Abstract
3-Nitrooxypropanol (3-NOP) is a nitrooxy compound that specifically targets methyl-coenzyme M reductase (MCR), ultimately resulting in a reduction in methane production. In this study, we undertook an in vitro investigation of the effects of different dosages of 3-NOP on ruminal fermentation parameters, methane production, and the microbial community. A single-factor completely randomized design was adopted, comprising a control treatment (C), where no 3-NOP was added to the fermentation substrate, and three 3-NOP treatments, where 0.025 mg (low-dose treatment, LD), 0.05 mg (medium-dose treatment, MD), or 0.1 mg (high-dose treatment, HD) was added to 1 g of fermentation substrate (DM basis), followed by incubation for 24 h in vitro. The results showed that, compared with the control treatment, the three dosages of 3-NOP reduced total gas production, methane production, and acetate production (all p < 0.01). In contrast, 3-NOP treatment increased H2 production and the molar proportions of propionate and butyrate (all p ≤ 0.02), resulting in a decrease in the acetate-propionate ratio (p < 0.01). Meanwhile, the microbial profiles were not altered by the treatments, but the relative abundances of Prevotella, Methanobrevibacter, and Ophryoscolex were increased by the MD and HD treatments (all p < 0.01), whereas those of Methanosarcina, Methanosaeta, Sphaerochaeta, RFN20, Entodinium, and Diplodinium were decreased by the HD treatment (all p ≤ 0.03). Moreover, the results of a correlation analysis showed that there was a certain correlation between these microorganisms and total gas production, methane production, H2 production, acetate, propionate, and butyrate. In summary, under in vitro conditions, the addition of 3-NOP to the diet affected the microbial community structure, thereby altering the ruminal fermentation pattern and reducing methane production. Our results indicated that 0.05 mg per g of dietary DM is the recommended inclusion ratio for 3-NOP in the diet of lambs.
1. Introduction
In addition to converting fibrous forage that cannot be directly used by monogastric animals into high-quality livestock products such as meat and milk, ruminants also emit greenhouse gases, such as methane (CH4), into the environment. Methanogenesis from ruminal fermentation not only contributes to global warming but also represents a 2% to 12% loss of the gross energy ingested by ruminants [1]. Thus, considerable research efforts have been undertaken worldwide aimed at reducing CH4 production by these animals. Over recent years, it has been found that 3-nitrooxypropanol (3-NOP), a structural analog of methyl-coenzyme M, can preferentially bind to methyl-coenzyme M reductase (MCR) [2] and catalyze the oxidation of nickel from Ni + 1 to Ni + 2, thereby temporarily inactivating MCR enzyme activity and specifically inhibiting ruminal CH4 production [3]. Some researchers observed significant dose-dependent effects of 3-NOP on CH4 reduction in both in vitro [4,5] and in vivo [6,7,8] studies. Furthermore, Dijkstra et al. [6] reported that the average 3-NOP doses administered to different groups of ruminants (beef cattle, dairy cows, and sheep) were determined to be 144, 81, and 111.2 mg/kg of feed DM, respectively, and that the inhibitory effect of 3-NOP fluctuated greatly, ranging from 26% to 76%. Likewise, subsequent research by Yu et al. [9] found that, when administered at the same additive dose, the efficacy of 3-NOP in inhibiting methane emissions was higher in dairy cows compared to beef cattle. In addition, the differential effect of 3-NOP on the rumen microbiota was also closely related to CH4 production. Some studies have shown that 3-NOP inhibits CH4 emissions by decreasing the copy numbers of methanogens [10,11,12] or reducing the relative abundance of Methanobrevibacter [13,14,15]; in contrast, others have reported that 3-NOP has no observable impact on the copy numbers of methanogens [16,17,18] or on the relative abundance of methanogens [19]. Liu et al. [20] even reported that the relative abundance of Methanobrevibacter_sp._AbM4 was increased by 3-NOP treatment, but that CH4 emissions still decreased. Previous studies preliminarily demonstrated that 3-NOP is an effective, long-acting inhibitor of methane production with no side effects in large adult ruminants [21,22]. These variations in 3-NOP in response to CH4 emission could possibly be due to differences in the type of animal used, dietary composition, dry matter intake, and 3-NOP dosage [23,24]. Additionally, the rumen microbial community in adult ruminants is highly stable [25,26]. Regulatory measures temporarily affect the mature rumen microbiota, which revert to their pretreatment state once the measures are discontinued [27]. Abecia et al. [28] and Meale et al. [29] found that early dietary regulation can alter archaeal colonization in the rumen, thereby impacting methane emissions from young ruminants. This may suggest that utilizing a lower dose of 3-NOP to regulate methane emissions in young ruminants could potentially become a viable strategy. However, not only is the literature on the effects of 3-NOP on methanogenesis in sheep very scarce, but the dose-response effects of a lower dose of 3-NOP on small young ruminants, especially lambs, are still unknown. Importantly, there are limited data available on how 3-NOP directly influences the diversity of protozoa, as they form an integral part of the rumen microecosystem and play a non-negligible role in ruminal methane production.
Based on the aforementioned information, we hypothesized that administering a lower dose of 3-NOP to young ruminants compared to adult ruminants (81–144 mg 3-NOP/kg of feed DM) could significantly affect rumen fermentation as well as the diversity of rumen microbiota, thereby inhibiting rumen CH4 production. Therefore, the objective of this study was to investigate the dose-response relationships of lower doses of 3-NOP with gas production, fermentation characteristics, and the diversity of rumen microbiota (including archaea, bacteria, and protozoa) through in vitro incubation using rumen fluid from lambs, ultimately aiming to provide a theoretical basis and reference for the application of lower doses of 3-NOP in reducing ruminal CH4 emissions from young ruminants.
2. Materials and Methods
2.1. Animals, Diet, and Experimental Design
Four healthy, weaned, and small-tailed Han lambs (2 months of age, BW: 16 ± 1.42 kg) served as ruminal fluid donors. All procedures involving animals were approved by the Animal Care and Use Committee of Inner Mongolia Minzu University (IMUN2022-56). Lambs were fed the same amount of total mixed ration (TMR) twice daily (08:00 and 18:00 h) and were allowed free access to fresh water. Before collecting rumen fluid, the donor lambs were adaptively fed for 14 days to ensure the stability and consistency of the rumen microbial community. According to China’s ‘Feeding Standard for Meat Sheep’ (NY/T 816-2021) [30], the basal TMR was formulated to meet the nutritional requirements for a lamb to achieve a daily weight gain of 100 g, with a concentrate-roughage ratio of 40:60. The ingredients and nutrient levels of the TMR are presented in Table 1, and the chemical composition of the individual ingredients that make up the diet is shown in Table 2.

Table 1.
Composition and nutrient levels of basal diets (DM basis).

Table 2.
Chemical compositions of the ingredients that make up the diet (%).
A single-factor and completely randomized design was used in this study, comprising a control (C) treatment, where no 3-NOP was added to the fermentation substrate, and three 3-NOP treatments, where 0.025 mg (low-dose treatment, LD), 0.05 mg (medium-dose treatment, MD), or 0.1 mg (high-dose treatment, HD) of 3-NOP (per gram, dry matter basis) was added to the fermentation substrate. 3-NOP was purchased from MedChemExpress LLC (Shanghai, China) and was supplied as a yellow transparent liquid with a purity of ≥98%. The diet fed to the rumen fluid donors was consistent with the substrate used for in vitro incubation, but it did not specifically include 3-NOP. For the experiment, the substrate was pulverized in a lab mill (JXFM110, Jingxin, Shanghai, China) to pass through a 40-mesh sieve and dried at 65 °C before use.
2.2. In Vitro Incubation
The ruminal fluid was collected via oral stomach tubing from four lambs 2 h after the morning feeding. A 500 mL syringe was used to pump at least 300 mL of rumen fluid from each lamb through a stainless steel probe (with a 2.5 mm screen). The rumen fluid was pooled in equal portions and strained through 12 layers of cheesecloth to remove feed particles, placed in a sealed, prewarmed 39 °C thermos flask filled with CO2, and then immediately transported to the laboratory and placed into a 39 °C constant-temperature water bath. A culture solution was prepared by mixing strained ruminal fluid with pre-warmed (39 °C) artificial saliva, prepared according to Menke et al. [31], at a ratio of 1:2 (ruminal fluid-buffer, v/v) under continual CO2 flow to ensure an anaerobic state. After mixing, 80 mL of the culture solution and 3-NOP were added to a 120 mL fermentation flask containing 1 g of fermentation substrate (DM basis). A silicone stopper was used to seal the fermentation flask, and a 200 mL aluminum foil gas bag was connected to the fermentation flask for the real-time collection of gases produced through fermentation. Finally, the sealed fermentation flask was transferred to an air bath shaking incubator (135 rpm) for up to 24 h at 39 °C. Each incubation was conducted every other day, and a total of five separate runs were performed under similar conditions and following the same procedure. For each incubation, each treatment was replicated using eight fermentation flasks, and an additional four blanks (containing only the culture solution) were also prepared to correct for gas production.
2.3. Sample Collection and Measurements
After 24 h of incubation, the flasks were placed in an ice-water bath to terminate the in vitro fermentation. The pH of the culture solution was immediately measured using a pH meter. A glass syringe was used to measure the volume of gas in the gas bag, which was considered the total gas production. Subsequently, a subsample of the gas was transferred to a 10 mL evacuated vacuum tube and analyzed for CH4 and H2 contents using a gas chromatograph equipped with a flame ionization detector and a thermal conductivity detector as described by Kim et al. [32]. The amounts of gas produced (CH4 or H2) were calculated based on the formula according to Soltan et al. [33,34]. A 5 mL sample of culture solution was acidified by adding 1 mL of 25% metaphosphoric acid and stored at −20 °C for volatile fatty acid (VFA) and ammonia-N (NH3-N) analysis. The concentrations of VFAs were determined by gas chromatography as described by Nolan et al. [35]. The NH3-N concentration was determined based on the phenol hypochlorite method [36]. Another 20 mL of culture solution was stored at −80 °C for DNA extraction and 16S rRNA gene sequencing. The fermentation residue was first filtered through a pre-weighed nylon bag (5 × 10 cm, 200-μm pore size), washed repeatedly with distilled water [37], and dried in a drying oven at 60 °C for 48 h for measurement of the dry matter degradability (DMD) and neutral detergent fiber degradability (NDFD) [38]. The contents of dry matter (DM), ether extract (EE), crude ash (CA), and crude protein (CP) in the feed samples were analyzed according to the method described by AOAC (1995) [39]. The contents of neutral detergent fiber (NDF) and acid detergent fiber (ADF) in the feed samples were determined using an A220 fiber analyzer (ANKOM Technology Corporation, Macedon, NY, USA) according to the method described by Van Soest et al. [40].
2.4. DNA Extraction, Real-Time PCR (qPCR) Analysis, Sequencing, and Diversity Analysis
An E.Z.N.A. Soil DNA Kit (D5625-01, Omega BIO-TEK, Norcross, GA, USA) was used to extract total DNA from the culture solution. For the identification of bacteria, the V3–V4 variable region of the 16S rRNA gene was amplified using the forward primer 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and the reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′) [41]. For the identification of archaea, the V4–V5 variable region of the 16S rRNA gene was amplified using the forward primer 524F (5′-TGYCAGCCGCCGCGGTAA-3′) and the reverse primer 958R (5′-YCCGGCGTTGAVTCCAATT-3′) [42]. For the identification of protozoa, the V4–V5 region of the 18S rRNA gene was amplified using the forward primer 316F (5′-GCTTTCGWTGGTAGTGTATT-3′) and the reverse primer 539R (5′-ACTTGCCCTCYAATCGTWCT-3′) [43]. The appropriate products were then purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA). Purified amplicons were pooled in equimolar amounts and underwent paired-end sequencing on the Illumina NovaSeq PE250 platform by Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China) using standard protocols. Bioinformatic analyses of the rRNA sequencing data were performed with QIIME2 (q2, v.2019.4) and the R package [44]. Raw sequences were demultiplexed by the q2-demux plug-in and quality filtered by the q2-cutadapt plug-in [45]. The DADA2 plug-in was employed for primer removal, quality filtering, denoising, merging, chimera removal, and yielding valid sequences [46]. Valid sequences were grouped into amplicon sequence variants (ASVs) at 100% similarity [47], which were then assigned to their respective taxa using the q2 (QIIME2)-feature-classifier plug-in based on the SILVA reference database (release 138) [48].
Total bacteria, methanogens, and protozoa based on the copy numbers of 16S/18S rRNA genes in the culture solution were estimated using qPCR as previously described by Haisan et al. [10].
2.5. Statistical Analysis
All high-throughput sequencing data were analyzed on the Shanghai Personal Gene Cloud Platform (https://www.genescloud.cn/hom, accessed on 16 May 2024). Chao 1, Shannon, and other alpha-diversity indices were calculated using mothur software (version 1.48.1, http://www.mothur.org/wiki/Calculators, accessed on 16 May 2024), and inter-group differences in alpha-diversity were analyzed by using the Kruskal–Wallis test. Similarities in microbial community structure between samples were tested using PCoA based on Bray–Curtis distances; linear discriminant effect size (LEfSe) analysis (http://huttenhower.sph.harvard.edu/LEfSe, accessed on 16 May 2024) was used to identify microbial groups displaying significant differences in abundance between different groups from the phylum to the genus level [49].
The analysis of all data in this study was the average of five incubation runs and executed using the MIXED procedure of SAS 9.4 software (SAS Institute, Inc., Cary, NC, USA). Data indices, including in vitro ruminal fermentation parameters, that were preliminarily collated in Excel 2021 were subjected to one-way analysis of variance using the ANOVA program, and orthogonal polynomial contrasts were used to evaluate the linear and quadratic effects of 3-NOP; relative abundances (%) of rumen microorganisms were determined using the Kruskal–Wallis test. The following model was used for data analysis:
where Yijk is the observation, μ is the overall mean, NOPi is the fixed effect of 3-NOP treatments (i = 0 mg/g of DM, 0.025 mg/g of DM, 0.05 mg/g of DM, or 0.1 mg/g of DM), τj is the random effect of incubation runs (j = 1, 2, 3, 4, or 5), εk is the random effect of sheep donors (k = 1, 2, 3, or 4), and eijk is the random residual error. p-values < 0.05 were considered significant, while tendencies were declared at 0.05 < p ≤ 0.10.
Yijk = μ + NOPi + τj + εk+ eijk
3. Results
3.1. The Effects of 3-NOP on Gas Production and the Degradability of the Substrate
As shown in Table 3, compared with the control treatment, the addition of 3-NOP did not affect the DMD (p = 0.67) or NDFD (p = 0.95); however, total gas production and methane production showed linear (both p < 0.01) and quadratic (both p < 0.01) decreases, while H2 production showed linear and quadratic increases (both p < 0.01), with an increase in the addition of 3-NOP. Of the three 3-NOP treatments, the HD treatment exhibited the lowest total gas production (p < 0.01) and methane production (p < 0.01), as well as the highest H2 production (p < 0.01).

Table 3.
Effect of 3-NOP on gas parameters and fermentation substrate degradability.
3.2. The Effects of 3-NOP on In Vitro Fermentation Characteristics and Microbial Profile
Compared with the C treatment, the molar proportions of propionate (p < 0.01) and butyrate (p = 0.02) were increased in the 3-NOP treatments, whereas that of acetate was decreased (p < 0.01) (Table 4). Consequently, the acetate-to-propionate ratio was also reduced (p < 0.01). The concentrations of total volatile fatty acids (TVFAs) in the incubation were lower (p < 0.01) in the MD and HD treatments than in the C treatment. Furthermore, linear and quadratic effects (both p ≤ 0.03) were also observed between the dosage of 3-NOP and volatile fatty acid profiles (TVFAs, acetate, propionate, butyrate, and acetate/propionate). No significant differences in incubation pH, NH3-N, or valerate contents were detected among the four treatments (p = 0.08, p = 0.90, and p = 0.96, respectively). The total copy numbers of bacteria, methanogens, and protozoa were also not affected by the treatment (p = 0.35, p = 0.36, and p = 0.36, respectively). The total copy numbers of methanogens and protozoa showed a tendency to decrease linearly, but no statistical difference was observed (p = 0.07 and p = 0.09, respectively).

Table 4.
Effects of adding different levels of 3-NOP on rumen fermentation parameters in vitro.
3.3. The Effects of 3-NOP on the Incubation Microbiota
The initially detected sequences were subjected to quality filtering, duplicate removal, clustering, and other steps using DADA2 and Vsearch. As shown in Figure 1A, a total of 1895 ASV taxonomic units were identified in the ruminal archaeal community. Of these, 339 were shared among all four treatments, accounting for 17.9% of the total ASVs; 575 were unique to the C treatment, accounting for 30.3% of the total ASVs; 335 were unique to the LD treatment, accounting for 17.7% of the total ASVs; 205 were unique to the MD treatment, accounting for 10.8% of the total ASVs; and 202 were unique to the HD treatment, accounting for 10.6% of the total ASVs. Alpha- and beta-diversity are typically used to reflect heterogeneity among different biological communities. As shown in Figure 2A, compared with the C treatment, the archaeal Chao1 index (p < 0.01) and the observed species count (p < 0.01) were decreased in the MD and HD treatments. Additionally, the Shannon index was lower (p = 0.01) in the LD and MD treatments than in the C treatment. PCoA based on the Bray–Curtis distance matrix is commonly used to reflect changes in beta-diversity in microbial communities. Principal coordinate 1 (PCo1) and PCo2 accounted for 32.9% and 21.3%, respectively, of the differences in archaeal community structures among the different samples (Figure 3A); however, the overlap of projections among the four treatments indicated that archaeal community structures were highly similar among the treatments, and none of the observed differences were significant. As shown in Table 5, which displays the relative abundance of the rumen microbiota at the phylum or genus level, the top 10 most abundant taxonomic groups are presented, while the remaining groups are combined and classified under the category ‘others’. At the phylum level, the most abundant archaeal group in all four treatments was Euryarchaeota, followed by Crenarchaeota. The relative abundance of Euryarchaeota (p = 0.03) was higher in MD and HD treatments than in the C treatment, and linear and quadratic effects (p = 0.01, p = 0.01) were also observed. At the genus level, the dominant archaeal genera across the four treatments were Methanobrevibacter, Methanosphaera, Methanosarcina, Methanimicrococcus, Methanoplanus, Methanobacterium, Methanosaeta, Candidatus_Nitrososphaera, Thermococcus, and Vadin CA11, which, collectively, represented over 98% of the total relative abundance. Compared to the C treatment, the relative abundance of the archaeon Methanobrevibacter (p < 0.01) was increased in the 3-NOP treatments, whereas that of Methanosarcina (p = 0.03) and Methanosaeta (p = 0.03) was decreased. Meanwhile, both linear and quadratic effects were also found between the dosage of 3-NOP and the relative abundance of the Methanobrevibacter (both p < 0.01) and Methanosarcina (p = 0.03, p = 0.01). LEfSe analysis is mainly used to identify differences in microbial marker taxa between different treatments and to evaluate the effect of different treatments. A taxonomic cladogram is used to show the taxonomic hierarchical distribution of marker taxa in each treatment of samples. As shown in Figure 4A, the results of the LEfSe analysis for ruminal archaea marker taxa showed that Methanomicrobia were enriched in the C treatment, while Methanobacteria, Methanobacteriaceae, and Methanobacteriales were enriched in the MD treatment; taxa such as Methanobrevibacter were enriched in the HD treatment, but no marker taxa were found in the LD treatment. Methanobacteria, Methanobacteriaceae, and Methanobacteriales in the MD treatment and Methanobrevibacter in the HD treatment were the only taxa with LDA scores > 2 (LDA value distribution histogram), indicating that these taxa had the greatest discriminating potential among the marker taxa for all of the treatments.
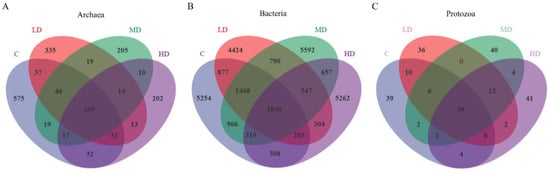
Figure 1.
Venn diagram representation of the ASVs shared among all of the treatments. (A) Venn diagram plot of archaea; (B) Venn diagram plot of bacteria; and (C) Venn diagram plot of protozoa.
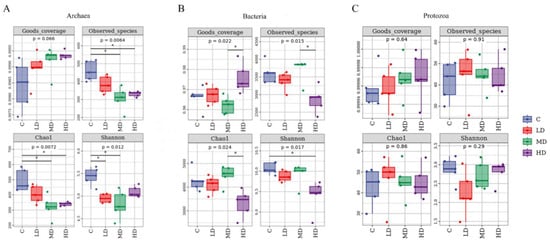
Figure 2.
Boxplots of alpha-diversity indices (good coverage, observed species, Chao1, and Shannon) in different treatments. Individual samples in each treatment are represented by dots of different colors; * means significant differences between different treatments, significance was determined at p < 0.05. (A) Alpha-diversity of archaea; (B) Alpha-diversity of bacteria; and (C) Alpha-diversity of protozoa.
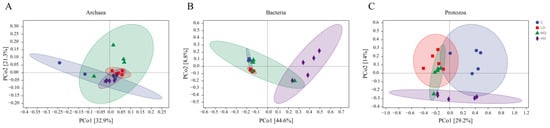
Figure 3.
Principal coordinate analysis (PCoA) of the rumen microbiota based on Bray–Curtis distances; each dot represents an individual sample marked with different shapes and colors in different treatments. (A) PCoA of archaeal microbiota; (B) PCoA of bacterial microbiota; and (C) PCoA of protozoal microbiota.

Table 5.
Effect of 3-NOP on the relative abundance of rumen microorganisms.
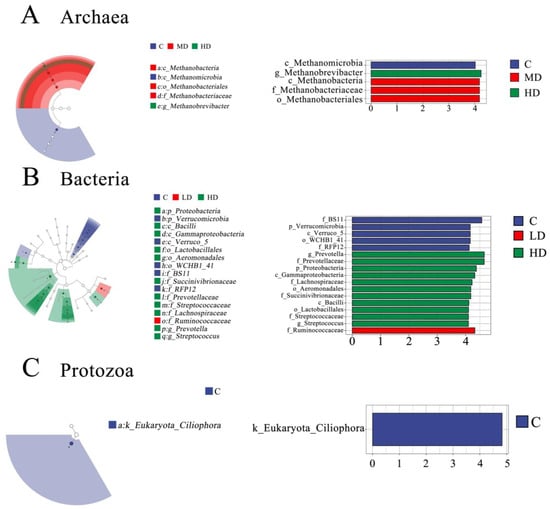
Figure 4.
LEfSe identification of ruminal microbial biomarkers among the treatments. In the LEfSe taxonomic cladogram, circles radiating from inside to outside represent taxonomic levels from the phylum to the genus, respectively. Each small circle represents a classification at the respective taxonomic level. The diameter of a small circle is directly proportional to the relative abundance. Colorless hollow nodes indicate taxa that do not differ significantly between treatments, colored solid nodes indicate taxa that do differ significantly between corresponding treatments, and different colors represent different treatments. In addition, the horizontal coordinate of the linear discriminant analysis (LDA) distribution histogram indicates the LDA value, while the vertical coordinate indicates the marker taxon/taxa in each treatment. The higher the score, the better the corresponding marker taxon can discriminate between treatments (LDA scores > 2.0, p < 0.05). (A) LEfSe analysis of archaea; (B) LEfSe analysis of bacteria; and (C) LEfSe analysis of protozoa.
As shown in Figure 1B, a total of 28,601 ASV taxonomic units were identified in the ruminal bacterial community, 1830 of which were shared among all the treatments, accounting for 6.4% of the total ASVs; meanwhile, 5254 were unique to the C treatment, accounting for 18.3% of the total ASVs; 4424 were unique to the LD treatment, accounting for 15.4% of the total ASVs; 5592 were unique to the MD treatment, accounting for 19.5% of the total ASVs; and 5262 were unique to the HD treatment, accounting for 18.3% of the total ASVs. The α-diversity of the bacterial community is shown in Figure 2B. The HD treatment had the highest bacterial coverage index (Good’s coverage, p = 0.022), which differed from that of the MD treatment. In contrast, the HD treatment had the lowest observed species (p = 0.015) and Chao1 indices (p = 0.024), which differed from those of the MD treatment. The HD treatment also had the lowest Shannon index (p = 0.017), reaching significance compared with the C treatment. A PCoA based on the Bray–Curtis distance matrix for bacteria is presented in Figure 3B; PCo1 and PCo2 explained 44.6% and 8.8%, respectively, of the differences in community structure among the different samples. Notably, the projection of the HD treatment was clearly separate ed from that of the other three treatments, indicating that the HD treatment significantly altered the ruminal bacterial community structure. The composition of the bacterial community is exhibited in Table 5. At the phylum level, the dominant phyla across the four treatments were Bacteroidetes, Firmicutes, Spirochaetes, Verrucomicrobia, Proteobacteria, Lentisphaerae, Tenericutes, Fibrobacteres, TM7, and Actinobacteria, which collectively accounted for over 98% of the total relative bacterial abundance. With an increase in the 3-NOP dose, the relative abundances of Spirochaetes (p = 0.02), Verrucomicrobia (p < 0.01), Lentisphaerae (p < 0.01), and TM7 (p < 0.01) exhibited both linear and quadratic decreasing effects, reaching the lowest levels in the HD treatment, whereas the relative abundances of Proteobacteria (p < 0.01) and Fibrobacteres (p < 0.01) demonstrated both linear and quadratic increasing effects, peaking at the highest levels in the HD treatment group. At the genus level, the dominant genera were Prevotella, Sphaerochaeta, BF311, Clostridiaceae_Clostridium, Succiniclasticum, RFN20, CF231, Streptococcus, YRC22, and Fibrobacter. Among bacteria, the relative abundances of Prevotella (p < 0.01), BF311 (p < 0.01), and Fibrobacter (p < 0.01) were increased, while those of Sphaerochaeta (p = 0.02) and RFN20 (p < 0.01) were decreased in the HD treatment compared with that in the C treatment. Further, the linear and quadratic effects (both p ≤ 0.02) were also observed between the dosage of 3-NOP and these bacteria (Prevotella, BF311, Fibrobacter, Sphaerochaeta, and RFN20). The relative abundance of Streptococcus tended to be higher than the control for the MD and HD treatments, but no statistical difference was observed (p = 0.09). Regarding ruminal bacterial marker taxa identification (Figure 4B), LEfSe analysis showed that taxa such as BS11, Verrucomicrobia, Verruco_5, WCHB1_41, and RFP12 were enriched in the C treatment; Ruminococcaceae were enriched in the LD treatment; and a total of 11 taxa, including Prevotellaceae and Prevotella, were enriched in the HD treatment. As shown in the histogram of LDA value distribution, all of the enriched taxa had LDA scores > 2.
As shown in Figure 1C, a total of 245 ASV taxonomic units were obtained for ruminal protozoa. Of these, 39 were shared among all four treatments, accounting for 15.9% of the total ASVs, while 39 were unique to the C treatment, accounting for 15.9% of the total ASVs; 36 were unique to the LD treatment, accounting for 14.6% of the total ASVs; 40 were unique to the MD treatment, accounting for 16.3% of the total ASVs; and 41 were unique to the HD treatment, accounting for 16.7% of the total ASVs. As shown in Figure 2C, compared with the control treatment, dietary supplementation with different doses of 3-NOP did not affect the coverage index, observed species, Chao1 index, or Shannon index of ruminal protozoa (all p ≥ 0.29). This suggests that dietary supplementation with 3-NOP does not impact protozoal diversity and richness. A PCoA for protozoa was also conducted based on the Bray–Curtis distance. The results are shown in Figure 3C. PCo1 and PCo2 explained 14% and 29.2%, respectively, of the differences in protozoal community structure among the different samples. The protozoal community was highly similar between the LD and MD treatments; however, the C treatment and the HD treatment were located in distinct regions on the coordinate axis, indicating that the protozoal community structure differed between these two treatments. At the order level for protozoa (Table 5), the dominant groups were Entodiniomorphida and Vestibuliferida, but no statistical difference was observed (p = 0.97 and p = 0.69, respectively). At the genus level, the dominant genera were Entodinium, Diplodinium, Ophryoscolex, Ostracodinium, Isotricha, Eudiplodinium, Epidinium, and Cycloposthium. Additionally, compared to the C treatment, the relative abundances of the protozoan genera Entodinium (p = 0.03) and Diplodinium (p < 0.01) decreased in the HD treatment, while that of Ophryoscolex (p < 0.01) exhibited an increase in the MD and HD treatments. Both linear and quadratic effects were found between the dosage of 3-NOP and Entodinium, Diplodinium, and Ophryoscolex. LEfSe analysis of ruminal protozoal marker taxa showed that Ciliophora was a marker taxon for the C treatment, exhibiting an LDA score > 2 (Figure 4C).
As shown in Figure 5, Methanobrevibacter, Prevotella, BF311, and Ophryoscolex exhibited a significant negative correlation with total gas production, CH4 production, acetate (except for BF311), TVFAs, and acetate-propionate ratio (all p < 0.05), while also showing a significant positive correlation with H2 production, propionate (except for Ophryoscolex), and butyrate (except for BF311, all p < 0.05). Conversely, both RFN20 and Diplodinium had a positive correlation with total gas production, CH4 production, acetate, and the acetate- propionate ratio (all p < 0.05), and a negative correlation with H2 production as well as butyrate (both p < 0.05). Meanwhile, Entodinium showed a positive correlation with CH4 production (p < 0.05) but a negative correlation with H2 production (p < 0.05).
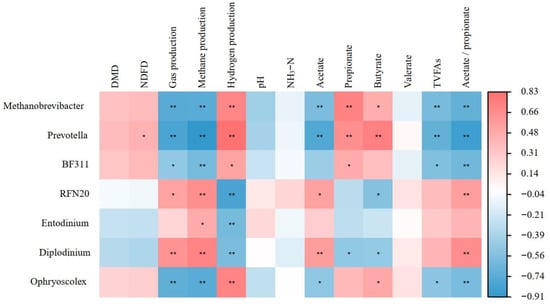
Figure 5.
Heatmap of correlations between rumen microbiota and rumen fermentation characteristics (* mean p < 0.05, and ** mean p < 0.01).
4. Discussion
4.1. The Effects of 3-NOP on Ruminal Fermentation Parameters
In vitro trials are typically used to evaluate the effects of feed substrates or additives on rumen fermentation. They can also directly reflect the activity of ruminal microorganisms and their utilization of fermentation products. In this study, we found that 3-NOP treatment had no significant effect on the DMD and NDFD, which is partly consistent with the results reported by Guyader et al. [50], Romero-Pérez et al. [11], Schilde et al. [5], and Liu et al. [19]. In agreement with the findings of previous reports, lower CH4 production but higher H2 production were found in the MD and HD treatments; however, it seems that H2 production could not fully compensate for the decrease in total gas volume. One possible reason for this might be that CO2 production has also decreased. Over recent years, relevant studies have shown that the extent of the suppressive effect of 3-NOP on ruminal methane production is closely related to feeding dose [5,7,8]. The results of the present study demonstrated that 3-NOP inclusion reduced fermentation-related methane production in vitro in a linear dose-dependent manner, with a maximum inhibition of approximately 97%. This was consistent with references [4,5,51], which reported that the inclusion of 3-NOP in the diet also reduced CH4 emission by up to 71.5%, 86.2%, and 97%, respectively. Furthermore, this was inconsistent with [24], which reported that the inclusion of 3-NOP in the diet reduces CH4 emission by approximately 30% on average. This discrepancy may be related to differences in animal breed and diet type, as well as the fact that this was an in vitro fermentation experiment. Compared with the in vivo observations, the stability of the ruminal microecosystem in vitro is relatively fragile, resulting in a stronger inhibitory effect. Additionally, the lack of effect of 3-NOP on the number of methanogens in the present study was partially in agreement with previous reports in which 3-NOP treatments did not reduce the total copy number of methanogens in the liquid phase as opposed to the solid phase [4,51]. Thus, it implies that the reduction in CH4 emissions might be related to a decrease in the number of methanogens in the solid phase.
Appropriate pH and NH3-N concentrations in the rumen are crucial for maintaining the normal growth and reproduction of ruminal microorganisms [52,53]. No significant differences in ruminal pH and NH3-N levels were observed between the C and 3-NOP treatments, which appears to align with the absence of a significant effect on DMD. Similar findings were also observed in previous reports [4,11,20]. VFAs are important indicators of ruminal metabolism as well as a primary energy source for ruminant animals and directly reflect the type of digestion, metabolism, and fermentation in rumens [54]. In the current study, supplementation with 3-NOP reduced the concentration of TVFAs but did not affect incubation pH. Similar observations were supported by the meta-analysis [55]. These observations could be explained by the high buffering capacity of the artificial saliva used in the incubation process. On the other hand, 3-NOP treatment increased the concentrations of propionic acid and butyric acid while decreasing that of acetic acid, leading to a significant decrease in the acetic acid-propionic acid ratio, which is consistent with previous findings [10,56,57,58]. Additionally, lower CH4 emissions associated with higher H2 production were found in 3-NOP treatment, and similar studies also reported that inhibiting methanogenesis was generally accompanied by the accumulation of H2 in liquid or gaseous phases [20,59,60,61]. Furthermore, it has been reported that propionic acid production can compete with methanogens for metabolic H, implying that increasing propionic acid production can help reduce methane production [19,62,63]. Similarly, the accumulation of H2 is typically related to an increase in the proportion of butyrate. This is because, in addition to alleviating the increase in rumen hydrogen partial pressure by promoting hydrogen consumption pathways (such as propionate synthesis), the butyrate production pathway releases fewer H+ ions compared to the acetate pathway [64], which also helps to slow down the rise in hydrogen partial pressure.
4.2. The Effects of Different Doses of 3-NOP on Ruminal Microbiota
In the present study, a very small fraction of the bacterial (6.4%), archaeal (17.9%), and protozoal (15.9%) communities was observed to be shared across all four treatments. This may be due to the fact that the ASV clustering was performed based on 100% similarity [47]. The results of this study indicated that 3-NOP treatment increased the relative abundance of Methanobrevibacter while decreasing that of Methanosarcina and Methanosaeta. This finding is not entirely consistent with previous studies [12,65], but aligns with the results reported by Liu et al. [20]. From a taxonomic perspective, except for the C treatment, the characteristic differential taxa in each 3-NOP treatment belonged to the same clade, and Methanobrevibacter was the primary characteristic differential taxon in the HD treatment. Moreover, previous studies indicated that certain methanogen species, such as Methanobrevibacter sp. AbM4, possess the ability to synthesize their own methyl-CoM and exhibit reduced sensitivity to methyl-CoM analogs, including 3-NOP [66,67]. Correspondingly, Pitta et al. [68] have reported that while MCR is present in all methanogens, variations in the distribution of mcr and mrt genes encoding MCR may also result in differential effects of 3-NOP on individual methanogenic lineages. This may explain why there was a higher relative abundance of Methanobrevibacter accompanied by a lower CH4 production in the current study. Simultaneously, the total count of methanogens showed a linear decreasing trend with increasing dosages of 3-NOP. Therefore, it is possible that the absolute abundance of Methanobrevibacter could be lower than in the C treatment, which might correlate with the observed decrease in CH4 production in 3-NOP treatments. On the other hand, ruminal microecological balance involves both synergistic and competitive relationships not only among ruminal microorganisms but also among different species of archaea [14,69,70]. Correspondingly, 3-NOP treatment led to a trend in alternating relative abundance among Methanosarcina (decreased), Methanosaeta (decreased), and Methanobrevibacter (increased). Shi et al. [71] found that a shift in the structure of the methanogenic community is also an important factor affecting CH4 production, especially when there is no difference in total methanogen density. Similar findings were also reported by Liu et al. [19]. This may be another reason for the reduction in CH4 production.
Bacteria are the most abundant microorganisms in the rumen and also play a crucial role in methane production in ruminants. In this study, the Chao1 and Shannon indices for ruminal bacteria were reduced in the HD treatment compared with those of the C treatment, which is inconsistent with the findings of Liu et al. [19,20]. This suggests that high doses of 3-NOP have a feedback inhibitory effect on species richness in the ruminal bacterial community, possibly due to differences in the dosage of 3-NOP added. Additionally, our results showed that increases in the relative abundance of Prevotella and propionate production were accompanied by lower CH4 production, which is consistent with the findings of Demeyer and van Nevel [72] as well as Denman et al. [73], who reported that in Prevotella, the production of propionate tends to increase when methane production is inhibited. Correspondingly, LEfSe analysis identified Prevotella as a marker taxon in the HD treatment. Furthermore, Aguilar-Marin et al. [74] and Betancur-Murillo et al. [75] reported that ruminants with lower CH4 emissions are typically abundant in Prevotella ruminicola, Prevotella bryantii, and Prevotella brevis, which possess the ability to consume metabolic hydrogen to produce propionate. However, in contrast to the findings of the present study, other studies have indicated that dietary supplementation with 3-NOP significantly reduce methane production, yet it does not affect the abundance of Prevotella. The reasons for this discrepancy may be closely related to different types of diets, animal species, and even the heritability of rumen bacteria. Jin et al. [76] reported that Prevotellaceae was the predominant heritable bacterial family in lambs and was more susceptible to modulation. Based on the results of the present study, it can be inferred that the addition of 3-NOP to the diet of lambs may partially inhibit methane production by stimulating the growth and colonization of propionate-producing Prevotella species. At the same time, this also suggests that when regulating ruminal methane production, it is necessary to consider both the host animal’s genetics and the composition of the microbiome. For example, Prevotella is a potential candidate for regulating methane emissions due to the strong correlation between its higher abundance and lower methane emissions [75].
Unlike for bacteria and archaea, 3-NOP treatment did not exert a significant effect on protozoal alpha-diversity. Previous studies have found that a reduction in CH4 production with 3-NOP treatment was not accompanied by an effect on the total population of protozoa [10,16,18], which was partially consistent with the present results. One possible explanation for this could be that 3-NOP reduced methane production by directly affecting methanogens or indirectly influencing specific species of rumen protozoa. Correspondingly, the relative abundances of Entodinium and Diplodinium tended to decrease in the MD or HD treatment, whereas that of Ophryoscolex showed a significant increase. Janssen and Kirs [77] demonstrated that Methanobrevibacter, a dominant archaeal genus symbiotically associated with ruminal protozoa, plays a crucial role in methane production. In particular, Kang et al. [78] emphasized that variations in methane emissions are not truly correlated with the total microbial biomass, but rather with fluctuations in the metabolic activity of specific key species. Thus, it is suggested that shifts in the dominant species of rumen protozoa might be closely associated with changes in the composition of the methanogenic community due to interspecies hydrogen transfer. Additionally, significant differences in beta-diversity were found between the HD treatment and the C treatment, indicating that a high dose of 3-NOP altered the ruminal protozoa community structure. In contrast, no significant differences in ruminal protozoal beta-diversity were detected in the LD and MD treatments. This may be due to the much larger size of ruminal protozoa relative to other microorganisms [79]; thus, the rumen protozoa might have more intracellular volume to dilute any 3-NOP that was taken up, and this lower 3-NOP concentration would be less inhibitory to endosymbiotic methanogens. On the other hand, LEfSe analysis did not identify a marker taxon for protozoa in the 3-NOP treatments, despite one having been identified for the control treatment. Based on these findings, we hypothesize that the efficiency of interspecies hydrogen transfer between ruminal protozoa and methanogenic archaea was much weaker in the 3-NOP treatments than in the C treatment, and rumen methane production was thus decreased. However, current knowledge about the effects of 3-NOP on the contribution of rumen protozoa to CH4 production remains limited. As such, further research is essential to investigate the effects of 3-NOP inclusion in the diet on the function and metabolic pathways of rumen protozoa, based on metagenomics combined with metabolomics.
In our study, Prevotella, Methanobrevibacter, and Ophryoscole showed a significant negative correlation with methane production. Additionally, Prevotella exhibited a significant positive correlation with propionate and butyrate. This suggests that the addition of 3-NOP to the diet not only effectively inhibits interspecies hydrogen transfer between Ophryoscolex and methanogenic bacteria but may also enhance the symbiotic relationship between Ophryoscolex and Prevotella involved in propionate production [80]. Furthermore, under the conditions of this experiment, the BF311 genus showed a very significant negative correlation with methane production but a significant positive correlation with propionate. Because BF311 and Prevotella both belong to the Prevotellaceae family, they may share similar metabolic pathways [81], implying that BF311 is also involved in the production and metabolism of propionate. Conversely, the RFN20 genus showed a very significant positive correlation with methane production and a significant positive correlation with acetate. RFN20 belongs to the phylum Firmicutes and is a major fiber-degrading bacterium in the rumen; this is consistent with the findings of Li et al. [82] and Wang et al. [83], where RFN20 is considered as a producer of acetate and hydrogen.
5. Conclusions
The addition of 3-NOP at levels of 0.025, 0.05, and 0.1 mg/g of feed DM was able to linearly reduce CH4 production and correspondingly increase H2 accumulation. It is noteworthy that both the 0.05 and 0.1 mg/g of feed DM doses achieved the maximum methane inhibition effect, yet no statistically significant difference was found between these two dosages. 3-NOP supplementation also shifted the in vitro fermentation toward producing less acetate but more propionate and butyrate without affecting the DMD and NDFD. Furthermore, the inclusion of 3-NOP in the diet did not change the populations of bacteria, archaea, or protozoa, but it did alter the composition of the rumen microbial community and had a notable effect on the abundance and diversity of specific microorganisms, such as Prevotella, Methanobrevibacter, Entodinium, Diplodinium, and Ophryoscolex. Notably, these results suggest that, in comparison to the inclusion levels of 3-NOP in existing similar studies, the administration of a lower dose of 3-NOP to young ruminants can also similarly achieve a significant reduction in CH4 emissions. However, further research is necessary to investigate the potential for achieving the same effect via a long-term in vivo experiment.
Author Contributions
T.X.: Writing—original draft; T.Z.: Investigation and data curation; T.L. (Tiyu Li): Conceptualization; B.W.: Investigation, Software; T.L. (Tailin Li) and W.B.: Methodology; W.Q.: Supervision, Project administration, Funding acquisition, Writing—review & editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Program for the National Natural Science Foundation of China [NO.31960679]; the Inner Mongolia Science and Technology Project [NO.2021GG0006]; the Natural Science Foundation of Inner Mongolian [NO.2021LHMS03011]; and the Tongliao Science and Technology Planning Project [NO.TL2023YF009].
Institutional Review Board Statement
The animal use protocol was approved by the Animal Care and Use Committee of the College of Animal Science and Technology of Inner Mongolia Minzu University (IMUN2022-56). This study was conducted in accordance with the local legislation and institutional requirements.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Johnson, K.A.; Johnson, D.E. Methane emissions from cattle. J. Anim. Sci. 1995, 73, 2483–2492. [Google Scholar] [CrossRef]
- Hristov, A.N.; Oh, J.; Giallongo, F.; Frederick, T.W.; Harper, M.T.; Weeks, H.L.; Branco, A.F.; Moate, P.J.; Deighton, M.H.; Williams, S.R.O.; et al. An inhibitor persistently decreased enteric methane emission from dairy cows with no negative effect on milk production. Proc. Natl. Acad. Sci. USA 2015, 11, 10663–10668. [Google Scholar] [CrossRef]
- Duin, E.C.; Wagner, T.; Shima, S.; Prakash, D.; Cronin, B.; Yáñez-Ruiz, D.R.; Duval, S.; Rümbeli, R.; Stemmler, R.T.; Thauer, R.K. Mode of action uncovered for the specific reduction of methane emissions from ruminants by the small molecule 3-nitrooxypropanol. Proc. Natl. Acad. Sci. USA 2016, 113, 6172–6177. [Google Scholar] [CrossRef]
- Romero-Pérez, A.; Okine, E.K.; Guan, L.L.; Duval, S.M.; Kindermann, M.; Beauchemin, K.A. Effects of 3-nitrooxypropanol on methane production using the rumen simulation technique (Rusitec). Anim. Feed Sci. Technol. 2015, 209, 98–109. [Google Scholar] [CrossRef]
- Schilde, M.; von Soosten, D.; Hüther, L.; Kersten, S.; Meyer, U.; Zeyner, A.; Dänicke, S. Dose–response effects of 3-nitrooxypropanol combined with low-and high-concentrate feed proportions in the dairy cow ration on fermentation parameters in a rumen simulation technique. Animals 2021, 11, 1784. [Google Scholar] [CrossRef]
- Dijkstra, J.; Bannink, A.; France, J.; Kebreab, E.; van Gastelen, S. Short communication: Antimethanogenic effects of 3-nitrooxypropanol depend on supplementation dose, dietary fiber content, and cattle type. J. Dairy Sci. 2018, 101, 9041–9047. [Google Scholar] [CrossRef]
- Melgar, A.; Welter, K.C.; Nedelkov, K.; Martins, C.M.M.R.; Harper, M.T.; Oh, J.; Räisänen, S.E.; Chen, X.J.; Cueva, S.F.; Duval, S.; et al. Dose-response effect of 3-nitrooxypropanol on enteric methane emissions in dairy cows. J. Dairy Sci. 2020, 103, 6145–6156. [Google Scholar] [CrossRef]
- Vyas, D.; McGinn, S.M.; Duval, S.M.; Kindermann, M.K.; Beauchemin, K.A. Optimal dose of 3-nitrooxypropanol for decreasing enteric methane emissions from beef cattle fed high-forage and high-grain diets. Anim. Prod. Sci. 2016, 58, 1049–1055. [Google Scholar] [CrossRef]
- Yu, G.; Beauchemin, K.A.; Dong, R. A review of 3-nitrooxypropanol for enteric methane mitigation from ruminant livestock. Animals 2021, 11, 3540. [Google Scholar] [CrossRef]
- Haisan, J.; Sun, Y.; Guan, L.L.; Beauchemin, K.A.; Iwaasa, A.; Duval, S.; Barreda, D.R.; Oba, M. The effects of feeding 3-nitrooxypropanol on methane emissions and productivity of Holstein cows in mid lactation. J. Dairy Sci. 2014, 97, 3110–3119. [Google Scholar] [CrossRef]
- Romero-Pérez, A.; Okine, E.K.; Guan, L.L.; Duval, S.M.; Kindermann, M.; Beauchemin, K.A. Rapid Communication: Evaluation of methane inhibitor 3-nitrooxypropanol and monensin in a high-grain diet using the rumen simulation technique (Rusitec). J. Anim. Sci. 2017, 95, 4072–4077. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Fernández, G.; Duval, S.; Kindermann, M.; Schirra, H.J.; Denman, S.E.; McSweeney, C.S. 3-NOP vs. halogenated compound: Methane production, ruminal fermentation and microbial community response in forage fed cattle. Front. Microbiol. 2018, 9, 1582. [Google Scholar] [CrossRef]
- Gruninger, R.J.; Zhang, X.M.; Smith, M.L.; Kung, L.J.; Vyas, D.; McGinn, S.M.; Kindermann, M.; Wang, M.; Tan, Z.L.; Beauchemin, K.A. Application of 3-nitrooxypropanol and canola oil to mitigate enteric methane emissions of beef cattle results in distinctly different effects on the rumen microbial community. Anim. Microbiome 2022, 4, 35. [Google Scholar] [CrossRef]
- Pitta, D.W.; Melgar, A.; Hristov, A.N.; Indugu, N.; Narayan, K.S.; Pappalardo, C. Temporal changes in total and metabolically active ruminal methanogens in dairy cows supplemented with 3-nitrooxypropanol. J. Dairy. Sci. 2021, 104, 8721–8735. [Google Scholar] [CrossRef]
- Pitta, D.W.; Indugu, N.; Melgar, A.; Hristov, A.N.; Challa, K.; Vecchiarelli, B.; Hennessy, M.L.; Narayan, K.S.; Duval, S.; Kindermann, M.; et al. The effect of 3-nitrooxypropanol, a potent methane inhibitor, on ruminal microbial gene expression profiles in dairy cows. Microbiome 2022, 10, 1–21. [Google Scholar] [CrossRef]
- Martínez-Fernández, G.; Abecia, L.; Arco, A.; Cantalapiedra-Hijar, G.; Martín-García, A.I.; Molina-Alcaide, E.; Kindermann, M.; Duval, S.; Yáñez-Ruíz, D.R. Effects of ethyl-3-nitrooxy propionate and 3-nitrooxypropanol on ruminal fermentation, microbial abundance, and methane emissions in sheep. J. Dairy Sci. 2014, 97, 3790–3799. [Google Scholar] [CrossRef]
- Romero-Pérez, A.; Okine, E.K.; McGinn, S.M.; Guan, L.L.; Oba, M.; Duval, S.; Kindermann, M.; Beauchemin, K.A. The potential of 3-nitrooxypropanol to lower enteric methane emissions from beef cattle. J. Anim. Sci. 2014, 92, 4682–4693. [Google Scholar] [CrossRef]
- Haisan, J.; Sun, Y.; Guan, L.; Beauchemin, K.A.; Iwaasa, A.; Duval, S.; Kindermann, M.; Barreda, D.R.; Oba, M. The effects of feeding 3-nitrooxypropanol at two doses on milk production, rumen fermentation, plasma metabolites, nutrient digestibility, and methane emissions in lactating Holstein cows. Anim. Prod. Sci. 2016, 57, 282–289. [Google Scholar] [CrossRef]
- Liu, Z.H.; Wang, K.; Nan, X.M.; Yang, L.G.; Wang, Y.; Zhang, F.; Cai, M.; Zhao, Y.G.; Xiong, B.H. Effects of combined addition of 3-nitrooxypropanol and vitamin B12 on methane and propionate production in dairy cows by in vitro-simulated fermentation. J. Dairy. Sci. 2023, 106, 219–232. [Google Scholar] [CrossRef]
- Liu, Z.H.; Wang, K.; Nan, X.M.; Cai, M.; Yang, L.; Xiong, B.H.; Zhao, Y.G. Synergistic effects of 3-Nitrooxypropanol with fumarate in the regulation of propionate formation and methanogenesis in dairy cows in vitro. Appl. Environ. Microbiol. 2022, 88, e0190821. [Google Scholar] [CrossRef]
- van Gastelen, S.; Burgers, E.E.A.; Dijkstra, J.; de Mol, R.; Muizelaar, W.; Walker, N.; Bannink, A. Long-term effects of 3-nitrooxypropanol on methane emission and milk production characteristics in Holstein Friesian dairy cows. J. Dairy Sci. 2024, 107, 5556–5573. [Google Scholar] [CrossRef]
- Romero-Pérez, A.; Okine, E.K.; McGinn, S.M.; Guan, L.L.; Oba, M.; Duval, S.; Kindermann, M.; Beauchemin, K.A. Sustained reduction in methane production from long-term addition of 3-nitrooxypropanol to a beef cattle diet. J. Anim. Sci. 2015, 93, 1780–1791. [Google Scholar] [CrossRef]
- Kim, H.; Lee, H.G.; Baek, Y.C.; Lee, S.; Seo, J. The effects of dietary supplementation with 3-nitrooxypropanol on enteric methane emissions, rumen fermentation, and production performance in ruminants: A meta-analysis. J. Anim. Sci. Technol. 2020, 62, 31–42. [Google Scholar] [CrossRef]
- Kebreab, E.; Bannink, A.; Pressman, E.M.; Walker, N.; Karagiannis, A.; van Gastelen, S.; Dijkstra, J. A meta-analysis of effects of 3-nitrooxypropanol on methane production, yield, and intensity in dairy cattle. J. Dairy Sci. 2023, 106, 927–936. [Google Scholar] [CrossRef]
- Jami, E.; Israel, A.; Kotser, A.; Mizrahi, I. Exploring the bovine rumen bacterial community from birth to adulthood. Isme J. 2013, 7, 1069–1079. [Google Scholar] [CrossRef]
- Morgavi, D.P.; Kelly, W.J.; Janssen, P.H.; Attwood, G.T. Rumen microbial (meta) genomics and its application to ruminant production. Animal 2013, 7, 184–201. [Google Scholar] [CrossRef]
- Yáñez-Ruiz, D.R.; Abecia, L.; Newbold, C.J. Manipulating rumen microbiome and fermentation through interventions during early life: A review. Front. Microbiol. 2015, 6, 1133. [Google Scholar] [CrossRef]
- Abecia, L.; Waddams, K.E.; Martínez-Fernandez, G.; Martín-García, A.I.; Ramos-Morales, E.; Newbold, C.J.; Yáñez-Ruiz, D.R. An antimethanogenic nutritional intervention in early life of ruminants modifies ruminal colonization by Archaea. Archaea 2014, 2014, 841463. [Google Scholar] [CrossRef]
- Meale, S.J.; Popova, M.; Saro, C.; Martin, C.; Bernard, A.; Lagrée, M.; Yáñez-Ruíz, D.R.; Boudra, H.; Duval, S.; Morgavi, D.P. Early life dietary intervention in dairy calves results in a long-term reduction in methane emissions. Sci. Rep. 2011, 11, 3003. [Google Scholar] [CrossRef]
- NY/T 816-2021; Nutrient Requirements of Meat-Type Sheep and Goat. Ministry of Agriculture of the PRC. China Agriculture Press: Beijing, China, 2021. (In Chinese)
- Menke, K.H.; Raab, L.; Salewski, A.; Steingass, H.; Fritz, D.; Schneider, W. The estimation of the digestibility and metabolizable energy content of ruminant feeding stuffs from the gas production when they are incubated with rumen liquor in vitro. J. Agric Sci. 1979, 93, 217–222. [Google Scholar] [CrossRef]
- Kim, W.Y.; Hanigan, M.D.; Lee, S.J.; Lee, S.M.; Kim, D.H.; Hyun, J.H.; Yeo, J.M. Effects of cordyceps militaris on the growth of rumen microorganisms and in vitro rumen fermentation with respect to methane emissions. J. Dairy Sci. 2014, 97, 7065–7075. [Google Scholar] [CrossRef] [PubMed]
- Morsy, A.S.; Soltan, Y.A.; Sallam, S.M.A.; Kreuzer, M.; Alencar, S.M.; Abdalla, A.L. Comparison of the in vitro efficiency of supplementary bee propolis of different origin in enhancing ruminal nutrient degradation and mitigating methane formation. Anim. Feed. Sci. Technol. 2015, 199, 51–60. [Google Scholar] [CrossRef]
- Soltan, Y.A.; Morsy, A.S.; Lucas, R.C.; Abdalla, A.L. Potential of mimosine of Leucaena leucocephala for modulating ruminal nutrient degradability and methanogenesis. Anim. Feed. Sci. Technol. 2017, 223, 30–41. [Google Scholar] [CrossRef]
- Nolan, J.V.; Hegarty, R.S.; Hegarty, J.; Godwin, I.R.; Woodgate, R. Effects of dietary nitrate on fermentation, methane production and digesta kinetics in sheep. Anim. Prod. Sci. 2010, 50, 801–806. [Google Scholar] [CrossRef]
- Broderick, G.A.; Kang, J.H. Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J. Dairy Sci. 1980, 63, 64–75. [Google Scholar] [CrossRef]
- Kim, H.; Jung, E.; Lee, H.G.; Kim, B.; Cho, S.; Lee, S.; Kwon, I.; Seo, J. Essential oil mixture on rumen fermentation and microbial community-an in vitro study. Asian Austral J. Anim. 2019, 32, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Shen, Y.B.; Ma, H.; Li, Y.; Lambo, M.T.; Dai, B.S.; Shen, W.Z.; Qu, Y.L.; Zhang, Y.G. Silibinin reduces in vitro methane production by regulating the rumen microbiome and metabolites. Front. Microbiol. 2023, 14, 1225643. [Google Scholar] [CrossRef] [PubMed]
- AOAC. Official Methods of Analysis, 18th ed.; Association of Analytical Chemists: Gaithersburg, MD, USA, 1995. [Google Scholar]
- van Soest, P.V.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Huse, S.M.; Dethlefsen, L.; Huber, J.A.; Welch, D.M.; Relman, D.A.; Sogin, M.L. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008, 4, e1000255. [Google Scholar] [CrossRef]
- Pires, A.C.C.; Cleary, D.F.R.; Almeida, A.; Cunha, Â.; Dealtry, S.; Mendonça-Hagler, L.C.; Smalla, K.; Gomes, N.C.M. Denaturing gradient gel electrophoresis and barcoded pyrosequencing reveal unprecedented archaeal diversity in mangrove sediment and rhizosphere samples. AEM 2012, 78, 5520–5528. [Google Scholar] [CrossRef]
- Sylvester, J.T.; Karnati, S.K.R.; Yu, Z.T.; Morrison, M.; Firkins, J.L. Development of an assay to quantify rumen ciliate protozoal biomass in cows using real-time PCR. J. Nutr. 2004, 134, 3378–3384. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic. Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.X.; Zhang, B.; Chen, Z.; Qin, W.T.; Wen, X.H. Sludge retention time affects the microbial community structure: A large-scale sampling of aeration tanks throughout China. Environ. Pollut. 2020, 261, 114140. [Google Scholar] [CrossRef] [PubMed]
- Guyader, J.; Ungerfeld, E.M.; Beauchemin, K.A. Redirection of metabolic hydrogen by inhibiting methanogenesis in the rumen simulation technique (RUSITEC). Front. Microbiol. 2017, 8, 393. [Google Scholar] [CrossRef]
- Romero-Pérez, A.; Okine, E.K.; Guan, L.L.; Duval, S.M.; Kindermann, M.; Beauchemin, K.A. Effects of 3-nitrooxypropanol and monensin on methane production using a forage-based diet in Rusitec fermenters. Anim. Feed. Sci. Technol. 2016, 220, 67–72. [Google Scholar] [CrossRef]
- Pereira, D.H.; Pereira, O.G.; da Silva, B.C.; Leão, M.I.; de Campos Valadares Filho, S.; Chizzotti, M.L.; García, R. Intake and total and partial digestibility of nutrients, ruminal pH and ammonia concentration and microbial efficiency in beef cattle fed with diets containing sorghum (Sorghum bicolor (L.) Moench) silage and concentrate in different ratios. Livest. Sci. 2007, 107, 53–61. [Google Scholar] [CrossRef]
- Ali, C.S.; Khaliq, T.; Sarwar, M.; Javaid, A.; Shahzad, M.A.; Nisa, M.; Zakir, S. Effect of various non protein nitrogen sources on in vitro dry matter digestibility, ammonia production, microbial growth and pH changes by rumen bacteria. Pak. Vet. J. 2009, 29, 25–30. Available online: https://www.researchgate.net/publication/26520131 (accessed on 16 May 2024).
- Bannink, A.; Kogut, J.; Dijkstra, J.; France, J.; Kebreab, E.; van Vuuren, A.M.; Tamminga, S. Estimation of the stoichiometry of volatile fatty acid production in the rumen of lactating cows. J. Theor. Biol. 2006, 238, 36–51. [Google Scholar] [CrossRef]
- Orzuna-Orzuna, J.F.; Godina-Rodríguez, J.E.; Garay-Martínez, J.R.; Granados-Rivera, L.D.; Maldonado-Jáquez, J.A.; Lara-Bueno, A. A Meta-Analysis of 3-Nitrooxypropanol Dietary Supplementation on Growth Performance, Ruminal Fermentation, and Enteric Methane Emissions of Beef Cattle. Fermentation 2024, 10, 273. [Google Scholar] [CrossRef]
- Melgar, A.; Harper, M.T.; Oh, J.; Giallongo, F.; Young, M.E.; Ott, T.L.; Duval, S.; Hristov, A.N. Effects of 3-nitrooxypropanol on rumen fermentation, lactational performance, and resumption of ovarian cyclicity in dairy cows. J. Dairy Sci. 2020, 103, 410–432. [Google Scholar] [CrossRef]
- Reynolds, C.K.; Humphries, D.J.; Kirton, P.; Kindermann, M.; Duval, S.; Steinberg, W. Effects of 3-nitrooxypropanol on methane emission, digestion, and energy and nitrogen balance of lactating dairy cows. J. Dairy Sci. 2014, 97, 3777–3789. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.M.; Smith, M.C.; Gruninger, R.J.; Kung, L.; Vyas, D.; McGinn, S.M.; Kindermann, M.; Wang, M.; Tan, Z.L.; Beauchemin, K.A. Combined effects of 3-nitrooxypropanol and canola oil supplementation on methane emissions, rumen fermentation and biohydrogenation, and total tract digestibility in beef cattle. J. Anim. Sci. 2021, 99, skab081. [Google Scholar] [CrossRef] [PubMed]
- Hungate, R.E. Hydrogen as an intermediate in the rumen fermentation. Arch. Mikrobiol. 1967, 59, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Trei, J.E.; Scott, G.C.; Parish, R.C. Influence of methane inhibition on energetic efficiency of lambs. J. Anim. Sci. 1972, 34, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sun, X.Z.; Janssen, P.H.; Tang, S.X.; Tan, Z.L. Responses of methane production and fermentation pathways to the increased dissolved hydrogen concentration generated by eight substrates in in vitro ruminal cultures. Anim. Feed. Sci. Technol. 2014, 194, 1–11. [Google Scholar] [CrossRef]
- Wang, K.; Nan, X.M.; Zhao, Y.G.; Tong, J.J.; Jiang, L.S.; Xiong, B.H. Effects of propylene glycol on in vitro ruminal fermentation, methanogenesis, and microbial community structure. J. Dairy Sci. 2021, 104, 2924–2934. [Google Scholar] [CrossRef]
- Wang, K.; Xiong, B.H.; Zhao, X. Could propionate formation be used to reduce enteric methane emission in ruminants? Sci. Total. Environ. 2023, 855, 158867. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, N.B.; Hvelplund, T.; Weisbjerg, M.R.; Nørgaard, P. Mikrobiel omsætning i formaverne. In Kvægets Ernæring og Fysiologi-Bind 1; Hvelplund, T., Nørgaard, P., Eds.; Danmarks Jordbrugsforskning, Ministeriet for Fødevarer, Landbrug og Fiskeri: Tjele, Denmark, 2003; pp. 211–235. [Google Scholar]
- Zhang, X.M.; Gruninger, R.J.; Alemu, A.W.; Wang, M.; Tan, Z.L.; Kindermann, M.; Beauchemin, K.A. 3-Nitrooxypropanol supplementation had little effect on fiber degradation and microbial colonization of forage particles when evaluated using the in situ ruminal incubation technique. J. Dairy. Sci. 2020, 103, 8986–8997. [Google Scholar] [CrossRef] [PubMed]
- Ungerfeld, E.M.; Rust, S.R.; Boone, D.R.; Liu, Y. Effects of several inhibitors on pure cultures of ruminal methanogens. J. Appl. Microbiol. 2004, 97, 520–526. [Google Scholar] [CrossRef]
- Leahy, S.C.; Kelly, W.J.; Li, D.; Li, Y.; Altermann, E.; Lambie, S.C.; Cox, F.; Attwood, G.T. The complete genome sequence of Methanobrevibacter sp. AbM4. Stand. Genomic. Sci. 2013, 8, 215–227. [Google Scholar] [CrossRef]
- Pitta, D.; Indugu, N.; Narayan, K.; Hennessy, M. Symposium review: Understanding the role of the rumen microbiome in enteric methane mitigation and productivity in dairy cows. J. Dairy Sci. 2022, 105, 8569–8585. [Google Scholar] [CrossRef] [PubMed]
- Söllinger, A.; Tveit, A.T.; Poulsen, M.; Noel, S.J.; Bengtsson, M.M.; Bernhardt, J.; Hellwing, A.L.F.; Lund, P.; Riedal, K.; Schleper, C. Holistic assessment of rumen microbiome dynamics through quantitative metatranscriptomics reveals multifunctional redundancy during key steps of anaerobic feed degradation. Msystems 2018, 3, e00038-18. [Google Scholar] [CrossRef] [PubMed]
- Söllinger, A.; Urich, T. Methylotrophic methanogens everywhere- physiology and ecology of novel players in global methane cycling. Biochem. Soc. Trans. 2019, 47, 1895–1907. [Google Scholar] [CrossRef]
- Shi, W.B.; Moon, C.D.; Leahy, S.C.; Kang, D.W.; Froula, J.; Kittelmann, S.; Fan, C.; Deutach, S.; Gagić, D.; Seedorf, H.; et al. Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome. Res. 2014, 24, 1517–1525. [Google Scholar] [CrossRef]
- Demeyer, D.I.; Van Nevel, C.J. Methanogenesis, an integrated part of carbohydrate fermentation and its control. Dig. Metab. Rumin. 1975, 366–382. Available online: https://www.cabidigitallibrary.org/doi/full/10.5555/19761433771 (accessed on 1 August 2024).
- Denman, S.E.; Fernandez, G.M.; Shinkai, T.; Mitsumori, M.; McSweeney, C.S. Metagenomic analysis of the rumen microbial community following inhibition of methane formation by a halogenated methane analog. Front. Microbiol. 2015, 6, 1087. [Google Scholar] [CrossRef]
- Aguilar-Marin, S.B.; Betancur-Murillo, C.L.; Isaza, G.A.; Mesa, H.; Jovel, J. Lower Methane Emissions Were Associated with Higher Abundance of Ruminal Prevotella in a Cohort of Colombian Buffalos. BMC Microbiol. 2020, 20, 364. [Google Scholar] [CrossRef] [PubMed]
- Betancur-Murillo, C.L.; Aguilar-Marín, S.B.; Jovel, J. Prevotella: A Key Player in Ruminal Metabolism. Microorganisms 2023, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.W.; Zhang, Z.; Zhang, G.H.; He, B.; Qin, Y.L.; Yang, B.; Yu, Z.T.; Wang, J.K. Maternal Rumen Bacteriota Shapes the Offspring Rumen Bacteriota, Affecting the Development of Young Ruminants. Microbiol. Spectr. 2023, 11, e03590-22. [Google Scholar] [CrossRef] [PubMed]
- Janssen, P.H.; Kirs, M. Structure of the archaeal community of the rumen. Appl. Environ. Microbiol. 2008, 74, 3619–3625. [Google Scholar] [CrossRef]
- Kang, S.H.; Evans, P.; Morrison, M.; McSweeney, C. Identification of metabolically active proteobacterial and archaeal communities in the rumen by DNA- and RNA-derived 16S rRNA gene. J. Appl. Microbiol. 2013, 115, 644–653. [Google Scholar] [CrossRef]
- Newbold, C.J.; De La Fuente, G.; Belanche, A.; Ramos-Morales, E.; McEwan, N.R. The role of ciliate protozoa in the rumen. Front. Microbiol. 2015, 6, 1313. [Google Scholar] [CrossRef]
- Emerson, E.L.; Weimer, P.J. Fermentation of model hemicelluloses by Prevotella strains and Butyrivibrio fibrisolvens in pure culture and in ruminal enrichment cultures. Appl. Microbiol. Biotechnol. 2017, 101, 4269–4278. [Google Scholar] [CrossRef]
- Bi, Y.L.; Zeng, S.Q.; Zhang, R.; Diao, Q.Y.; Tu, Y. Effects of dietary energy levels on rumen bacterial community composition in Holstein heifers under the same forage to concentrate ratio condition. BMC Microbiol. 2018, 18, 69. [Google Scholar] [CrossRef]
- Li, N.; Xue, Y.G.; Chen, S.S.; Takahashi, J.; Dai, L.L.; Dai, X.H. Methanogenic population dynamics regulated by bacterial community responses to protein-rich organic wastes in a high solid anaerobic digester. Chem. Eng. J. 2017, 317, 444–453. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, J.Z.; Zhou, Y.L.; Almeida, A.; Bateman, A.; Danchin, A.; He, L.S. Phylogenomics of expanding uncultured environmental Tenericutes provides insights into their pathogenicity and evolutionary relationship with Bacilli. BMC Genom. 2020, 21, 408. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).