022, 68, 319–342. [Google Scholar] [CrossRef]
- Jiang, W.; Lei, Z.; Gao, H.; Jiang, Y.; Lin, C.S.K.; Zhang, W.; Xin, F.; Jiang, M. Biodetoxification of Lignocellulose Hydrolysate for Direct Use in Succinic Acid Production. Biodes. Res. 2024, 6, 0044. [Google Scholar] [CrossRef]
- Infante-Neta, A.A.; de Carvalho, Á.A.O.; D’Almeida, A.P.; Gonçalves, L.R.B.; de Albuquerque, T.L. Xylitol production from passion fruit peel hydrolysate: Optimization of hydrolysis and fermentation processes. Bioresour. Technol. 2024, 414, 131628. [Google Scholar] [CrossRef]

| Plasmid | Description | Source |
|---|---|---|
| pEZ15A | Shuttle vector containing Z. mobilis origin and E. coli origin p15A, SpeR | |
| PL2R | CRISPR expression plasmid used in Zymomonas mobilis for efficient genome engineering by repurposing the endogenous Type I-f CRISPR-Cas system, SpeR | [26] |
| pXA | pEZ15A carrying XDH genes driven by strong promoter Ppdc, and SpeR | This work |
| pL2R-ZMO0038-Ppdc-XDH | pL2R used to knockout ZMO0038 and replace it with Ppdc-XDH; SpeR | This work |
| pL2R-ZMO1650-Ppdc-XDH | pL2R used to knockout ZMO1650 and replace it with Ppdc-XDH; SpeR | This work |
| pL2R-ZMO1094-Ppdc-XDH | pL2R used to knockout ZMO1094 and replace it with Ppdc-XDH; SpeR | This work |
| pL2R-ZMO1547-Ppdc-XDH | pL2R used to knockout ZMO1547 and replace it with Ppdc-XDH; SpeR | This work |
| pL2R-ZMO1577-Ppdc-XDH | pL2R used to knockout ZMO1577 and replace it with Ppdc-XDH; SpeR | This work |
| pL2R-ΔZMO0976 | pL2R used to knockout ZMO0976; SpeR | This work |
| pEZ15A-Peno-xylE | pEZ15A carrying xylE genes driven by strong promoter Peno; kana | This work |
| pEZ15A-Peno-xylFGH | pEZ15A carrying xylFGH genes driven by strong promoter Peno; kana | This work |
| pEZ15A-Peno-nadK | pEZ15A carrying nadK genes driven by strong promoter Peno; kana | This work |
| pEZ15A-Peno-noxE | pEZ15A carrying nadK genes driven by strong promoter Peno; kana | This work |
| pEZ15A-Peno-ndh | pEZ15A carrying ndh genes driven by strong promoter Peno; kana | This work |
| E. coli DH5α | E. coli strain for plasmid construction | Lab stock |
| E. coli Trans 110 | E. coli strain for plasmid demethylation | Lab stock |
| Z. mobilis ZM4 | Z. mobilis wild-type strain | Lab stock |
| EXA | E. coli DH5α strain containing pEZ15A-Pgap(6M)-XDH | This work |
| XA1 | Z. mobilis ZM4 strain containing pXA | This work |
| XA2 | ZMO0038 of ZM4 replaced with Ppdc-xdh | This work |
| XA3 | ZMO1650 of XA2 replaced with Ppdc-xdh | This work |
| XA4 | ZMO0976 of XA2 deleted | This work |
| XA5 | ZMO0976 of XA3 deleted | This work |
| XA6 | Strain XA3 containing plasmid pXA | This work |
| XA7 | ZMO1094 of XA3 replaced with Ppdc-xdh | This work |
| XA8 | ZMO1547 of XA7 replaced with Ppdc-xdh | This work |
| XA9 | ZMO1577 of XA8 replaced with Ppdc-xdh | This work |
| XA10 | XA8 strain containing pEZ15A-Peno-xylE | This work |
| XA11 | XA8 strain containing pEZ15A-Peno-xylFGH | This work |
| XA12 | XA8 strain containing pEZ15A-Peno-nadK | This work |
| XA13 | XA8 strain containing pEZ15A-Peno-noxE | This work |
| XA14 | XA8 strain containing pEZ15A-Peno-ndh | This work |
| Volume | Xylose Consumed (g/L) | Ethanol Concentration (g/L) | Xylonic Acid Production | ||
|---|---|---|---|---|---|
| Titer (g/L) | Yield * (g/g) | Productivity (g/L/h) | |||
| 20% | 48.99 ± 0.0 | 6.3 ± 1.1 | 45.4 ± 0.1 | 0.89 | 0.45 |
| 50% | 43.84 ± 0.0 | 8.7 ± 0.1 | 38.5 ± 0.2 | 0.88 | 0.40 |
| 80% | 26.50 ± 0.1 | 8.7 ± 0.1 | 23.3 ± 0.2 | 0.80 | 0.24 |
| Strain | Media | Glucose Consumed (g/L) | Xylose Consumed (g/L) | Ethanol Concentration (g/L) | Xylonic Acid Production | ||
|---|---|---|---|---|---|---|---|
| Titer (g/L) | Yield * (g/g) | Productivity (g/L/h) | |||||
| XA3 | G2X5 | 17.774 | 45.6 | 6.36 | 35.2 ± 0.4 | 0.77 | 0.37 |
| XA7 | G2X5 | 17.774 | 45.6 | 6.64 | 39.5 ± 0.4 | 0.86 | 0.41 |
| XA8 | G2X5 | 17.792 | 49.8 | 7.51 | 48.5 ± 1.1 | 0.97 | 0.51 |
| XA9 | G2X5 | 19.267 | 47.5 | 7.80 | 49.4 ± 0.1 | 1.04 | 0.51 |
| XA9 | G5X5 | 51.745 | 47.5 | 20.03 | 51.9 ± 0.1 | 1.10 | 0.54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruan, B.; Yan, X.; He, Z.; He, Q.; Yang, S. Metabolic Engineering of Zymomonas mobilis for Xylonic Acid Production from Lignocellulosic Hydrolysate. Fermentation 2025, 11, 141. https://doi.org/10.3390/fermentation11030141
Ruan B, Yan X, He Z, He Q, Yang S. Metabolic Engineering of Zymomonas mobilis for Xylonic Acid Production from Lignocellulosic Hydrolysate. Fermentation. 2025; 11(3):141. https://doi.org/10.3390/fermentation11030141
Chicago/Turabian StyleRuan, Banrui, Xiongying Yan, Zhaoqing He, Qiaoning He, and Shihui Yang. 2025. "Metabolic Engineering of Zymomonas mobilis for Xylonic Acid Production from Lignocellulosic Hydrolysate" Fermentation 11, no. 3: 141. https://doi.org/10.3390/fermentation11030141
APA StyleRuan, B., Yan, X., He, Z., He, Q., & Yang, S. (2025). Metabolic Engineering of Zymomonas mobilis for Xylonic Acid Production from Lignocellulosic Hydrolysate. Fermentation, 11(3), 141. https://doi.org/10.3390/fermentation11030141







