Gut-Derived Lactic Acid Bacteria from Cotton Bollworm Exhibit Efficient Gossypol Degradation and Probiotic Potential During Solid-State Fermentation of Cottonseed Meal
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Isolation of Gossypol-Degrading Bacteria from Helicoverpa Armigera Larval Gut
2.3. Identification of Gossypol-Degrading Bacteria from Helicoverpa armigera Gut
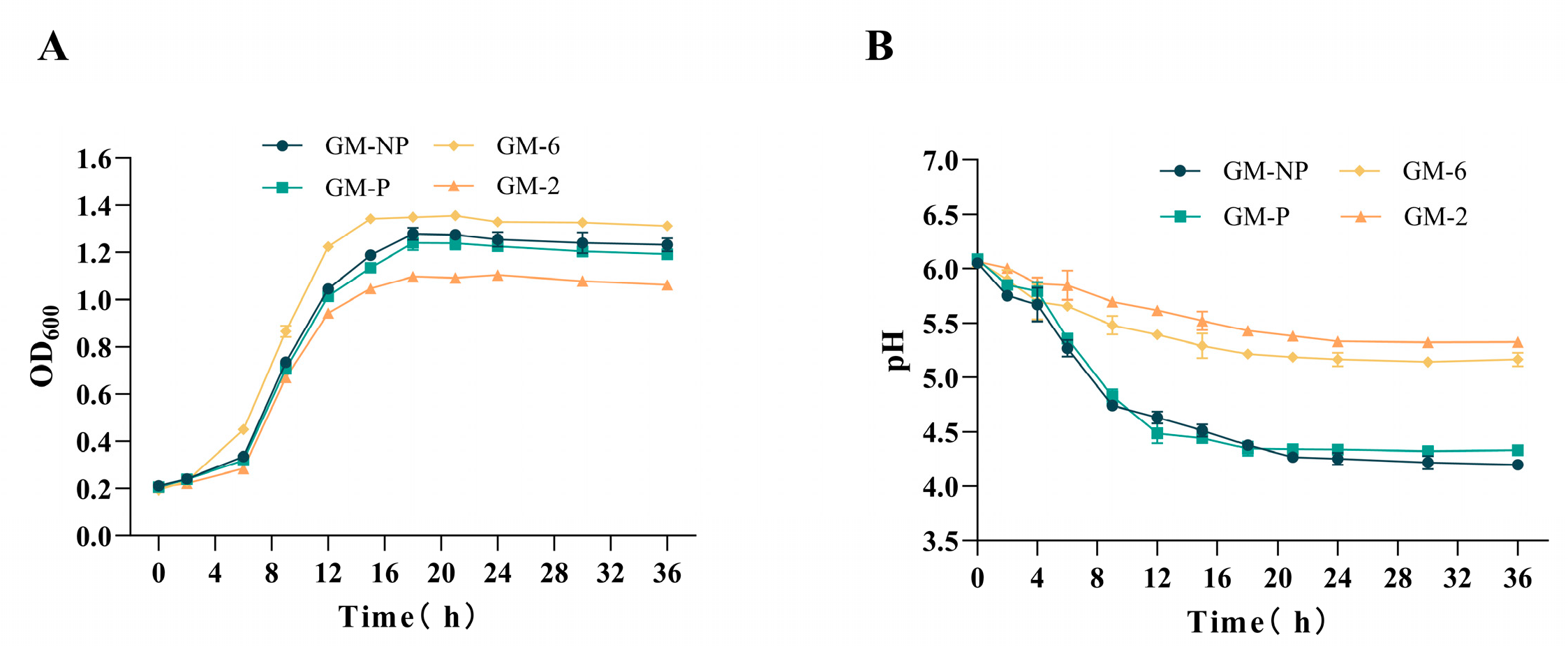
2.4. Determination of the Growth Curve and Free Gossypol Degradation Rate for the Target Strain
2.5. Artificial Gastric Juice and Bile Salt Tolerance
2.6. Temperature Tolerance Assessment
2.7. Evaluation of Cell Surface Properties
2.8. Auto-Aggregation Assessment
2.9. Hemolytic and Antioxidant Assessment
2.10. Assessment of Antibacterial Activity
2.11. Antibiotic Susceptibility Testing
2.12. Solid-State Fermentation of Cottonseed Meal by the Target Strain
2.13. Statistical Analysis
3. Results
3.1. Preliminary Characterisation of Isolated Bacteria
3.2. 16S rDNA Sequencing and Phylogenetic Analysis of Bacterial Strains
3.3. Physiological and Biochemical Characteristics of the Strain
3.4. Growth Curves and Acid Production Curves of Four Strains of Lactic Acid Bacteria
3.5. Tolerance of Four Strains of Lactic Acid Bacteria to Artificial Gastrointestinal Environments
3.6. Thermal Tolerance of the Four Lactic Acid Bacterial Strains
3.7. Auto-Aggregation and Surface Hydrophobicity of Four Lactic Acid Bacterial Strains
3.8. Antibacterial Activity of Four Lactic Acid Bacterial Strains
3.9. Hemolytic Activity and Antioxidant Capacity of Four Lactic Acid Bacterial Strains
3.10. Drug Sensitivity of Four Strains of Lactic Acid Bacteria
3.11. Solid-State Fermentation of Cottonseed Meal and Gossypol Detoxification, as Well as Nutritional Quality Indicators
4. Discussion
4.1. Isolation and Gossypol Degradation Capacity of Lactic Acid Bacteria Strains
4.2. Gastrointestinal Tolerance and Heat Resistance
4.3. Auto-Aggregation and Cell Surface Hydrophobicity
4.4. Antioxidant and Antimicrobial Activities
4.5. Safety Assessment: Antibiotic Susceptibility and Hemolytic Activity
4.6. Solid-State Fermentation and Nutritional Improvement of CSM
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rakita, S.; Kokić, B.; Manoni, M.; Mazzoleni, S.; Lin, P.; Luciano, A.; Ottoboni, M.; Cheli, F.; Pinotti, L. Cold-Pressed Oilseed Cakes as Alternative and Sustainable Feed Ingredients: A Review. Foods 2023, 12, 432. [Google Scholar] [CrossRef] [PubMed]
- Ninkuu, V.; Liu, Z.; Zhou, Y.; Sun, X. The Nutritional and Industrial Significance of Cottonseeds and Genetic Techniques in Gossypol Detoxification. Plants People Planet 2024, 6, 271–286. Available online: https://nph.onlinelibrary.wiley.com/doi/10.1002/ppp3.10433 (accessed on 20 August 2025). [CrossRef]
- Tao, A.; Wang, J.; Luo, B.; Liu, B.; Wang, Z.; Chen, X.; Zou, T.; Chen, J.; You, J. Research Progress on Cottonseed Meal as a Protein Source in Pig Nutrition: An Updated Review. Anim. Nutr. 2024, 18, 220–233. [Google Scholar] [CrossRef]
- Yu, J.; Yang, H.; Wang, J.; Chen, S.; Huang, Z.; Wang, J.; Wang, Z. Effects of Gossypol Acetate on Growth, Serum Biochemical Parameters, and Intestinal Health of Goslings. Poult. Sci. 2024, 103, 104025. [Google Scholar] [CrossRef]
- Lim, W.; Ham, J.; Park, S.; Bae, H.; You, S.; Song, G. Gossypol Induces Disruption of Spermatogenesis and Steroidogenesis in Male Mice. J. Agric. Food Chem. 2019, 67, 2075–2085. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yang, H.; Wang, J.; Huang, Z.; Chen, S.; Zhao, H.; Wang, J.; Wang, Z. Comprehensive Analysis of Histophysiology, Transcriptomics and Metabolomics in Goslings Exposed to Gossypol Acetate: Unraveling Hepatotoxic Mechanisms. Front. Vet. Sci. 2025, 12, 1527284. [Google Scholar] [CrossRef]
- Huang, X.; Hu, Y.; Li, Z.; Jiao, B.; Ma, X.; Guo, Q.; Wang, Q. Dephenolization Methods, Quality Characteristics, Applications, and Advancements of Dephenolized Cottonseed Protein: Review. Foods 2025, 14, 628. [Google Scholar] [CrossRef]
- Aguieiras, E.C.G.; de Barros, D.S.N.; Fernandez-Lafuente, R.; Freire, D.M.G. Production of Lipases in Cottonseed Meal and Application of the Fermented Solid as Biocatalyst in Esterification and Transesterification Reactions. Renew. Energy 2019, 130, 574–581. [Google Scholar] [CrossRef]
- Stecher, B.; Hardt, W.-D. Mechanisms Controlling Pathogen Colonization of the Gut. Curr. Opin. Microbiol. 2011, 14, 82–91. [Google Scholar] [CrossRef]
- Li, J.; Gao, T.; Hao, Z.; Guo, X.; Zhu, B. Anaerobic Solid-State Fermentation with Bacillus subtilis for Digesting Free Gossypol and Improving Nutritional Quality in Cottonseed Meal. Front. Nutr. 2022, 9, 1017637. [Google Scholar] [CrossRef]
- Lv, L.; Xiong, F.; Liu, Y.; Pei, S.; He, S.; Li, S.; Yang, H. The Rumen-Derived Lact. Mucosae LLK-XR1 Exhibited Greater Free Gossypol Degradation Capacity during Solid-State Fermentation of Cottonseed Meal and Probiotic Potential. BMC Microbiol. 2024, 24, 15. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, F.; Chen, H.; Li, S.; Cao, Z.; Wang, W.; Yang, H. Integrative Omics and Gene Knockout Analyses Suggest a Possible Gossypol Detoxification Mechanism and Potential Key Regulatory Genes of a Ruminal Lactobacillus rhamnosus Strain. J. Agric. Food Chem. 2025, 73, 1619–1629. [Google Scholar] [CrossRef]
- Wang, S.; Liang, Q.; Zhan, Y.; Mukhtar, H.; Fu, X.; Zhang, F.; Wang, Y.; Mou, H. A Novel Gossypol-Degradation Approach by Meyerozyma guilliermondii WST-M1 and Its Application in the Development of Cottonseed Meal as Feed Resource. Ind. Crops Prod. 2024, 220, 119299. [Google Scholar] [CrossRef]
- Ceja-Navarro, J.A.; Vega, F.E.; Karaoz, U.; Hao, Z.; Jenkins, S.; Lim, H.C.; Kosina, P.; Infante, F.; Northen, T.R. Gut Microbiota Mediate Caffeine Detoxification in the Primary Insect Pest of Coffee. Nat. Commun. 2015, 6, 7618, Erratum in Nat. Commun. 2023, 14, 6306. [Google Scholar] [CrossRef] [PubMed]
- Kaltenpoth, M.; Engl, T. Defensive Microbial Symbionts in Hymenoptera. Funct. Ecol. 2014, 28, 315–327. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The Gut Microbiota of Insects—Diversity in Structure and Function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef]
- Krempl, C.; Sporer, T.; Reichelt, M.; Ahn, S.-J.; Heidel-Fischer, H.; Vogel, H.; Heckel, D.G.; Joußen, N. Potential Detoxification of Gossypol by UDP-Glycosyltransferases in the Two Heliothine Moth Species Helicoverpa armigera and Heliothis virescens. Insect Biochem. Mol. Biol. 2016, 71, 49–57. [Google Scholar] [CrossRef]
- Frago, E.; Dicke, M.; Godfray, H.C.J. Insect Symbionts as Hidden Players in Insect-Plant Interactions. Trends Ecol. Evol. 2012, 27, 705–711. [Google Scholar] [CrossRef]
- Zhao, P.; Yao, X.; Cai, C.; Li, R.; Du, J.; Sun, Y.; Wang, M.; Zou, Z.; Wang, Q.; Kliebenstein, D.J.; et al. Viruses Mobilize Plant Immunity to Deter Nonvector Insect Herbivores. Sci. Adv. 2019, 5, eaav9801. [Google Scholar] [CrossRef]
- Dar, M.A.; Shaikh, A.F.; Pawar, K.D.; Xie, R.; Sun, J.; Kandasamy, S.; Pandit, R.S. Evaluation of Cellulose Degrading Bacteria Isolated from the Gut-System of Cotton Bollworm, Helicoverpa armigera and Their Potential Values in Biomass Conversion. PeerJ 2021, 9, e11254. [Google Scholar] [CrossRef]
- Gracy, R.G.; Malathi, V.M.; Jalali, S.K.; Jose, V.L.; Thulasi, A. Variation in Larval Gut Bacteria between Insecticide-Resistant and -Susceptible Populations of Helicoverpa armigera (Hübner) (Lepidoptera: Noctuidae). Phytoparasitica 2016, 44, 477–490. Available online: https://link.springer.com/article/10.1007/s12600-016-0547-9 (accessed on 21 August 2025). [CrossRef]
- Woo, I.-K.; Hyun, J.-H.; Jang, H.J.; Lee, N.-K.; Paik, H.-D. Probiotic Pediococcus acidilactici Strains Exert Anti-Inflammatory Effects by Regulating Intracellular Signaling Pathways in LPS-Induced RAW 264.7 Cells. Probiotics Antimicrob. Proteins 2024. [Google Scholar] [CrossRef]
- Jabbari, V.; Khiabani, M.S.; Mokarram, R.R.; Hassanzadeh, A.M.; Ahmadi, E.; Gharenaghadeh, S.; Karimi, N.; Kafil, H.S. Lactobacillus plantarum as a Probiotic Potential from Kouzeh Cheese (Traditional Iranian Cheese) and Its Antimicrobial Activity. Probiotics Antimicrob. Proteins 2017, 9, 189–193. [Google Scholar] [CrossRef]
- Ning, Z.; Xue, B.; Wang, H. Evaluation of the Adhesive Potential of Bacteria Isolated from Meat-Related Sources. Appl. Sci. 2021, 11, 10652. Available online: https://www.mdpi.com/2076-3417/11/22/10652 (accessed on 21 August 2025). [CrossRef]
- Alp, D.; Kuleaşan, H. Adhesion Mechanisms of Lactic Acid Bacteria: Conventional and Novel Approaches for Testing. World J. Microbiol. Biotechnol. 2019, 35, 156. Available online: https://link.springer.com/article/10.1007/s11274-019-2730-x (accessed on 21 August 2025). [CrossRef]
- Dinçer, E.; Kıvanç, M. In Vitro Evaluation of Probiotic Potential of Enterococcus faecium Strains Isolated from Turkish Pastırma. Arch. Microbiol. 2021, 203, 2831–2841. [Google Scholar] [CrossRef]
- Li, X.; Li, W.; Zhao, L.; Li, Y.; He, W.; Ding, K.; Cao, P. Characterization and Assessment of Native Lactic Acid Bacteria from Broiler Intestines for Potential Probiotic Properties. Microorganisms 2024, 12, 749. Available online: https://www.mdpi.com/2076-2607/12/4/749 (accessed on 21 August 2025). [CrossRef] [PubMed]
- Comunian, R.; Daga, E.; Dupré, I.; Paba, A.; Devirgiliis, C.; Piccioni, V.; Perozzi, G.; Zonenschain, D.; Rebecchi, A.; Morelli, L.; et al. Susceptibility to Tetracycline and Erythromycin of Lactobacillus paracasei Strains Isolated from Traditional Italian Fermented Foods. Int. J. Food Microbiol. 2010, 138, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Elnar, A.G.; Kim, G.-B. Probiotic Potential and Safety Assessment of Bacteriocinogenic Enterococcus faecalis CAUM157. Front. Microbiol. 2025, 16, 1563444. [Google Scholar] [CrossRef] [PubMed]
- Sara, P.; Ledala, N.; Rezaul, K.; Lin, Q.; Zhou, Y.; Provatas, A.A.; Bennett, E.; Lindberg, T.; Caimano, M.; Matson, A.P. Full Article: Cytotoxin-Producing Klebsiella oxytoca in the Preterm Gut and Its Association with Necrotizing Enterocolitis. Emerg. Microbes Infect. 2020, 9, 1321–1329. Available online: https://www.tandfonline.com/doi/full/10.1080/22221751.2020.1773743 (accessed on 3 September 2025).
- Gismene, C.; González, J.E.H.; de Freitas Calmon, M.; Nascimento, A.F.Z.; Santisteban, A.R.N.; Calil, F.A.; da Silva, A.D.T.; Rahal, P.; Góes, R.M.; Arni, R.K.; et al. Necrotic Activity of ExhC from Mammaliicoccus sciuri Is Mediated by Specific Amino Acid Residues. Int. J. Biol. Macromol. 2024, 254, 127741. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-K.; Li, W.-J.; Wu, Q.-C.; Wang, Y.-L.; Li, S.-L.; Yang, H.-J. Isolation and Identification of a Rumen Lactobacillus Bacteria and Its Degradation Potential of Gossypol in Cottonseed Meal during Solid-State Fermentation. Microorganisms 2021, 9, 2200. [Google Scholar] [CrossRef]
- Wang, W.-K.; Yang, H.-J.; Wang, Y.-L.; Yang, K.-L.; Jiang, L.-S.; Li, S.-L. Gossypol Detoxification in the Rumen and Helicoverpa armigera Larvae: A Review. Anim. Nutr. (Zhongguo Xu Mu Shou Yi Xue Hui) 2021, 7, 967–972. [Google Scholar] [CrossRef]
- Yousefi, M.; Shariatifar, N.; Tajabadi Ebrahimi, M.; Mortazavian, A.M.; Mohammadi, A.; Khorshidian, N.; Arab, M.; Hosseini, H. In Vitro Removal of Polycyclic Aromatic Hydrocarbons by Lactic Acid Bacteria. J. Appl. Microbiol. 2019, 126, 954–964. [Google Scholar] [CrossRef]
- Abbasiliasi, S.; Tan, J.S.; Ibrahim, T.A.T.; Ramanan, R.N.; Vakhshiteh, F.; Mustafa, S.; Ling, T.C.; Rahim, R.A.; Ariff, A.B. Isolation of Pediococcus acidilactici Kp10 with Ability to Secrete Bacteriocin-like Inhibitory Substance from Milk Products for Applications in Food Industry. BMC Microbiol. 2012, 12, 260. [Google Scholar] [CrossRef] [PubMed]
- Dunne, C.; O’Mahony, L.; Murphy, L.; Thornton, G.; Morrissey, D.; O’Halloran, S.; Feeney, M.; Flynn, S.; Fitzgerald, G.; Daly, C.; et al. In Vitro Selection Criteria for Probiotic Bacteria of Human Origin: Correlation with in Vivo Findings. Am. J. Clin. Nutr. 2001, 73, 386S–392S. [Google Scholar] [CrossRef]
- Li, S.; Tang, R.; Yi, H.; Cao, Z.; Sun, S.; Liu, T.-X.; Zhang, S.; Jing, X. Neutral Processes Provide an Insight into the Structure and Function of Gut Microbiota in the Cotton Bollworm. Front. Microbiol. 2022, 13, 849637. [Google Scholar] [CrossRef]
- Santivarangkna, C.; Kulozik, U.; Foerst, P. Inactivation Mechanisms of Lactic Acid Starter Cultures Preserved by Drying Processes. J. Appl. Microbiol. 2008, 105, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Baccouri, O.; Boukerb, A.M.; Farhat, L.B.; Zébré, A.; Zimmermann, K.; Domann, E.; Cambronel, M.; Barreau, M.; Maillot, O.; Rincé, I.; et al. Probiotic Potential and Safety Evaluation of Enterococcus faecalis OB14 and OB15, Isolated from Traditional Tunisian Testouri Cheese and Rigouta, Using Physiological and Genomic Analysis. Front. Microbiol. 2019, 10, 881. [Google Scholar] [CrossRef]
- Krausova, G.; Hyrslova, I.; Hynstova, I. In Vitro Evaluation of Adhesion Capacity, Hydrophobicity, and Auto-Aggregation of Newly Isolated Potential Probiotic Strains. Fermentation 2019, 5, 100. Available online: https://www.mdpi.com/2311-5637/5/4/100 (accessed on 21 August 2025). [CrossRef]
- Racioppo, A.; Accettulli, A.; d’Amelio, A.; Corbo, M.R.; Sinigaglia, M.; Speranza, B.; Bevilacqua, A. Effects of Different Environmental Stresses on Cell Surface Hydrophobicity of Lactobacilli, Bifidobacteria and Propionibacteria. BMC Microbiol. 2025, 25, 409. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Wang, J. Oxidative Stress Tolerance and Antioxidant Capacity of Lactic Acid Bacteria as Probiotic: A Systematic Review. Gut Microbes 2020, 12, 1801944. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-T.; Kim, J.-W.; Kim, S.-I.; Kim, S.; Nguyen, T.H.; Kang, C.-H. Antioxidant and Anti-Inflammatory Effect and Probiotic Properties of Lactic Acid Bacteria Isolated from Canine and Feline Feces. Microorganisms 2021, 9, 1971. [Google Scholar] [CrossRef] [PubMed]
- Williams, N.T. Probiotics. Am. J. Health-Syst. Pharm. 2010, 67, 449–458. [Google Scholar] [CrossRef]
- Kim, S.-K.; Guevarra, R.B.; Kim, Y.-T.; Kwon, J.; Kim, H.; Cho, J.H.; Kim, H.B.; Lee, J.-H. Role of Probiotics in Human Gut Microbiome-Associated Diseases. J. Microbiol. Biotechnol. 2019, 29, 1335–1340. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Zhao, J.; Zhang, X.; Liu, S.; Qi, H.; Qiao, F.; Yao, H. Isolation and Evaluation of Pediococcus acidilactici YH-15 from Cat Milk: Potential Probiotic Effects and Antimicrobial Properties. Heliyon 2024, 10, e39539. [Google Scholar] [CrossRef]
- Wenger, A.; Bär, C.; Portmann, R.; Schmidt, R.S.; Eugster, E.; Weisskopf, L.; Irmler, S. The Aminotransferase Aat Initiates 3-Phenyllactic Acid Biosynthesis in Pediococcus acidilactici. Front. Microbiol. 2023, 14, 1150425. [Google Scholar] [CrossRef]
- Duche, R.T.; Singh, A.; Wandhare, A.G.; Sangwan, V.; Sihag, M.K.; Nwagu, T.N.T.; Panwar, H.; Ezeogu, L.I. Antibiotic Resistance in Potential Probiotic Lactic Acid Bacteria of Fermented Foods and Human Origin from Nigeria. BMC Microbiol. 2023, 23, 142. [Google Scholar] [CrossRef]
- Mathur, S.; Singh, R. Antibiotic Resistance in Food Lactic Acid Bacteria—A Review. Int. J. Food Microbiol. 2005, 105, 281–295. [Google Scholar] [CrossRef]
- Yilmaz, N.; Ozogul, Y.; Coşkun Dağgeçen, E.; Akyol, İ.; Bhojraj Rathod, N.; Reddy Surasani, V.K.; Ozogul, F. Isolation, Characterization and Antibiotic Resistance of Lactic Acid Bacteria from Dairy and Seafood Sources. Food Biosci. 2025, 64, 105895. [Google Scholar] [CrossRef]
- Sethuvel, D.P.M.; Bakthavatchalam, Y.D.; Karthik, M.; Irulappan, M.; Shrivastava, R.; Periasamy, H.; Veeraraghavan, B. β-Lactam Resistance in ESKAPE Pathogens Mediated through Modifications in Penicillin-Binding Proteins: An Overview. Infect. Dis. Ther. 2023, 12, 829–841. [Google Scholar] [CrossRef]
- Tarrah, A.; da Silva Duarte, V.; de Castilhos, J.; Pakroo, S.; Lemos Junior, W.J.F.; Luchese, R.H.; Fioravante Guerra, A.; Rossi, R.C.; Righetto Ziegler, D.; Corich, V.; et al. Probiotic Potential and Biofilm Inhibitory Activity of Lactobacillus casei Group Strains Isolated from Infant Feces. J. Funct. Foods 2019, 54, 489–497. [Google Scholar] [CrossRef]
- Zhao, H.-M.; Guo, X.-N.; Zhu, K.-X. Impact of Solid State Fermentation on Nutritional, Physical and Flavor Properties of Wheat Bran. Food Chem. 2017, 217, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Wu, D.; Liang, X.; Wang, Z.; Zou, H.; Wu, F.; Li, H.; Jiang, Y.; Peng, Q.; Xiao, J.; et al. Solid State Fermentation Improves the Utilization Value of Cotton Stalk. Ind. Crops Prod. 2025, 230, 121113. [Google Scholar] [CrossRef]
- Ramakrishnan, V.; Goveas, L.C.; Narayan, B.; Halami, P.M. Comparison of Lipase Production by Enterococcus Faecium MTCC 5695 and Pediococcus acidilactici MTCC 11361 Using Fish Waste as Substrate: Optimization of Culture Conditions by Response Surface Methodology. ISRN Biotechnol. 2013, 2013, 980562. [Google Scholar] [CrossRef]
- Fusco, V.; Quero, G.M.; Cho, G.-S.; Kabisch, J.; Meske, D.; Neve, H.; Bockelmann, W.; Franz, C.M.A.P. The Genus Weissella: Taxonomy, Ecology and Biotechnological Potential. Front. Microbiol. 2015, 6, 155. [Google Scholar] [CrossRef]
- Handa, C.L.; de Lima, F.S.; Guelfi, M.F.; da Silva Fernandes, M.; Georgetti, S.R.; Ida, E.I. Parameters of the Fermentation of Soybean Flour by Monascus purpureus or Aspergillus oryzae on the Production of Bioactive Compounds and Antioxidant Activity. Food Chem. 2019, 271, 274–283. [Google Scholar] [CrossRef]
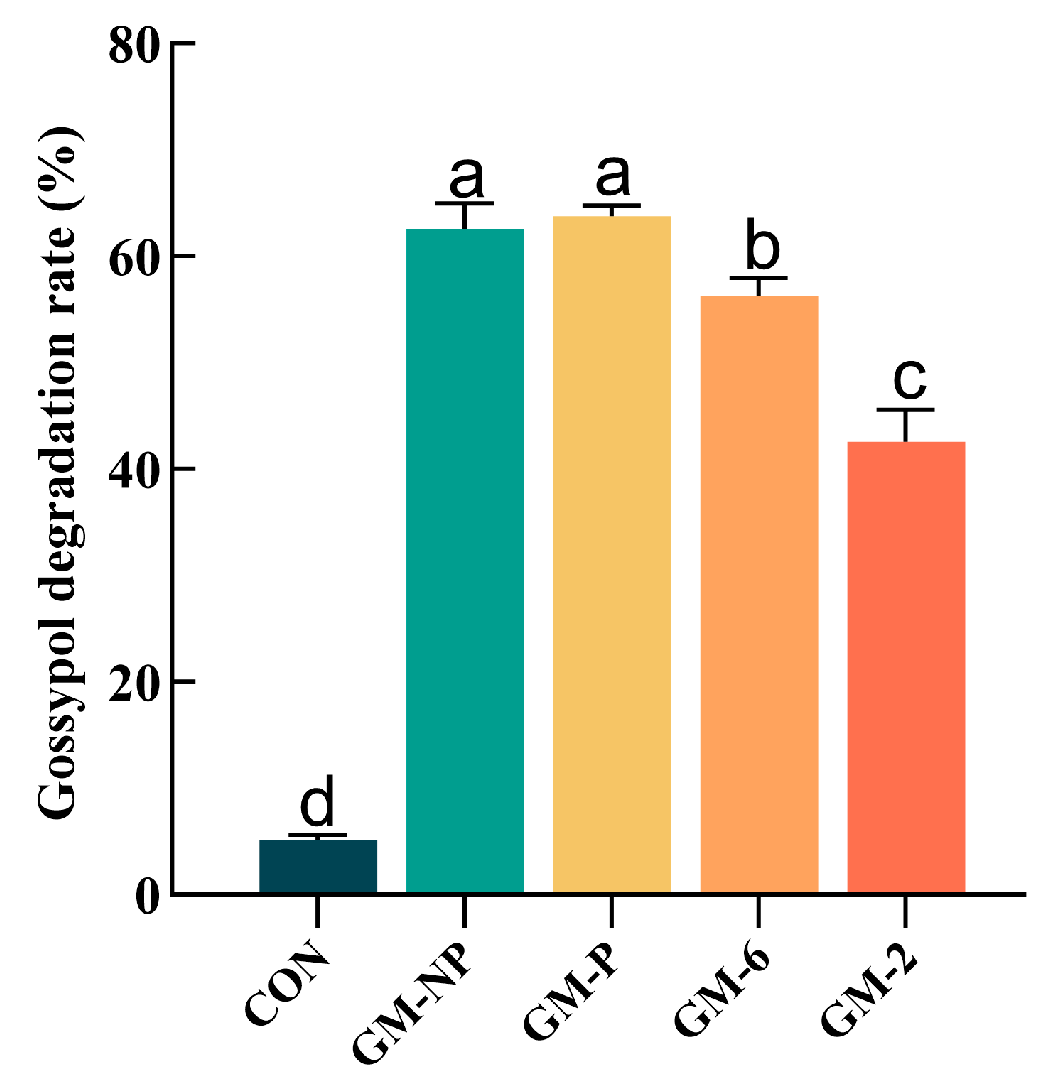
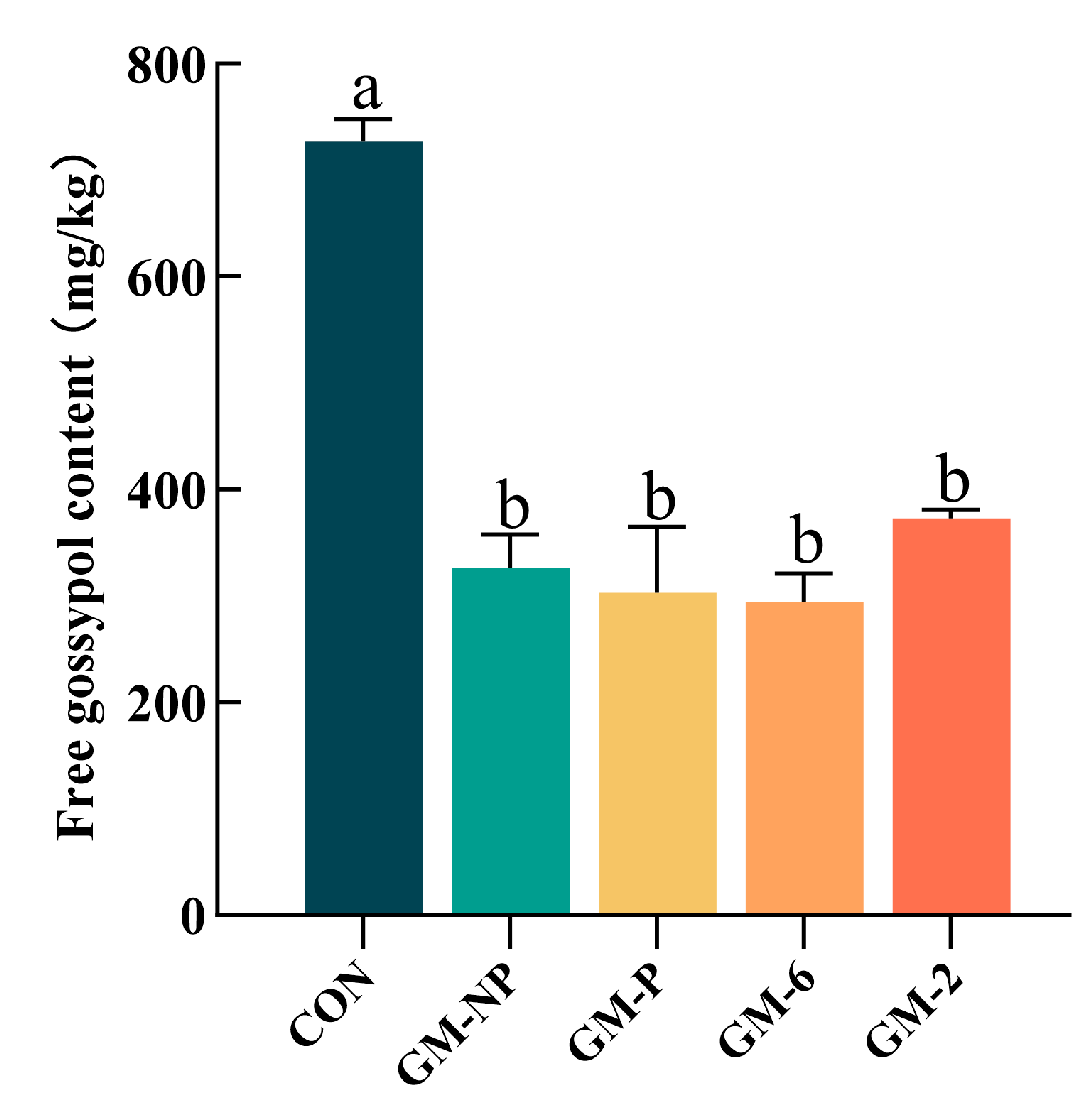
| Media | Composition |
|---|---|
| LB | 1%yeast extract, 2%peptone, 1%NaCl, 2% agar |
| MRS | 1%peptone, 0.8%Beef extract, 0.4%Yeast, 2%glucose, 0.2%K2HPO4, 0.2%(NH4)2HC6H5O7, 0.5%CH3COONa, 0.02%MgSO4, 0.004%MgSO4, 0.1%Tween 80 |
| MRSA | 1%peptone, 0.8%Beef extract, 0.4%Yeast, 2%glucose, 0.2%K2HPO4, 0.2%(NH4)2HC6H5O7, 0.5%CH3COONa, 0.02%MgSO4, 0.004%MgSO4, 0.1%Tween 80, 2%agar |
| GMRS | 0.1%gossypol, 1%peptone, 0.8%Beef extract, 0.4%Yeast, 0.2%K2HPO4, 0.2%(NH4)2HC6H5O7, 0.5%CH3COONa, 0.02%MgSO4, 0.004%MgSO4, 0.1%Tween 80 |
| GMRSA | 0.2%gossypol, 1%peptone, 0.8%Beef extract, 0.4%Yeast, 0.2%K2HPO4, 0.2%(NH4)2HC6H5O7, 0.5%CH3COONa, 0.02%MgSO4, 0.004%MgSO4, 0.1%Tween 80, 2% agar |
| Strains | Closest Match | GenBank Accession No. | Similarity/% |
|---|---|---|---|
| GM-NP | Pediococcus acidilactici | NR042057.1 | 99.184% |
| GM-P | Pediococcus acidilactici | NR042057.1 | 99.515% |
| GM-2 | Weissella confusa | NR040816.1 | 99.861% |
| GM-3 | Mammaliicoccus sciuri | NR041327.1 | 99.859% |
| GM-4 | Enterococcus innesii | NR181755.1 | 99.718% |
| GM-5 | Enterococcus innesii | NR181755.1 | 99.858% |
| GM-6 | Enterococcus faecalis | NR115765.1 | 100.000% |
| GM-7 | Klebsiella oxytoca | NR118853.1 | 99.861% |
| GM-8 | Klebsiella oxytoca | NR118853.1 | 99.645% |
| GM-9 | Klebsiella oxytoca | NR118853.1 | 99.363% |
| GM-10 | Klebsiella oxytoca | NR118853.1 | 99.153% |
| Item | P. acidilactici GM-NP | P. acidilactici GM-P | E. faecalis GM-6 | W. confusa GM-2 |
|---|---|---|---|---|
| Aesculin | + | + | + | − |
| 1%Sodium Hippurat | + | + | + | + |
| Cellobiose | − | − | + | + |
| Maltose | − | − | + | + |
| Mannose | − | − | + | + |
| Salicin | − | − | + | + |
| Sorbitol | − | − | + | + |
| Sucrose | − | − | + | + |
| Raffinose | − | − | + | + |
| Inulin | − | − | − | + |
| Lactose | − | − | + | + |
| Items | Cell Numbers (Log CFU/mL) | |||
|---|---|---|---|---|
| P. acidilactici GM-NP | P. acidilactici GM-P | E. faecalis GM-6 | W. confusa GM-2 | |
| Artificial gastric juice tolerance | ||||
| Initial cell number | 9.71 ± 0.03 | 9.36 ± 0.02 | 8.28 ± 0.05 | 8.52 ± 0.05 |
| pH2.5, 0.3% pepsin, 1.5 h | 6.98 ± 0.10 | 6.95 ± 0.09 | 5.91 ± 0.20 | 5.82 ± 0.17 |
| Survival rate (%) | 71.83 a ± 1.10 | 74.28 a ± 1.06 | 71.35 a ± 2.40 | 68.31 b ± 2.07 |
| Artificial intestinal fluid tolerance | ||||
| Initial cell number | 9.43 ± 0.02 | 9.40 ± 0.07 | 8.62 ± 0.02 | 8.37 ± 0.01 |
| pH6.8, 1% trypsin, three h | 8.48 ± 0.16 | 8.52 ± 0.07 | 7.68 ± 0.07 | 7.48 ± 0.07 |
| Survival rate (%) | 89.93 a ± 1.50 | 90.64 a ± 0.86 | 89.10 a ± 0.86 | 85.98 b ± 0.53 |
| Artificial bile salt tolerance | ||||
| Initial cell number | 8.66 ± 0.03 | 8.69 ± 0.02 | 8.56 ± 0.03 | 8.53 ± 0.02 |
| 0.3% Ox Gall, four h | 6.78 ± 0.13 | 7.36 ± 0.20 | 5.87 ± 0.05 | 6.26 ± 0.15 |
| Survival rate (%) | 78.11 b ± 1.32 | 84.76 a ± 2.16 | 68.54 d ± 0.78 | 73.46 c ± 1.79 |
| Items | Survival Rate (%) | |||
|---|---|---|---|---|
| P. acidilactici GM-NP | P. acidilactici GM-P | E. faecalis GM-6 | W. confusa GM-2 | |
| Heat treatment time/min | ||||
| 60 °C, 5 min | 67.23 b ± 0.54 | 66.36 bc ± 0.64 | 72.20 a ± 0.47 | 60.94 c ± 0.40 |
| 60 °C, 10 min | 60.29 b ± 0.9 | 57.44 c ± 0.97 | 63.06 a ± 0.87 | 52.51 d ± 0.42 |
| 60 °C, 15 min | 48.56 c ± 0.86 | 46.72 c ± 0.81 | 53.83 a ± 1.26 | 43.96 d ± 0.35 |
| 70 °C, 5 min | 57.74 b ± 0.46 | 52.53 c ± 0.74 | 60.86 a ± 0.58 | 51.64 c ± 0.48 |
| 70 °C, 10 min | 52.14 c ± 0.69 | 49.01 d ± 0.99 | 56.91 a ± 1.45 | 44.69 e ± 0.32 |
| 70 °C, 15 min | 33.56 b ± 1.14 | 32.79 b ± 0.72 | 39.04 a ± 0.41 | 31.31 b ± 0.38 |
| 80 °C, 5 min | 34.81 b ± 0.38 | 31.87 c ± 1.00 | 37.97 a ± 0.91 | 25.75 d ± 0.64 |
| 80 °C, 10 min | 6.68 bc ± 0.40 | 5.17 c ± 0.22 | 9.04 a ± 0.87 | 7.10 bc ± 0.82 |
| 80 °C, 15 min | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| 90 °C, 5 min | 17.72 b ± 0.69 | 15.87 bc ± 0.33 | 21.49 a ± 1.14 | 15.32 c ± 0.53 |
| 90 °C, 10 min | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| 90 °C, 15 min | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Items | Strains | |||
|---|---|---|---|---|
| P. acidilactici GM-NP | P. acidilactici GM-P | E. faecalis GM-6 | W. confusa GM-2 | |
| Assessment of auto-aggregation ability (%) | ||||
| 8 h | 48.60 a ± 1.01 | 46.77 a ± 0.55 | 37.21 c ± 1.11 | 43.39 b ± 1.27 |
| 24 h | 82.96 a ± 0.95 | 81.10 a ± 1.80 | 72.89 b ± 1.76 | 74.34 b ± 0.73 |
| Cell surface properties (%) | 54.45 b ± 1.62 | 51.63 bc ± 1.95 | 47.92 c ± 1.91 | 41.60 d ± 2.29 |
| Pathogens | Antimicrobial Activities | |||
|---|---|---|---|---|
| P. acidilactici GM-NP | P. acidilactici GM-P | E. faecalis GM-6 | W. confusa GM-2 | |
| E. coli ATCC25922 | ★★★ | ★★★★ | ★★ | ★ |
| S. aureus ATCC6538 | ★★★ | ★★★ | ★★ | ★ |
| Salmonella ATCC14028 | ★★★★ | ★★★★ | ★★ | - |
| P. aeruginosa ATCC9027 | ★★★ | ★★★ | ★★ | ★ |
| Items | Strains | |||
|---|---|---|---|---|
| P. acidilactici GM-NP | P. acidilactici GM-P | E. faecalis GM-6 | W. confusa GM-2 | |
| Hemolytic activity | γ | γ | γ | γ |
| Free radical scavenging rate (%) (DPPH) | 43.81 a ± 1.11 | 45.29 a ± 1.26 | 28.73 c ± 1.32 | 37.62 b ± 0.50 |
| Antibiotic Susceptibility | Strains | |||
|---|---|---|---|---|
| P. acidilactici GM-NP | P. acidilactici GM-P | E. faecalis GM-6 | W. confusa GM-2 | |
| Cefalotin | S | S | I | S |
| Clindamycin | S | S | R | S |
| Chloramphenicol | I | S | I | S |
| Penicillin | R | R | S | S |
| Rifampicin | S | S | S | S |
| Erythromycin | S | S | I | S |
| Ampicillin | S | S | I | S |
| Tetracycline | S | S | I | S |
| Levofloxacin | R | R | I | S |
| Gentamicin | R | R | I | S |
| Streptomycin | R | R | I | R |
| Polymyxin B | R | R | R | R |
| Enrofloxacin | R | R | S | I |
| Ciprofloxacin | R | R | I | R |
| Ofloxacin | R | R | I | I |
| Items | CON | P. acidilactici GM-NP | P. acidilactici GM-P | E. faecalis GM-6 | W. confusa GM-2 |
|---|---|---|---|---|---|
| CP (%) | 49.77 b ± 0.16 | 53.43 a ± 1.05 | 53.39 a ± 0.77 | 54.35 a ± 0.56 | 52.83 a ± 0.65 |
| ASP (%) | 3.49 b ± 0.21 | 6.17 a ± 0.13 | 6.10 a ± 0.06 | 6.08 a ± 0.07 | 6.48 a ± 0.72 |
| NDF (%) | 36.76 a ± 1.52 | 28.94 cd ± 0.71 | 30.01 cd ± 1.29 | 29.40 cd ± 1.13 | 31.97 bc ± 0.59 |
| ADF (%) | 27.53 a ± 0.73 | 19.87 d ± 1.44 | 18.83 cd ± 0.27 | 22.31 bc ± 0.75 | 23.28 b ± 0.78 |
| Crude fat (%) | 1.30 a ± 0.06 | 0.71 b ± 0.04 | 0.51 b ± 0.13 | 0.44 b ± 0.03 | 0.59 b ± 0.10 |
| Ash (%) | 6.72 ± 0.58 | 7.26 ± 0.49 | 6.41 ± 0.11 | 6.99 ± 0.32 | 7.17 ± 0.14 |
| Ca (%) | 0.32 ± 0.02 | 0.37 ± 0.03 | 0.36 ± 0.03 | 0.34 ± 0.04 | 0.30 ± 0.08 |
| P (%) | 0.70 ± 0.05 | 0.68 ± 0.04 | 0.79 ± 0.07 | 0.70 ± 0.01 | 0.72 ± 0.09 |
| pH | 6.39 a ± 0.03 | 5.99 b ± 0.22 | 5.58 C ± 0.02 | 5.92 b ± 0.04 | 6.11 b ± 0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Deng, S.; Zhang, P.; Lu, Q.; Pu, W.; Ma, M.; Li, S.; Zhang, W.; Chen, C. Gut-Derived Lactic Acid Bacteria from Cotton Bollworm Exhibit Efficient Gossypol Degradation and Probiotic Potential During Solid-State Fermentation of Cottonseed Meal. Fermentation 2025, 11, 598. https://doi.org/10.3390/fermentation11100598
Li S, Deng S, Zhang P, Lu Q, Pu W, Ma M, Li S, Zhang W, Chen C. Gut-Derived Lactic Acid Bacteria from Cotton Bollworm Exhibit Efficient Gossypol Degradation and Probiotic Potential During Solid-State Fermentation of Cottonseed Meal. Fermentation. 2025; 11(10):598. https://doi.org/10.3390/fermentation11100598
Chicago/Turabian StyleLi, Sijin, Shangya Deng, Peng Zhang, Qicheng Lu, Wei Pu, Mingyu Ma, Shu Li, Wenju Zhang, and Cheng Chen. 2025. "Gut-Derived Lactic Acid Bacteria from Cotton Bollworm Exhibit Efficient Gossypol Degradation and Probiotic Potential During Solid-State Fermentation of Cottonseed Meal" Fermentation 11, no. 10: 598. https://doi.org/10.3390/fermentation11100598
APA StyleLi, S., Deng, S., Zhang, P., Lu, Q., Pu, W., Ma, M., Li, S., Zhang, W., & Chen, C. (2025). Gut-Derived Lactic Acid Bacteria from Cotton Bollworm Exhibit Efficient Gossypol Degradation and Probiotic Potential During Solid-State Fermentation of Cottonseed Meal. Fermentation, 11(10), 598. https://doi.org/10.3390/fermentation11100598








