Secondary Metabolites from Actinokineospora spp.: Insights into a Sparsely Studied Genus of Actinomycetes
Abstract
1. Introduction
2. Overview of the Genus Actinokineospora
| Species | Strain | Analyzed Assembly | Putative BGC Similarity to Known Ones from MiBIG Database * | ||||
|---|---|---|---|---|---|---|---|
| High | Medium | Low | No | Total | |||
| Actinokineospora alba | DSM 45114 | GCF_004362515 | 1 | 0 | 7 | 15 | 23 |
| Actinokineospora auranticolor | DSM 44650 | GCF_041897805 | 4 | 4 | 10 | 18 | 36 |
| Actinokineospora baliensis | DSM 45656 | GCF_016907695 | 3 | 6 | 8 | 24 | 41 |
| Actinokineospora cianjurensis | DSM 45657 | GCF_003663795 | 4 | 3 | 13 | 16 | 36 |
| Actinokineospora bangkokensis | 44EHW | GCF_001940455 | 3 | 2 | 12 | 14 | 31 |
| Actinokineospora diospyrosa | DSM 44255 | GCF_024171925 | 3 | 4 | 15 | 21 | 43 |
| Actinokineospora globicatena | DSM 44256 | GCF_024171945 | 3 | 4 | 11 | 19 | 37 |
| Actinokineospora inagensis | DSM 44258 | GCF_000482865 | 3 | 4 | 11 | 22 | 40 |
| Actinokineospora terrae | DSM 44260 | GCF_900111175 | 4 | 2 | 15 | 16 | 37 |
| Actinokineospora enzanensis | DSM 44649 | GCF_000374445 | 5 | 5 | 10 | 18 | 38 |
| Actinokineospora fastidiosa | JCM 3276 | GCF_014648415 | 3 | 2 | 9 | 24 | 38 |
| Actinokineospora guangxiensis | CGMCC 4.7154 | GCF_042657845 | 2 | 3 | 8 | 16 | 29 |
| Actinokineospora pegani | TRM 65233 | GCF_009745975 | 4 | 2 | 16 | 33 | 55 |
| Actinokineospora soli | JCM 17695 | JBHTEY010000000 | 1 | 4 | 9 | 16 | 30 |
| Actinokineospora spheciospongiae | CECT 8578 | GCF_003182415 | 6 | 2 | 11 | 19 | 38 |
| Actinokineospora xionganensis | HBU206404 | GCF_014323725 | 1 | 1 | 8 | 11 | 21 |
| Actinokineospora sp. | G85 | GCF_049672885 | 5 | 3 | 13 | 12 | 33 |
| Actinokineospora sp. | HUAS TT18 | GCF_051364635 | 1 | 2 | 8 | 22 | 33 |
| Actinokineospora sp. | NBRC 105648 | GCF_030269645 | 4 | 4 | 19 | 25 | 52 |
| Actinokineospora sp. | PR83 | GCF_021056305 | 4 | 4 | 18 | 39 | 65 |
| Actinokineospora sp. | UTMC 2448 | GCF_024760565 | 3 | 2 | 10 | 24 | 39 |
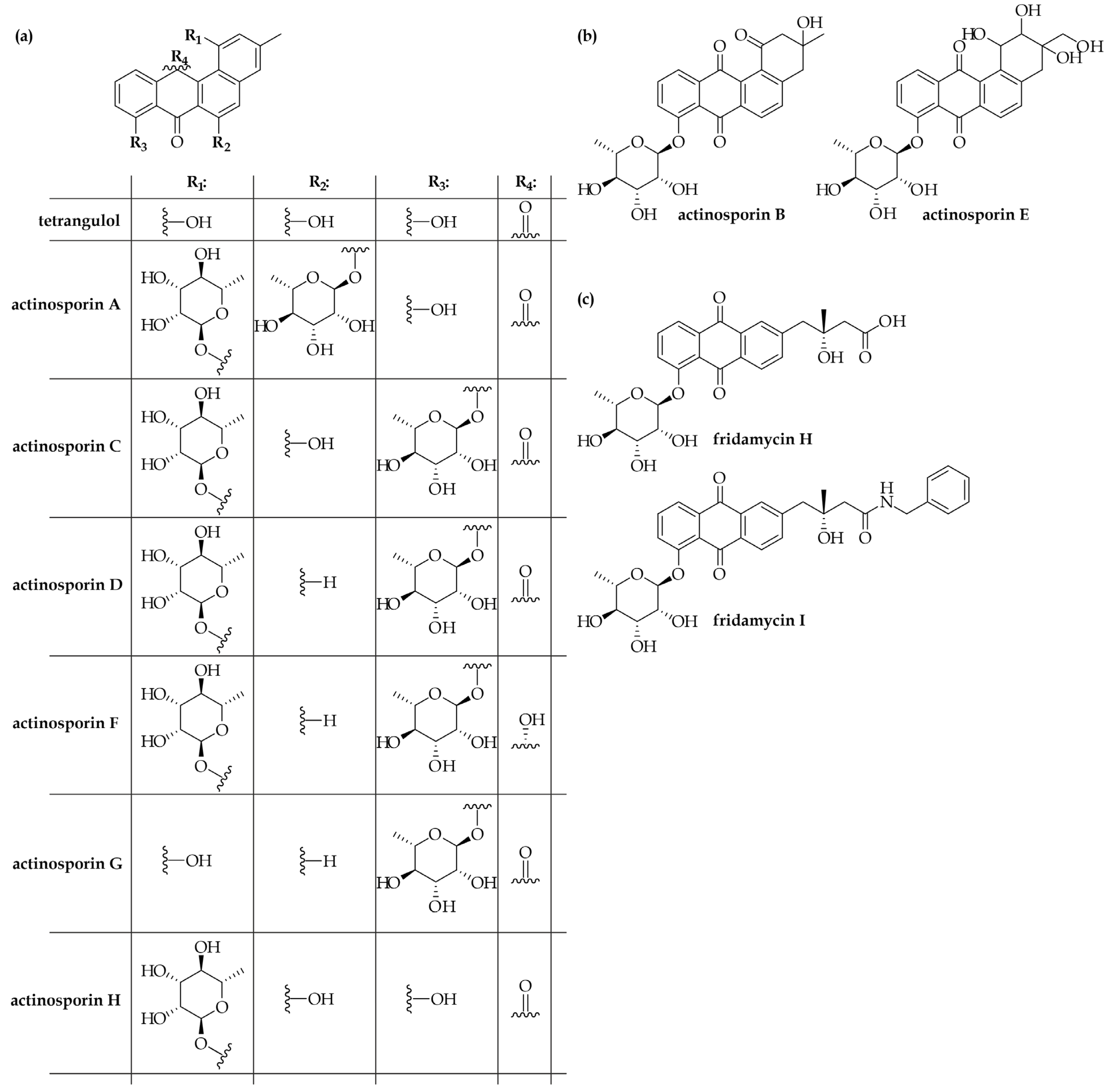
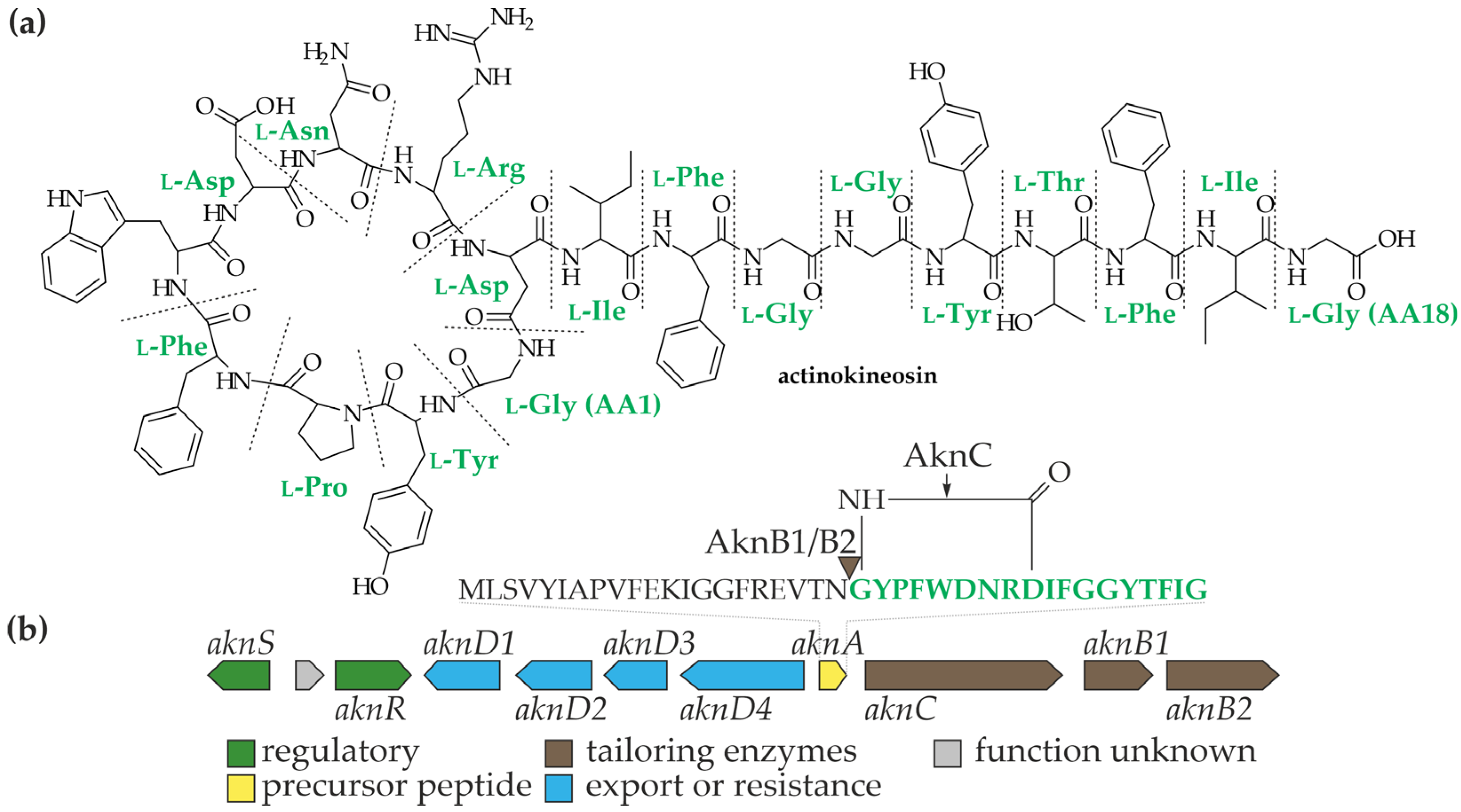
3. Actinokineospora spheciospongiae EG49T: One Strain—Many Compounds
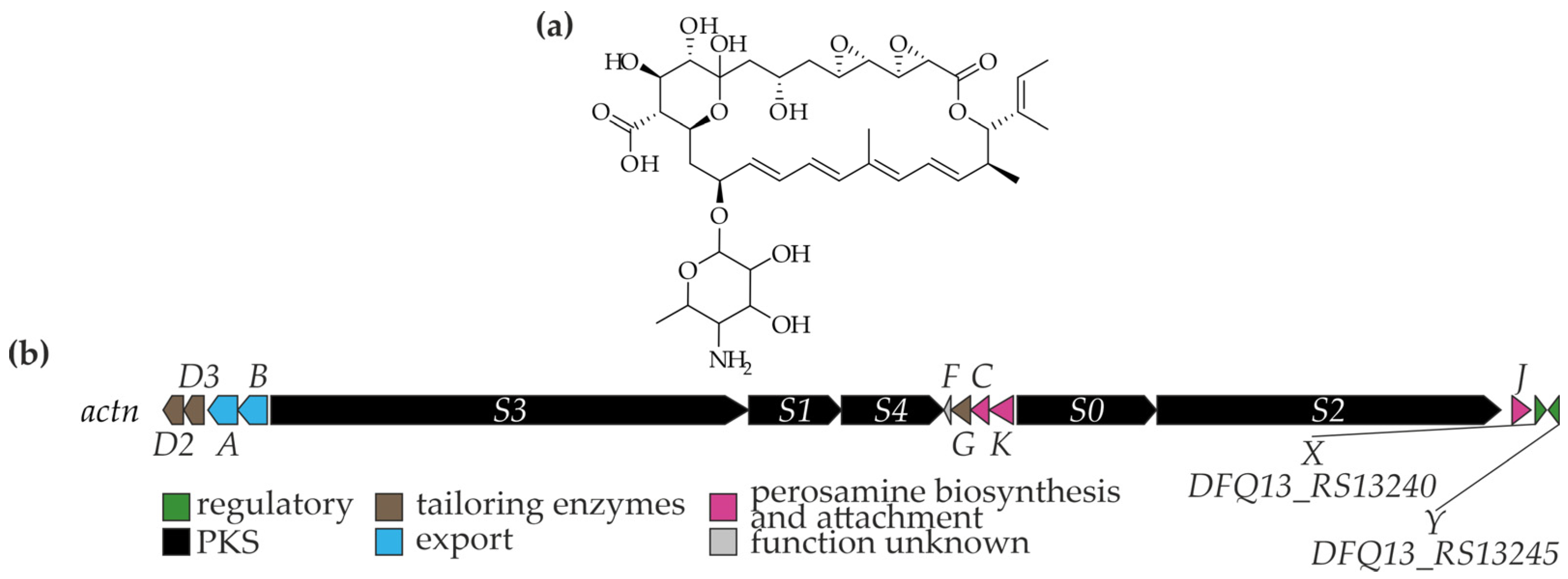
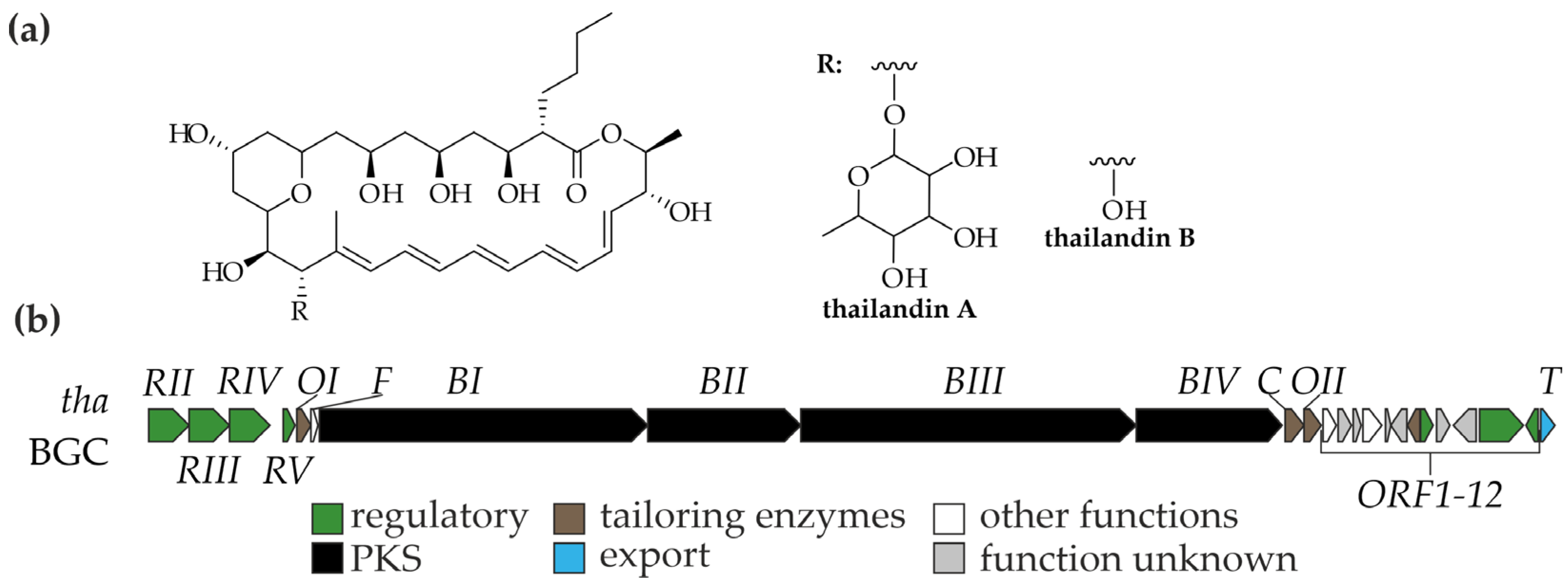
4. Thailandins: Polyene Antifungals from Actinokineospora bangkokensis 44EHWT
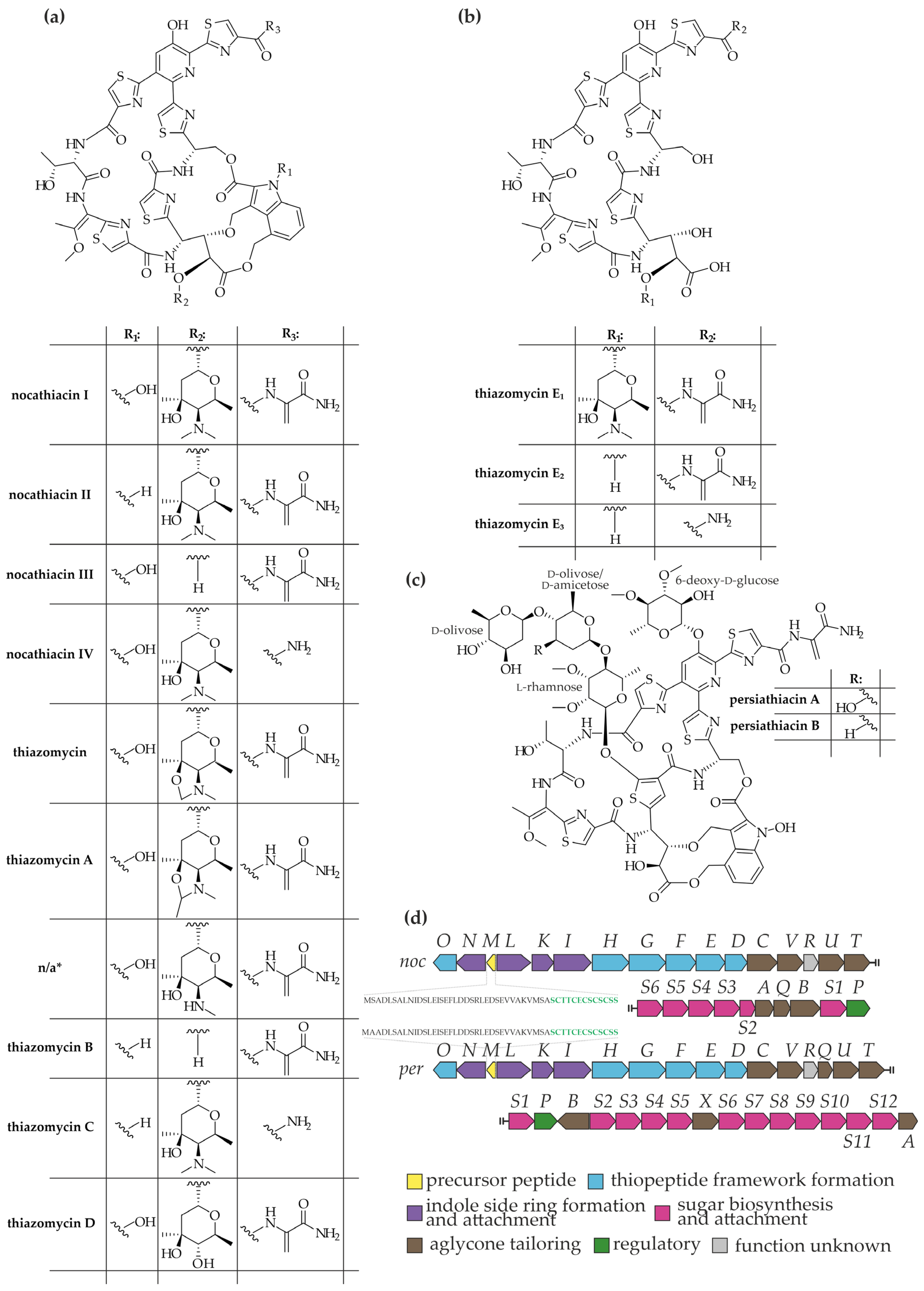
5. Glycosylated Thiopeptides from Actinokineospora spp.
6. Kineomicins: Novel Glycopeptide Antibiotic Complex from Actinokineospora auranticolor DSM 44650T
7. Conclusions and Outlook
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Waksman, S.A.; Woodruff, H.B. The soil as a source of microorganisms antagonistic to disease-producing bacteria. J. Bacteriol. 1940, 40, 581–600. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, M.; Truman, A.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Donald, L.; Pipite, A.; Subramani, R.; Owen, J.; Keyzers, R.A.; Taufa, T. Streptomyces: Still the biggest producer of new natural secondary metabolites, a current perspective. Microbiol. Res. 2022, 13, 418–465. [Google Scholar] [CrossRef]
- Parra, J.; Beaton, A.; Seipke, R.F.; Wilkinson, B.; Hutchings, M.I.; Duncan, K.R. Antibiotics from rare actinomycetes, beyond the genus Streptomyces. Curr. Opin. Microbiol. 2023, 76, 102385. [Google Scholar] [CrossRef]
- Tiwari, K.; Gupta, R.K. Rare actinomycetes: A potential storehouse for novel antibiotics. Crit. Rev. Biotechnol. 2012, 32, 108–132. [Google Scholar] [CrossRef]
- Zdouc, M.M.; Blin, K.; Louwen, N.L.L.; Navarro, J.; Loureiro, C.; Bader, C.D.; Bailey, C.B.; Barra, L.; Booth, T.J.; Bozhüyük, K.A.J.; et al. MIBiG 4.0: Advancing biosynthetic gene cluster curation through global collaboration. Nucleic Acids Res. 2025, 53, D678–D690. [Google Scholar] [CrossRef]
- González-Salazar, L.A.; Quezada, M.; Rodríguez-Orduña, L.; Ramos-Aboites, H.; Capon, R.J.; Souza-Saldívar, V.; Barona-Gomez, F.; Licona-Cassani, C. Biosynthetic novelty index reveals the metabolic potential of rare actinobacteria isolated from highly oligotrophic sediments. Microb. Genom. 2023, 9, mgen000921. [Google Scholar] [CrossRef]
- Abdelmohsen, U.R.; Cheng, C.; Viegelmann, C.; Zhang, T.; Grkovic, T.; Ahmed, S.; Quinn, R.J.; Hentschel, U.; Edrada-Ebel, R.A. Dereplication strategies for targeted isolation of new antitrypanosomal actinosporins A and B from a marine sponge associated-Actinokineospora sp. EG49. Mar. Drugs 2014, 12, 1220–1244. [Google Scholar] [CrossRef]
- Tang, Y.; Zhang, C.; Cui, T.; Lei, P.; Guo, Z.; Wang, H.; Liu, Q. The discovery of actinospene, a new polyene macrolide with broad activity against plant fungal pathogens and pathogenic yeasts. Molecules 2021, 26, 7020. [Google Scholar] [CrossRef]
- Takasaka, N.; Kaweewan, I.; Ohnishi-Kameyama, M.; Kodani, S. Isolation of a new antibacterial peptide actinokineosin from Actinokineospora spheciospongiae based on genome mining. Lett. Appl. Microbiol. 2017, 64, 150–157. [Google Scholar] [CrossRef]
- Intra, B.; Greule, A.; Bechthold, A.; Euanorasetr, J.; Paululat, T.; Panbangred, W. Thailandins A and B, New polyene macrolactone compounds isolated from Actinokineospora bangkokensis strain 44EHWT, possessing antifungal activity against anthracnose fungi and pathogenic yeasts. J. Agric. Food Chem. 2016, 64, 5171–5179. [Google Scholar] [CrossRef]
- Li, W.; Leet, J.E.; Ax, H.A.; Gustavson, D.R.; Brown, D.M.; Turner, L.; Brown, K.; Clark, J.; Yang, H.; Fung-Tomc, J.; et al. Nocathiacins, new thiazolyl peptide antibiotics from Nocardia sp. I. Taxonomy, fermentation and biological activities. J. Antibiot. 2003, 56, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Dashti, Y.; Mohammadipanah, F.; Zhang, Y.; Cerqueira Diaz, P.M.; Vocat, A.; Zabala, D.; Fage, C.D.; Romero-Canelon, I.; Bunk, B.; Spröer, C.; et al. Discovery and biosynthesis of persiathiacins: Unusual polyglycosylated thiopeptides active against multidrug resistant tuberculosis. ACS Infect. Dis. 2024, 10, 3378–3391. [Google Scholar] [CrossRef] [PubMed]
- Yushchuk, O.; Berini, F.; Zhong, L.; Rückert-Reed, C.; Bernasconi, E.; Bartolone, L.; Busche, T.; Kalinowski, J.; Süssmuth, R.D.; Marinelli, F. A rare peptide scaffold in kineomicins, the glycopeptide antibiotics produced by Actinokineospora auranticolor DSM 44650. Commun. Chem. 2025, 8, 134. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, T. Actinokineospora: A new genus of the Actinomycetales. Actinomycetologica 1988, 2, 31–45. [Google Scholar] [CrossRef]
- Dahal, R.H.; Shim, D.S.; Kim, J. Description of Actinokineospora acnipugnans sp. nov., an actinomycete isolated from soil, showing potential uses in cosmetics. Int. J. Syst. Evol. Microbiol. 2017, 67, 3043–3049. [Google Scholar] [CrossRef]
- Yuan, L.J.; Zhang, Y.Q.; Yu, L.Y.; Liu, H.Y.; Guan, Y.; Lee, J.C.; Kim, C.J.; Zhang, Y.Q. Alloactinosynnema album gen. nov., sp. nov., a member of the family Actinosynnemataceae isolated from soil. Int. J. Syst. Evol. Microbiol. 2010, 60, 39–43. [Google Scholar] [CrossRef]
- Otoguro, M.; Hayakawa, M.; Yamazaki, T.; Tamura, T.; Hatano, K.; Iimura, Y. Numerical phenetic and phylogenetic analyses of Actinokineospora isolates, with a description of Actinokineospora auranticolor sp. nov. and Actinokineospora enzanensis sp. nov. Actinomycetologica 2001, 15, 30–39. [Google Scholar] [CrossRef]
- Lisdiyanti, P.; Otoguro, M.; Ratnakomala, S.; Lestari, Y.; Hastuti, R.D.; Triana, E.; Katsuhiko, A.; Widyastuti, Y. Actinokineospora baliensis sp. nov., Actinokineospora cibodasensis sp. nov. and Actinokineospora cianjurensis sp. nov., isolated from soil and plant litter. Int. J. Syst. Evol. Microbiol. 2010, 60, 2331–2335. [Google Scholar] [CrossRef]
- Intra, B.; Matsumoto, A.; Inahashi, Y.; Omura, S.; Takahashi, Y.; Panbangred, W. Actinokineospora bangkokensis sp. nov., isolated from rhizospheric soil. Int. J. Syst. Evol. Microbiol. 2013, 63, 2655–2660. [Google Scholar] [CrossRef]
- Tamura, T.; Hayakawa, M.; Nonomura, H.; Yokota, A.; Hatano, K. Four new species of the genus Actinokineospora: Actinokineospora inagensis sp. nov., Actinokineospora globicatena sp. nov., Actinokineospora terrae sp. nov., and Actinokineospora diospyrosa sp. nov. Int. J. Syst. Bacteriol. 1995, 45, 371–378. [Google Scholar] [CrossRef]
- Labeda, D.P.; Price, N.P.; Tan, G.Y.A.; Goodfellow, M.; Klenk, H.P. Emended description of the genus Actinokineospora Hasegawa 1988 and transfer of Amycolatopsis fastidiosa Henssen et al. 1987 as Actinokineospora fastidiosa comb. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 1444–1449. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Liu, B. Actinokineospora guangxiensis sp. nov isolated from soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 4650–4654. [Google Scholar] [CrossRef] [PubMed]
- Aouiche, A.; Bouras, N.; Mokrane, S.; Zitouni, A.; Schumann, P.; Spröer, C.; Sabaou, N.; Klenk, H.P. Actinokineospora mzabensis sp. nov., a novel actinomycete isolated from Saharan soil. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2015, 107, 291–296. [Google Scholar] [CrossRef]
- Lei, Y.J.; Xia, Z.F.; Luo, X.X.; Zhang, L.L. Actinokineospora pegani sp. nov., an endophytic actinomycete isolated from the surface-sterilized root of Peganum harmala L. Int. J. Syst. Evol. Microbiol. 2020, 70, 4358–4363. [Google Scholar] [CrossRef]
- Tang, X.; Zhou, Y.; Zhang, J.; Ming, H.; Nie, G.X.; Yang, L.L.; Tang, S.K.; Li, W.J. Actinokineospora soli sp. nov., a thermotolerant actinomycete isolated from soil, and emended description of the genus Actinokineospora. Int. J. Syst. Evol. Microbiol. 2012, 62, 1845–1849. [Google Scholar] [CrossRef]
- Kämpfer, P.; Glaeser, S.P.; Busse, H.J.; Abdelmohsen, U.R.; Ahmed, S.; Hentschel, U. Actinokineospora spheciospongiae sp. nov., isolated from the marine sponge Spheciospongia vagabunda. Int. J. Syst. Evol. Microbiol. 2015, 65, 879–884. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, J.; Liu, T.; Zhang, Y.; Zhang, L.; Zhang, X. Actinokineospora xionganensis sp. nov., a filamentous actinomycete isolated from the lakeside soil of Baiyangdian. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2021, 114, 487–496. [Google Scholar] [CrossRef]
- Meena, B.; Rajan, L.A.; Vinithkumar, N.V.; Kirubagaran, R. Novel marine actinobacteria from emerald Andaman & Nicobar Islands: A prospective source for industrial and pharmaceutical byproducts. BMC Microbiol. 2013, 13, 145. [Google Scholar] [CrossRef]
- Arn, F.; Frasson, D.; Kroslakova, I.; Rezzonico, F.; Pothie, J.F.; Riedl, R.; Sievers, M. Isolation and identification of actinomycetes strains from Switzerland and their biotechnological potential. Chimia 2020, 74, 382–390. [Google Scholar] [CrossRef]
- Oberhofer, M.; Hess, J.; Leutgeb, M.; Gössnitzer, F.; Rattei, T.; Wawrosch, C.; Zotchev, S.B. Exploring Actinobacteria associated with rhizosphere and endosphere of the native alpine medicinal plant Leontopodium nivale subspecies alpinum. Front. Microbiol. 2019, 10, 2531. [Google Scholar] [CrossRef]
- Frederico, J.; Rodrigues, M.; Tackmann, J.; Malfertheiner, L.; Patsch, D.; Perez-Molphe-Montoya, E.; Näpflin, N.; Gaio, D.; Rot, G.; Danaila, M.; et al. The MicrobeAtlas database: Global trends and insights into Earth’s microbial ecosystems. bioRxiv 2025. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Vader, L.; Szenei, J.; Reitz, Z.L.; Augustijn, H.E.; Cediel-Becerra, J.D.D.; De Crécy-Lagard, V.; Koetsier, R.A.; Williams, S.E.; et al. AntiSMASH 8.0: Extended gene cluster detection capabilities and analyses of chemistry, enzymology, and regulation. Nucleic Acids Res. 2025, 53, W32–W38. [Google Scholar] [CrossRef]
- Morin, L.M.C.; Dekoninck, K.; Sridhar, V.; Disney-McKeethen, S.; Proctor, T.; Eng, A.Y.; Traxler, M.F. Why do filamentous Actinomycetota produce such a vast array of specialized metabolites? Annu. Rev. Microbiol. 2025, 79, 753–772. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, U.R.; Pimentel-Elardo, S.M.; Hanora, A.; Radwan, M.; Abou-El-Ela, S.H.; Ahmed, S.; Hentschel, U. Isolation, phylogenetic analysis and anti-infective activity screening of marine sponge-associated actinomycetes. Mar. Drugs 2010, 8, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Hoet, S.; Opperdoes, F.; Brun, R.; Quetin-Leclercq, J. Natural products active against African trypanosomes: A step towards new drugs. Nat. Prod. Rep. 2004, 21, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Llewellyn, T.; Nowell, R.W.; Aptroot, A.; Temina, M.; Prescott, T.A.K.; Barraclough, T.G.; Gaya, E. Metagenomics shines light on the evolution of “sunscreen” pigment metabolism in the Teloschistales (lichen-forming Ascomycota). Genome Biol. Evol. 2023, 15, evad002. [Google Scholar] [CrossRef]
- Frandsen, R.J.N.; Khorsand-Jamal, P.; Kongstad, K.T.; Nafisi, M.; Kannangara, R.M.; Staerk, D.; Okkels, F.T.; Binderup, K.; Madsen, B.; Møller, B.L.; et al. Heterologous production of the widely used natural food colorant carminic acid in Aspergillus nidulans. Sci. Rep. 2018, 8, 12853. [Google Scholar] [CrossRef]
- Stalman, M.; Koskamp, A.M.; Luderer, R.; Vernooy, J.H.J.; Wind, J.C.; Wullems, G.J.; Croes, A.F. Regulation of anthraquinone biosynthesis in cell cultures of Morinda citrifolia. J. Plant Physiol. 2003, 160, 607–614. [Google Scholar] [CrossRef]
- Cheemalamarri, C.; Batchu, U.R.; Thallamapuram, N.P.; Katragadda, S.B.; Reddy Shetty, P. A review on hydroxy anthraquinones from bacteria: Crosstalk’s of structures and biological activities. Nat. Prod. Res. 2022, 36, 6186–6205. [Google Scholar] [CrossRef]
- Fujioka, K.; Shimazu, A.; Hayakawa, Y.; Seto, H.; Furihata, K. Isolation and characterization of atramycin A and atramycin B, new isotetracenone type antitumor antibiotics. J. Antibiot. 1991, 44, 1025–1028. [Google Scholar] [CrossRef]
- Ramos, A.; Lombó, F.; Braña, A.F.; Rohr, J.; Méndez, C.; Salas, J.A. Biosynthesis of elloramycin in Streptomyces olivaceus requires glycosylation by enzymes encoded outside the aglycon cluster. Microbiology 2008, 154, 781–788. [Google Scholar] [CrossRef] [PubMed]
- Kojiri, K.; Arakawa, H.; Okura, A.; Suda, H.; Okanishi, M.; Satoh, F.; Kawamura, K. New antitumor substances, Be-12406A and Be-12406B, produced by a streptomycete I. Taxonomy, fermentation, isolation, physico-chemical and biological properties. J. Antibiot. 1991, 44, 1054–1060. [Google Scholar] [CrossRef] [PubMed]
- Murenets, N.V.; Kudinova, M.K.; Klyev, N.A. Galtamycin: Galtamycinone structure. Antibiot. I Med. Biotechnol. 1986, 31, 431–434. [Google Scholar]
- Kuntsmann, M.P.; Mitscher, L.A. The structural characterization of tetrangomycin and tetrangulol. J. Org. Chem. 1966, 31, 2920–2925. [Google Scholar] [CrossRef]
- Alhadrami, H.A.; Thissera, B.; Hassan, M.H.A.; Behery, F.A.; Ngwa, C.J.; Hassan, H.M.; Pradel, G.; Abdelmohsen, U.R.; Rateb, M.E. Bio-guided isolation of antimalarial metabolites from the coculture of two red sea sponge-derived Actinokineospora and Rhodococcus spp. Mar. Drugs 2021, 19, 109. [Google Scholar] [CrossRef]
- Grkovic, T.; Abdelmohsen, U.R.; Othman, E.M.; Stopper, H.; Edrada-Ebel, R.; Hentschel, U.; Quinn, R.J. Two new antioxidant actinosporin analogues from the calcium alginate beads culture of sponge-associated Actinokineospora sp. strain EG49. Bioorganic Med. Chem. Lett. 2014, 24, 5089–5092. [Google Scholar] [CrossRef]
- Dashti, Y.; Grkovic, T.; Abdelmohsen, U.R.; Hentschel, U.; Quinn, R.J. Actinomycete metabolome induction/suppression with N-acetylglucosamine. J. Nat. Prod. 2017, 80, 828–836. [Google Scholar] [CrossRef]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef]
- Tawfike, A.; Attia, E.Z.; Desoukey, S.Y.; Hajjar, D.; Makki, A.A.; Schupp, P.J.; Edrada-Ebel, R.A.; Abdelmohsen, U.R. New bioactive metabolites from the elicited marine sponge-derived bacterium Actinokineospora spheciospongiae sp. nov. AMB Express 2019, 9, 12. [Google Scholar] [CrossRef]
- Gerber, N.N.; Lechevalier, M.P. Novel benzo[α]naphthacene quinones from an actinomycete, Frankia G-2 (ORS 020604). Can. J. Chem. 1984, 62, 2818–2821. [Google Scholar] [CrossRef]
- Künzel, E.; Faust, B.; Oelkers, C.; Weissbach, U.; Bearden, D.W.; Weitnauer, G.; Westrich, L.; Bechthold, A.; Rohr, J. Inactivation of the urdGT2 gene, which encodes a glycosyltransferase responsible for the C-glycosyltransfer of activated d-olivose, leads to formation of the novel urdamycins I, J, and K. J. Am. Chem. Soc. 1999, 121, 11058–11062. [Google Scholar] [CrossRef]
- Naoki, A.; Yasukazu, N.; Takehiko, N.; Nobuyasu, E.; Hideaki, U.; Masanobu, M. Novel antitumor antibiotics, saptomycins I. Taxonomy of the producing organism, fermentation, HPLC analysis and biological activities. J. Antibiot. 1993, 46, 1530–1535. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Harjes, J.; Ryu, T.; Abdelmohsen, U.R.; Moitinho-Silva, L.; Horn, H.; Ravasi, T.; Hentschel, U. Draft genome sequence of the antitrypanosomally active sponge-associated bacterium Actinokineospora sp. strain EG49. Genome Announc. 2014, 2, e00160-14. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, J.F.; Caffrey, P.; Gil, J.A.; Zotchev, S.B. Polyene antibiotic biosynthesis gene clusters. Appl. Microbiol. Biotechnol. 2003, 61, 179–188. [Google Scholar] [CrossRef]
- Yushchuk, O.; Ostash, I.; Mösker, E.; Vlasiuk, I.; Deneka, M.; Rückert, C.; Busche, T.; Fedorenko, V.; Kalinowski, J.; Süssmuth, R.D.; et al. Eliciting the silent lucensomycin biosynthetic pathway in Streptomyces cyanogenus S136 via manipulation of the global regulatory gene adpA. Sci. Rep. 2021, 11, 3507. [Google Scholar] [CrossRef]
- Sheng, Y.; Ou, Y.; Hu, X.; Deng, Z.; Bai, L.; Kang, Q. Generation of tetramycin B derivative with improved pharmacological property based on pathway engineering. Appl. Microbiol. Biotechnol. 2020, 104, 2561–2573. [Google Scholar] [CrossRef]
- Kimura, K.I.; Kanou, F.; Takahashi, H.; Esumi, Y.; Uramoto, M.; Yoshihama, M. Propeptin, a new inhibitor of prolyl endopeptidase produced by Microbispora. I. Fermentation, isolation and biological properties. J. Antibiot. 1997, 50, 373–378. [Google Scholar] [CrossRef]
- Greule, A.; Intra, B.; Flemming, S.; Rommel, M.G.E.; Panbangred, W.; Bechthold, A. The draft genome sequence of Actinokineospora bangkokensis 44EHWT reveals the biosynthetic pathway of the antifungal thailandin compounds with unusual butylmalonyl-CoA extender units. Molecules 2016, 21, 1607. [Google Scholar] [CrossRef]
- Pandey, R.C.; Narasimhachari, N.; Rinehart, K.L.; Millington, D.S. Polyene antibiotics. IV. Structure of chainin. J. Am. Chem. Soc. 1972, 94, 4306–4310. [Google Scholar] [CrossRef]
- Goodfellow, M.; Williams, S.T.; Alderson, G. Transfer of Chainia species to the genus Streptomyces with emended description of species. Syst. Appl. Microbiol. 1986, 8, 55–60. [Google Scholar] [CrossRef]
- Just-Baringo, X.; Albericio, F.; Álvarez, M. Thiopeptide antibiotics: Retrospective and recent advances. Mar. Drugs 2014, 12, 317–351. [Google Scholar] [CrossRef] [PubMed]
- Just-Baringo, X.; Albericio, F.; Álvarez, M. Thiopeptide engineering: A multidisciplinary effort towards future drugs. Angew. Chem. Int. Ed. 2014, 53, 6602–6616. [Google Scholar] [CrossRef] [PubMed]
- Jayasuriya, H.; Herath, K.; Ondeyka, J.G.; Zhang, C.; Zink, D.L.; Brower, M.; Gailliot, F.P.; Greene, J.; Birdsall, G.; Venugopal, J.; et al. Isolation and structure elucidation of thiazomycin—A potent thiazolyl peptide antibiotic from Amycolatopsis fastidiosa. J. Antibiot. 2007, 60, 554–564. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Leet, J.E.; Li, W.; Ax, H.A.; Matson, J.A.; Huang, S.; Huang, R.; Cantone, J.L.; Drexler, D.; Dalterio, R.A.; Lam, K.S. Nocathiacins, new thiazolyl peptide antibiotics from Nocardia sp. II. Isolation, characterization, and structure determination. J. Antibiot. 2003, 56, 232–242. [Google Scholar] [CrossRef]
- Benazet, F.; Cartier, M.; Florent, J.; Godard, C.; Jung, G.; Lunel, J.; Mancy, D.; Pascal, C.; Renaut, J.; Tarridec, P.; et al. Nosiheptide, a sulfur-containing peptide antibiotic isolated from Streptomyces actuosus 40037. Experientia 1980, 36, 414–416. [Google Scholar] [CrossRef]
- Zhang, C.; Zink, D.L.; Ushio, M.; Burgess, B.; Onishi, R.; Masurekar, P.; Barrett, J.F.; Singh, S.B. Isolation, structure, and antibacterial activity of thiazomycin A, a potent thiazolyl peptide antibiotic from Amycolatopsis fastidiosa. Bioorganic Med. Chem. 2008, 16, 8818–8823. [Google Scholar] [CrossRef]
- Zhang, C.; Herath, K.; Jayasuriya, H.; Ondeyka, J.G.; Zink, D.L.; Occi, J.; Birdsall, G.; Venugopal, J.; Ushio, M.; Burgess, B.; et al. Thiazomycins, thiazolyl peptide antibiotics from Amycolatopsis fastidiosa. J. Nat. Prod. 2009, 72, 841–847. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, D.; Lin, J.; Liao, R.; Tong, W.; Xu, Z.; Liu, W. Characterization of NocL involved in thiopeptide nocathiacin I biosynthesis: A [4Fe-4S] cluster and the catalysis of a radical S-adenosylmethionine enzyme. J. Biol. Chem. 2011, 286, 21287–21294. [Google Scholar] [CrossRef]
- Pucci, M.J.; Bronson, J.J.; Barrett, J.F.; Denbleyker, K.L.; Discotto, L.F.; Fung-Tomc, J.C.; Ueda, Y. Antimicrobial evaluation of nocathiacins, a thiazole peptide class of antibiotics. Antimicrob. Agents Chemother. 2004, 48, 3697–3701. [Google Scholar] [CrossRef]
- Singh, S.B.; Xu, L.; Meinke, P.T.; Kurepina, N.; Kreiswirth, B.N.; Olsen, D.B.; Young, K. Thiazomycin, nocathiacin and analogs show strong activity against clinical strains of drug-resistant Mycobacterium tuberculosis. J. Antibiot. 2017, 70, 671–674. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Singh, S.B.; Occi, J.; Jayasuriya, H.; Herath, K.; Motyl, M.; Dorso, K.; Gill, C.; Hickey, E.; Overbye, K.M.; Barrett, J.F.; et al. Antibacterial evaluations of thiazomycin—A potent thiazolyl peptide antibiotic from Amycolatopsis fastidiosa. J. Antibiot. 2007, 60, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Woodgate, J.; Sumang, F.A.; Salliss, M.E.; Belousoff, M.; Ward, A.C.; Challis, G.L.; Zenkin, N.; Errington, J.; Dashti, Y. Mode of action and mechanisms of resistance to the unusual polyglycosylated thiopeptide antibiotic persiathiacin A. ACS Infect. Dis. 2025, 11, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Yushchuk, O.; Binda, E.; Marinelli, F. Glycopeptide antibiotic resistance genes: Distribution and function in the producer actinomycetes. Front. Microbiol. 2020, 11, 1173. [Google Scholar] [CrossRef]
- Yushchuk, O.; Ostash, B. Glycopeptide antibiotics: Genetics, chemistry, and new screening approaches. In Natural Products from Actinomycetes: Diversity, Ecology and Drug Discovery; Springer: Singapore, 2022; pp. 411–444. [Google Scholar] [CrossRef]
- Yushchuk, O.; Homoniuk, V.; Ostash, B.; Marinelli, F.; Fedorenko, V. Genetic insights into the mechanism of teicoplanin self-resistance in Actinoplanes teichomyceticus. J. Antibiot. 2020, 73, 255–259. [Google Scholar] [CrossRef]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Nicolaou, K.C.; Boddy, C.N.C.; Bräse, S.; Winssinger, N. Chemistry, biology, and medicine of the glycopeptide antibiotics. Angew. Chem. Int. Ed. 1999, 38, 2096–2152. [Google Scholar] [CrossRef]
| Species | Strain | Validly Published | Genome Sequence | Biotope of Isolation | Isolation Approach | Reference |
|---|---|---|---|---|---|---|
| Actinokineospora acnipugnans | R434 | yes | n/a * | Reclaimed grassland soil, South Korea | Soil sample inoculated on Reasoner’s 2A medium (R2A), no pretreatment | [16] |
| Actinokineospora alba | 03-9939 | yes | GCA_004362515 (DSM 45114) GCA_900099755 (CPCC 201030) GCA_900104505 (IBRC-M 10655) | Forest soil sample, China | Czapek’s agar | [17] |
| Actinokineospora auranticolor | YU 961-1 | yes | GCA_002934265 (YU 961-1) GCA_041897805 (DSM 44650) | Fallen leaves in a level-land forest, Japan | Dry CaCO3 enrichment treatment, Humic acid-vitamin (HV) agar, 10 mg/L nalidixic acid, 20 mg/L trimethoprim | [18] |
| Actinokineospora baliensis | ID03-0561 | yes | GCA_016907695 (DSM 45656) | Soil, Indonesia | Differential centrifugation for selective isolation of motile actinomycetes, HV agar | [19] |
| Actinokineospora cianjurensis | ID03-0810 | yes | (GCA_003663795) DSM 45657 | Leaf-litter of the botanical garden, Indonesia | Differential centrifugation for selective isolation of motile actinomycetes, HV agar | [19] |
| Actinokineospora cibodasensis | ID03-0748 | yes | n/a | Litter of the botanical garden, Indonesia | Differential centrifugation for selective isolation of motile actinomycetes, HV agar | [19] |
| Actinokineospora bangkokensis | 44EHW | yes | GCA_001940455 (4EHW) | Rhizospheric soil under Colocasia esculenta, Thailand | Dry heat pretreatment at 120 °C for 1 h, water-proline agar, 25 µg/mL nalidixic acid, 50 µg/mL cycloheximide | [20] |
| Actinokineospora diospyrosa | YU8-1 | yes | GCA_024171925 (DSM 44255) GCA_039534305 (JCM 9921) | Fallen persimmon leaves, Japan | Dessication/rehydration pretreatment, HV agar | [21] |
| Actinokineospora globicatena | YU6-1 | yes | GCA_024171945 (DSM 44256) GCA_039534325 (JCM 9922) | Soil around a pond, Japan | Dessication/rehydration pretreatment, HV agar | [21] |
| Actinokineospora inagensis | YU4-1 | yes | GCA_00048286 (DSM 44258) | Fallen leaves of a lakeside, Japan | Dessication/rehydration pretreatment, HV agar | [21] |
| Actinokineospora terrae | YU6-3 | yes | GCA_900111175 (DSM 44260) | Soil around a pond, Japan | Dessication/rehydration pretreatment, HV agar | [21] |
| Actinokineospora enzanensis | YU 924-101 | yes | GCA_000374445 (DSM 44649) | Level-land forest soil, Japan | Dry CaCO3 enrichment treatment, Humic acid-vitamin (HV) agar | [18] |
| Actinokineospora fastidiosa | NRRL B-16697 | yes | GCA_014648415 (JCM 3276) | Soil, Egypt | n/a | [22] |
| Actinokineospora guangxiensis | Gk-6 | yes | GCA_042657845 (CGMCC 4.7154) | Soil, China | Soil sample inoculated on HV agar, no pretreatment | [23] |
| Actinokineospora mzabensis | PAL84 | yes (heterotypic synonym of Actinokineospora spheciospongiae) | n/a | Saharan soil from a palm grove, Algeria | Chitin-vitamin agar, 80 µg/mL cycloheximide, 25 µg/mL rifampicin, no pretreatment | [24] |
| Actinokineospora pegani | TRM 65233 | yes | GCA_009745975 (TRM 65233) | Surface-sterilized root of Peganum harmala, China | Starch casein agar at pH 9.0 supplemented with K2Cr2O7, 50 µg/mL nystatin, 50 µg/mL cycloheximide, 80 µg/mL nalidixic acid | [25] |
| Actinokineospora riparia | C-39162 | yes | n/a | Soil, the Ado River, Japan | n/a | [15] |
| Actinokineospora soli | YIM 75948 | yes | GCA_042666155 (JCM 17695) | Soil, China | Soil sample inoculated on ISP2 agar, no pretreatment | [26] |
| Actinokineospora spheciospongiae | EG49 | yes | GCA_000564855 (EG49) GCA_003182415 (CECT 8578) | Marine sponge Spheciospongia vagabunda from the Red Sea, Egypt | M1, ISP2, Oligotrophic medium, M1 plus, Actinomycete Isolation Agar, Marine Agar, Glycerol Asparagine Agar, R2A Agar, 100 μg/mL cycloheximide, 25 μg/mL nystatin, 25 μg/mL nalidixic acid, no pretreatment | [27] |
| Actinokineospora xionganensis | HBU206404 | no | GCA_014323725 (HBU206404) | Lakeside soil, China | Soil extract medium, 1% (v/v) recombinant resuscitation-promoting factor, no pretreatment | [28] |
| Actinokineospora sp. | G85 | no | GCA_049672885 (G85) | Rock surface, Tunisia | n/a | n/a |
| Actinokineospora sp. | HUAS TT18 | no | GCA_051364635 (HUAS TT18) | Soil, China | n/a | n/a |
| Actinokineospora sp. | NBRC 105648 | no | GCA_030269645 (NBRC 105648) | Soil, Japan | n/a | n/a |
| Actinokineospora sp. | PR83 | no | GCA_021056305 (PR83) | Sediment, Mexico | n/a | [7] |
| Actinokineospora sp. | UTMC 2448 | no | GCA_024760565 (UTMC 2448) | Coastal area, Iran | n/a | [13] |
| Species | Strain | Bioactivities Identified upon Identification Against | Identified Specialized Metabolites | Reference |
|---|---|---|---|---|
| Actinokineospora acnipugnans | R434 | Propionibacterium acnes, Staphylococcus epidermidis | n/a * | [16] |
| Actinokineospora alba | 03-9939 | Staphylococcus aureus CPCC 100051 Pseudomonas aeruginosa CPCC 100109 | n/a | [17] |
| Actinokineospora auranticolor | YU 961-1 | n/a | Kineomicin | [18] |
| Actinokineospora bangkokensis | 44EHW | n/a | Thailandin | [20] |
| Actinokineospora fastidiosa | NRRL B-16697 | Various Gram-positive bacteria | n/a | [22] |
| Actinokineospora guangxiensis | Gk-6 | No antibacterial activity was observed against Staphylococcus aureus and Pseudomonas aeruginosa | n/a | [23] |
| Actinokineospora riparia | C-39162 | Antimycoplasmal activity | n/a | [15] |
| Actinokineospora spheciospongiae | EG49 | n/a | Actinosporins, actinospene, actinokineosin | [27] |
| Actinokineospora sp. | G85 | Broad-spectrum antimicrobial activity | n/a | n/a |
| Actinokineospora sp. | PR83 | Bacillus subtilis, carcinoma cells (NCIH460) | n/a | [7] |
| Actinokineospora sp. | UTMC 2448 | Staphylococcus aureus | Persiathiacin A and B | [13] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yushchuk, O. Secondary Metabolites from Actinokineospora spp.: Insights into a Sparsely Studied Genus of Actinomycetes. Fermentation 2025, 11, 663. https://doi.org/10.3390/fermentation11120663
Yushchuk O. Secondary Metabolites from Actinokineospora spp.: Insights into a Sparsely Studied Genus of Actinomycetes. Fermentation. 2025; 11(12):663. https://doi.org/10.3390/fermentation11120663
Chicago/Turabian StyleYushchuk, Oleksandr. 2025. "Secondary Metabolites from Actinokineospora spp.: Insights into a Sparsely Studied Genus of Actinomycetes" Fermentation 11, no. 12: 663. https://doi.org/10.3390/fermentation11120663
APA StyleYushchuk, O. (2025). Secondary Metabolites from Actinokineospora spp.: Insights into a Sparsely Studied Genus of Actinomycetes. Fermentation, 11(12), 663. https://doi.org/10.3390/fermentation11120663






