Abstract
The distinct microbiological and physicochemical characteristics of sheep milk enable the production of cheeses with unique organoleptic properties. The scenario of sheep cheeses in Brazil is still uncertain, lacking data and regulations. This study aims to characterize the diversity of bacterial groups with potential pathogenic and/or harmful properties to milk technology and correlate the presence of these bacteria with the physicochemical properties of sheep cheese. Additionally, it investigates the presence of virulence genes, resistance genes, and toxins in reference genomes. The main steps were (a) sample preparation and fractionation; (b) physicochemical analysis; (c) analysis of the free fatty acids profile by GC-FID; (d) analysis of the volatile compounds profile by GC-MS; (e) molecular analysis—Next Generation Sequencing of the 16S rRNA gene (V3-V4 region); and (f) in silico analysis—Genomics. A total of 1,061,392 sequences were identified, corresponding to 45 genera and 57 species. Of the total abundance, 95.7% were considered beneficial, while 3.9% were classified as bacteria with pathogenic potential (BPP) and/or bacteria harmful to milk technology (BHMT). Five phyla were identified: Firmicutes, Proteobacteria, Actinobacteriota, Bacteroidota, and Fusobacteriota. The genera Acinetobacter, Pseudomonas, and Staphylococcus stood out in the BPP and BHMT groups, showing higher abundance. Alpha diversity of the cheese samples showed that the cheese origin significantly influences the richness, evenness, and abundance of bacterial species. Some physicochemical parameters, fatty acids, and volatile organic compounds showed a negative correlation with Staphylococcus. Reference genomes of two species exhibited a higher number of resistance and virulence genes. This reinforces the need to monitor bacteria considered of lesser relevance to prevent the transmission, through food, of potentially resistant and virulent pathogens.
1. Introduction
The world’s production of sheep’s milk has grown in recent years, and in 2022 more than 10 million tons were recorded, with Asia being the largest producer (44%) and China standing out by producing around 1 million tons annually. The Americas represent just 1% of this production, with countries such as Mexico, Bolivia, and Ecuador totaling more than 80 thousand tons of sheep’s milk that year [1]. In Brazil, production data are scarce, with records showing the production of 840 thousand liters of milk in 2016, primarily from the states of Santa Catarina (SC) and Rio Grande do Sul (RS), with 315 thousand and 270 thousand liters, respectively [2].
Sheep’s milk has physicochemical characteristics desirable for the production of dairy products such as fresh or aged cheese [3]. Fresh cheeses are known for their ease of digestion [4,5] such as Labneh cheese (“yogurt cheese”) [6,7] and Feta [8]. Among the matured cheeses produced with raw sheep’s milk, the most traditional are Pecorino Romano and Fiore Sardo [9], which are white in color and have a hard consistency and spiciness [10].
Global production data for fresh sheep’s cheese in 2021 indicate a total of 690,710.14 tons, with Europe (58.1%—401,6135.60 t) and Asia (34.1%—235,433.94 t) responsible for 92.2% of this total [1]. Although 80% of sheep dairy farms in Brazil process the cheese on-site, there are no specific records of this productivity in the country [2]. The production is concentrated in the states of Santa Catarina (SC), Rio Grande do Sul (RS), Rio de Janeiro (RJ), and Minas Gerais (MG) [11]. Currently, at least ten types of cheeses, including Pecorino Toscano, Labneh, Boursin, and colonial cheese, are produced and marketed in the states of SC and RS [12].
The quality of these cheeses is influenced by the raw material, manufacturing processes, geographic region, diet, animal breed, and sanitary management. Good manufacturing practices, the use of starter cultures, and the ripening process are essential in the industry to ensure the distinctive organoleptic characteristics and quality of the final product. [13]. The physicochemical characterization (PC), including analyses of fatty acids (FA) and volatile organic compounds (VOC), is essential for understanding their nutritional and sensory properties. These cheeses are rich in fatty acids, such as conjugated linoleic acid (CLA), which are beneficial to health [14]. The interactions between the microbiota, PC, and VOC are widely studied, highlighting cultures such as Lactococcus lactis subsp. lactis, which adds buttery flavor notes and creaminess [15]. The analysis of VOC also assesses the quality of the production process, being considered a “fingerprint” of the cheese [16,17], reflecting production technology and quality aspects.
These characteristics are related to the fermentation process and the microbial, bacterial, and fungal composition of the dairy matrix. The majority of lactic acid bacteria (LAB) dominate this microbial flora, but within this population, spoilage bacteria, with pathogenic potential (BPP) and/or harmful to milk technology (BHMT), may be present [18,19]. Foodborne pathogens commonly described in legislation and reported in cheeses include Listeria monocytogenes, which has a psychotropic character [18,20]; Staphylococcus aureus, known for enterotoxin production and osmotolerance [21]; Shiga toxin-producing Escherichia coli, which causes intestinal complications [22,23]; and Salmonella spp., which induces gastroenteritis [24,25]. Most studies focus on specific foodborne pathogens, but it is also crucial to consider the dynamics of other emerging pathogens to implement control measures and ensure product quality [18]. Studies highlight the lack of attention to the presence of emerging genera due to the absence of correlation with the matrix or production process [26,27]. Additionally, there is an observed increase in antibiotic-resistant microorganisms and the acquisition of virulence genes, creating uncertainties and risks to public health [28]. In the food production scenario, there is concern about these resistant microorganisms in foods originating from environmental contamination [29].
The contamination of microorganisms in cheese must also be seen from another perspective, based on the presence of BHMT. This group of bacteria produces metabolites that are not initially harmful to the health of the consumer but harm and alter the sensorial quality of the cheese, making it less appealing to consumers [30,31]. However, Wang et al. [32] correlated the presence of bacterial metabolites, such as pigments, with human illnesses in their study. The genus Pseudomonas can produce proteases and lipases that interfere with the ripening process and cause sensory changes in cheese [30,31]. Another important genus in this context is Acinetobacter, which can influence the color of the cheese by producing a brown pigment [33].
The application of bioinformatics, in addition to sequencing, accelerates and facilitates microbial identification compared to current methods [34] and enhances taxonomic classification through the use of specific genetic biomarkers [35]. This approach to analyzing bacterial community composition has already been applied in several studies with different types of foods [36,37,38,39,40,41,42,43]. The identification of the microbial profile of the food matrix allows us to understand the “history” of the product [44], and omics and biomolecular techniques have gained prominence in this characterization. Metagenomics emerges as one of these techniques, focusing on the direct analysis of genetic material present in a given habitat without the need to isolate and culture each microorganism, providing a more comprehensive view of the genetic diversity and microbial potential present [45,46,47].
From the identification of the microbiota, several approaches seek to better understand the cheese ecosystem and clarify population and metabolic interactions to assist in cheese manufacturing [48]. In this same context, bioinformatics tools emerged, such as the pangenome, aimed at identifying and characterizing genes and proteins with potential application in the diagnosis, prevention, and therapy of infectious diseases [49]. The pangenome approach focuses on analyzing all the genes found in the metagenome sequencing, characterizing those present in all microorganisms (core genome), those that appear in some but not in all microorganisms (accessory genome), and species-specific ones (singletons or unique genomes) [50]. This approach allows for the selection of proteins with specific subcellular localization essential for the survival of microorganisms [51], thus enabling the development of techniques for rapid detection and creating mechanisms and technologies for inhibiting pathogens by improving quality control [49,52].
This study aimed to characterize the diversity of bacterial groups present in sheep cheese, including bacteria with potential pathogenic and/or harmful effects on dairy technology, as well as to correlate the presence of these bacteria with the PC characteristics, the production of VOCs, and the FA profile of the cheese. Additionally, we sought to investigate and prospect, through genomic techniques, the presence of virulence, resistance, and toxin genes in reference genomes available in databases. The importance of this study for the dairy production chain and for consumer health is notable, revealing innovative information regarding the presence, in sheep’s cheese, of potential pathogens and bacteria that can harm milk technology, through modern molecular biology techniques.
2. Materials and Methods
The experiment consisted of the following steps: (a) research and selection of sheep’s cheese production and trade centers inspected in three states (Paraná—PR, Santa Catarina—SC, and Rio Grande do Sul—RS) in the southern region of Brazil; (b) sample collection (n = 13) between September and November 2023; (c) sample preparation and fractionation; (d) physicochemical analysis; (e) analysis of the free fatty acids profile by GC-FID; (f) analysis of the volatile compounds profile by GC-MS; (g) molecular analysis—Next Generation Sequencing (NGS) of the 16S rRNA gene (V3-V4 region); (h) survey of bacteria with pathogenic potential (BPP) and/or bacteria harmful to milk technology (BHMT); (i) in silico analysis—Genomics, which involved 16S rRNA gene phylogeny among 168 NGS sequences, similarity analysis between 39 16S rRNA gene sequences (19 NGS sequences and 20 reference sequences), prospecting for virulence genes and antimicrobial resistance (20 complete reference genomes), and functional annotation of genes (20 complete reference genomes); and (j) statistical analysis—Spearman’s non-parametric correlation test and Principal Component Analysis (PCA).
2.1. Sample Characterization and Collection
Thirteen samples of commercial sheep’s cheese were collected in the southwestern regions of Paraná (PR), western Santa Catarina (SC), and Rio Grande do Sul (RS), located in southern Brazil. Information such as type of cheese, type of milk treatment, maturation time, origin of collection, production, and inspection service is described in Table 1. The cheeses were produced by formal agribusinesses registered with the Federal Inspection Service (SIF) and/or in the Brazilian Animal Products Inspection System (SISBI).

Table 1.
Information about sheep’s cheese sold in the southern region of Brazil.
The samples were collected in two stages: (a) September—SC and PR, and (b) November 2023—RS. After collection, they were kept in their original packaging, placed in an isothermal box under refrigeration, and transported to the Milk Quality Laboratory (LabQuaL—CNPq: Special Dairy Technology) of the Department of Food Technology and Science (DTCA) of the Federal University of Santa Maria (UFSM). Subsequently, the samples were portioned aseptically in a laminar flow hood, and approximately 30 g of cheese, duly identified and frozen at −20 °C, was sent refrigerated to a service provider laboratory (GoGenetic—Paraná) for sequencing analysis of the genetic material.
2.2. Characterization of Sheep Cheese
2.2.1. Physicochemical Analyses
The humidity analysis of the cheeses was conducted by gravimetry, according to the direct drying method in an oven at 105 °C, as per the Instituto Adolfo Lutz [53]. Aliquots of 3 g of the samples were previously homogenized and weighed, then subjected to drying in an oven at 105 °C until a constant weight was achieved. Data are expressed as % humidity (w/w). The determination of mineral residue (ash) was performed using the gravimetric method, where 3 g of the sample was weighed in a porcelain crucible, dried at 105 °C for 6 h, and incinerated at 550 °C overnight, followed by weighing the mineral content [54]. The fat content was determined using direct reading in a Gerber butyrometer, involving treatment of the sample with sulfuric acid and isoamyl alcohol, followed by reading in the butyrometer, and calculation of fat in dry extract (FDE) based on humidity data [54]. The protein content was determined by the micro-Kjeldahl method with 0.2 g of the sample, which includes digestion with sulfuric acid and a catalytic mixture, distillation with NaOH, and collection of the distillate in boric acid, followed by titration with hydrochloric acid to calculate total nitrogen and convert to % protein using a correction factor of 6.38 [54]. Total carbohydrates were calculated by difference according to Tontisirin et al. [55]. All analyses were performed in triplicate to ensure the accuracy of the results.
2.2.2. Fatty Acid Profile Analysis by GC-FID
The extraction of lipid fractions from the samples followed the modified Bligh and Dyer protocol [56]. Initially, 8 mL of chloroform, 16 mL of methanol, and 8 mL of distilled water were mixed with 2 g of the sample and shaken for 30 min. Subsequently, an additional 8 mL of chloroform and 8 mL of anhydrous sodium sulfate solution were added, and the mixture was shaken and centrifuged at 4000× g rpm for 3 min. The lower phase (chloroform + fat) was transferred to a tube containing anhydrous sodium sulfate, shaken, filtered, and the filtrate evaporated in a vacuum oven at 45 °C until a constant weight was achieved.
The derivatization followed the method of Hartman and Lago [57]. First, 1 mL of potassium hydroxide in methanol solution (0.4 mol/L) was added to the lipid solution and heated at 100 °C for 10 min. Then, 3 mL of sulfuric acid in methanol (1 mol/L) was added, and the mixture was maintained at 100 °C for an additional 10 min. After cooling, 2 mL of isooctane was added, and the upper layer containing fatty acid methyl esters (FAMEs) was collected for chromatographic analysis. Additionally, 250 µL of methyl tricosanoate in isooctane (4 mg/mL; Sigma-Aldrich, USA) was added as an internal standard. The analysis was conducted using a Shimadzu GC-2010 Plus gas chromatograph, equipped with an FID detector and an AOC-20i (Shimadzu, Kyoto, Japan) autosampler. A Zebron ZB-WAXplus column (60 m × 0.25 mm, 0.25 µm film) was used, with helium as the carrier gas at 1.21 mL/min. A 1 µL sample was injected with a split ratio of 50:1. The injector was set to 250 °C, and the oven temperature program started at 50 °C, increased to 160 °C at a rate of 8 °C/min, followed by a rise to 240 °C at a rate of 5 °C/min, and held at 240 °C for 25 min. The detector temperature was maintained at 240 °C.
The determination of the AF profile followed the method of Visentainer [58], identifying FAME peaks by comparison with reference standards (Supelco 37 Component FAME Mix, Sigma-Aldrich, St. Louis, MI, USA). Quantification was performed based on the FID response factor and the internal standard methyl tricosanoate (Sigma-Aldrich, St. Louis, MI, USA). The analyses were conducted in triplicate.
2.2.3. Volatile Compounds Profile Analysis by GC-MS
The extraction of VOCs from the samples was performed by solid-phase microextraction in headspace (HS-SPME), using a divinylbenzene/carboxen/polydimethylsiloxane 50/30 μm fiber (DVB/CAR/PDMS, Supelco®, Bellefonte, PA, USA). The analysis was conducted using a gas chromatograph (Agilent 6890, Little Falls, DE, USA) coupled with a mass spectrometer (Agilent 5975) (GC-MS) and based on the methodology described by Zianni et al. [59]. Samples were weighed (4.0 g) in a 20 mL SPME vial with headspace along with 50 μL of internal standard solution of 1-octanol (1 mg/L in methanol; Merck KGaA, Darmstadt, Germany), sealed with a cap equipped with a silicone/PTFE septum (Supelco, Bellefonte, PA, USA), and conditioned for 5 min at 40 °C under continuous agitation. The fiber was then exposed to the vial headspace for 45 min, agitated at 400 rpm, and heated to 50 °C. After this period, the fiber was retracted and inserted into the chromatographic injector, in splitless mode, for 1.5 min for the desorption of volatile compounds, with the aid of a carrier gas (helium), and transferred directly to the chromatographic column (SPB-5, 60 m × 0.32 mm, film thickness df = 1 μm, 5% diphenyl–95% dimethyl polysiloxane, Supelco, Bellefonte, PA, USA) at 50 mL/min for 5.8 min, 250 °C. The GC oven conditions were set as follows: 40 °C (held for 5 min) and 220 °C (rate of 4 °C/min, held for 10 min). The total cycle time was 60 min. The transfer line, ion source, and quadrupole MS temperatures were 250, 230, and 150 °C, respectively. The MS detector operated in electron ionization mode set to 70 eV and in a scan range of 50–550 m/z.
The analytes were provisionally identified using linear retention indices (LRIs) and confirmed using the National Institute of Standards and Technology (NIST V2.2, Gaithersburg, MD, USA) library database [60]. Agilent Chem Station (Agilent Technologies, Santa Clara, CA, USA) was used for data collection and processing. Compounds were identified based on their LRI, mass spectra from the NIST library [60], or authentic standards measured under the same conditions. Compounds with a probability greater than 50% were selected for identification. To improve the precision of compound identification, only substances that provided a match factor greater than 700 and a match factor versus a reverse match factor ratio greater than 0.8 were selected for data processing [61,62].
2.3. 16S Amplicon Sequencing and Analysis
To identify the profile and bacterial population, NGS was used—16S rRNA amplicon sequencing with a cover of 30 thousand reads. DNA extraction, library preparation, and sequencing were performed by GoGenetic (Curitiba, Brazil), with Illumina Nextseq P1–600 in paired-end. Initially, DNA was extracted from 500 mg of the sample using the Wizard® Magnetic DNA Purification System for Food kit (Promega Corp., Madison, WI, USA), according to the manufacturer’s recommendations. To determine the concentration of extracted DNA, Nanodrop 1000 (Thermo, Wilmington, NC, USA) was used. Amplification of the genetic material was performed using the 16S-specific primers (16S_357F TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG and 16S_805R GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC) targeting the 16S V3 and V4 regions. Subsequently, the sequences were generated by the Illumina NextSeq platform, and the data obtained were processed using Qiime2 (v.2022.2) [63].
The sequences were imported into the program in the Casava 1.8 demultiplexed (paired) format, using the “qiime tools import” function. After this process, the primers used in sequencing were removed using the Cutadapt tool. Subsequently, the Dada2 tool was used to remove low-quality sequences and correct sequencing errors, using trimming mechanisms (–p-trim-left-f 0, –p-trim-left-r 0, –p-trunc-len-f and –p-trunc-len-r), linked with the “–p-chimera-method” flag to remove chimeras, and tables were generated with the data, sequences, and the frequency of amplicon sequence variants (ASVs) used in our work. The taxonomy was assigned to ASVs using the naïve–Bayes approach implemented through the scikit-learn Python library with default parameters and the GTDB database (v. 207). Relative abundance and the alpha diversity index were calculated using the R program (v. 3.6.1) (https://www.R-project.org/ (accessed on 17 January 2024)) with the reshape2 packages (v. 1.4.3) [64], phyloseq (v. 1.14.0) [65] and vegan (v. 2.4.1) [66].
As a negative control, one water sample was used in parallel for every 48 samples and the extraction product was used as a template for the Polymerase Chain Reaction (PCR). The positive control used was Zymo Mock Control (ZymoBIOMICS™ Microbial Community Standard II, D6310, Zymo Research Corporation, Irvine, CA, USA) for every 400 samples.
2.4. Pangenomic Analysis
2.4.1. Comparative Analysis of Genomes
Phylogenetic Analysis
The sequences of the 16S rRNA genes identified by NGS in the cheese samples were used to infer the phylogenetics tree. After removing the duplicate sequences obtained in sequencing, alignment was carried out using the MUSCLE alignment method [67] through the MEGA program, version 11 [68], and the result was exported in “.nexus” format. Subsequently, this file was used as input in the IQ-TREE program, version 1.6.12 [69]. The phylogenetics tree was generated using the maximum-likelihood method, with 1000 bootstraps, and visualized in cladogram format with the FigTree program, version 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 16 January 2024)).
The VITCOMIC2 tool (http://vitcomic.org/ (accessed on 18 January 2024)) was used to generate a circular map representing the taxonomic composition of microbial communities and their similarity to reference sequences in a database. As an input method in the program, 16S rRNA sequences were used [70].
Complete Reference Genomes for In Silico Analysis
Species identified as BPP and BHMT were selected for gene similarity analysis, research into virulence and antimicrobial resistance genes, and functional annotation of the genes. For this process, the complete genomes of the lineages considered as references by the NCBI itself of each of the species here were downloaded from the REFSEQ database [71] in “.fna”, “.gbff”, and “.faa” format. Furthermore, the 16S genes from each of these reference strains were downloaded separately in “.fna” format.
Genomic Similarity Analysis between 16S rRNA Genes and Reference Genes
For this analysis, the 16S rRNA genes found by amplicon sequencing and the 16S genes from the reference species, which were downloaded separately in the previous step, were used. This analysis was carried out using Gegenees software, version 3.1 [72]. This software is based on a similarity analysis methodology, where initially the genomes are fragmented into segments of a predefined size. In this case, 200 base pairs (bp) and a 100 bp reading matrix are created for analysis. After this, the shared content between the analyzed data was subtracted from all the analyzed genes, resulting in the variable content between them, used to calculate the percentage. The percentages were then plotted on the heatmap, ranging from low similarity (red) to high similarity (green). Subsequently, using the BLASTn method, a distance matrix was calculated with the result of comparing all fragments with each other, which was then plotted in a heatmap. In the heatmap, the “auto-sort” tool was used to rearrange the closest percentages, and the final result was exported in “.html” format. All programs were used with their default parameters.
2.4.2. Research of Virulence and Antimicrobial Resistance Genes
Prospecting for virulence and antimicrobial resistance genes in all complete genomes of reference species was conducted using Pan Virulence and Resistance Analysis software (PanVita), version 1 [73], a program focused on searching for resistance genes by comparing the input genome with the Comprehensive database Antibiotic Resistance Database (CARD), and in the search for virulence genes through comparison with the Virulence Factor Database (VFDB). The “-vfdb” and “-card” flags were used to compare the input files (NCBI “.gbff” format) with the VFDB and CARD databases, and the “-png” flag was used to export the result in “.png” format.
2.5. Statistical Analysis
GraphPad Software (version 8.2, Inc., La Jolla, CA, USA) was used to perform the statistical analysis of Spearman’s nonparametric correlation test. In addition, PCA was also performed, which is a statistical technique for multivariate analysis using the Excel program (XLSTAT 2023.2.0.1411). For PCA analysis, data standardization was carried out.
3. Results and Discussion
3.1. Bacterial Population Profile in Sheep’s Cheese
The NGS sequencing of the 16S rRNA gene allowed the characterization of the bacterial microbiota of 13 sheep’s cheese samples. For this, 1,061,392 bacterial sequences were obtained, emphasizing samples QJ188 and VF712, which presented 155,791 and 145,942 sequences, respectively. Of the total sequences, 3732 were not identified and/or classified as bacteria. From reading the sequences, five phyla were identified: Firmicutes, Proteobacteria, Actinobacteriota, Bacteroidota, and Fusobacteriota, with the first two being the most abundant, with 95.7% and 3.4%, respectively (Figure 1). The analysis carried out in VITCOMIC confirmed this result, which showed high similarity (>90%) of the majority of sequences with the two phyla mentioned above (Figure 2). Considering the phylum Proteobacteria, the most abundant class was Gammaproteobacteria (Figure 1B and Figure 2A). Other studies with sheep’s cheese report the predominance of the phyla Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria, highlighting the phylum Proteobacteria as it includes families of potential human pathogens, such as Enterobacteriaceae and Moraxellaceae [44,74,75,76].
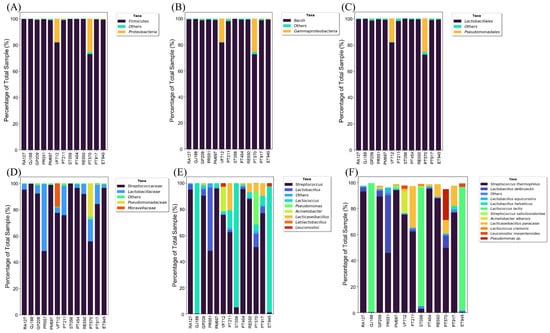
Figure 1.
Relative abundance of bacterial taxa identified in sheep’s cheese samples, sold in the southern region of Brazil, with more than 1% abundance. (A) Phyla; (B) Class; (C) Order; (D) Family; (E) Genus; and (F) Species.
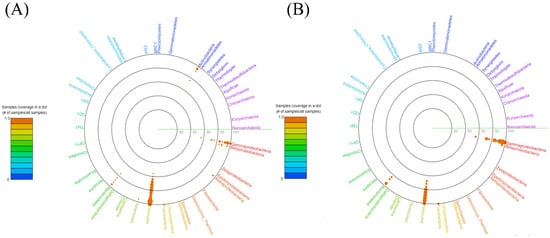
Figure 2.
Abundance and similarity of bacterial groups of sheep’s cheese 16S rRNA genes using the VITCOMIC circular diagram. (A) Phylum-level similarity of the general population of bacteria; (B) phylum-level similarity of the BPP/BHMT population. The color of each group varies according to the legend in the left corner of the figure, ranging from more presence (reddish) to less presence (bluish/purple). The circles filled in orange represent the abundance according to the percentage of similarity with the sequences in the database. The symbol # is related to number of reads in each of the orange dots in the image.
The relative abundance of phyla is directly influenced by factors such as care with raw materials, materials, and utensils until obtaining the final product [77], as well as by the severe thermal processing of milk, which ends up inhibiting part of the microbial population and, consequently, diversity decreases [77]. Therefore, to make up for the absence of autonomous fermenting bacteria, which are responsible for guaranteeing the organoleptic characteristics of the cheese, cultures containing LABs were used, which were the basis of the microbiota of the dairy product [78].
From the taxonomic classification, it was possible to identify 45 genera and 57 bacterial species in the 13 sheep’s cheese samples. Of these, we can observe the predominance of LABs, especially the genus Streptococcus, which represents around 60% of the total abundance, followed by Lactococcus, Lactobacillus, and Lacticaseibacillus, which account for 34.7%. The genera Pseudomonas and Acinetobacter represent 2.0% and 1.4% of the total sequences, respectively. Considering these last two genera, sample VF712 stands out with 17.8% of Acinetobacter abundance and PT570 with 25.6% of Pseudomonas. The remaining genera had abundances below 1% each.
The phylogenetic relationship of the bacterial sequences identified in sheep’s cheese sold in southern Brazil can be understood in Figure 3. Initially, the tree was divided into two branches: the first branch with a clade (dark green—right and central region) consisting of species from the genera Acinetobacter, Moraxella, and Psychrobacter, and the second branch with other genera/species. This result highlights the evolutionary distance of the species present in the dark green clade in relation to the rest of the cheese population.
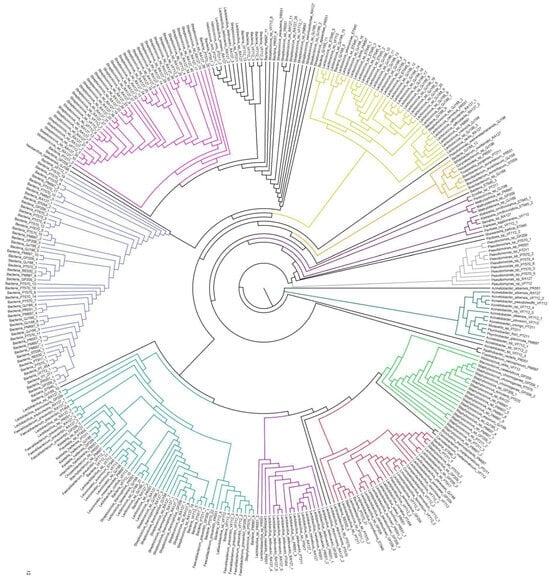
Figure 3.
Phylogenetic tree of the bacterial population identified in samples of sheep’s cheese sold in the southern region of Brazil. The circular phylogenetics tree was constructed using the maximum-likelihood method using 1000 bootstraps in the IQ-TREE program and visualized using the FigTree program. Each of the branches represents the evolutionary distance of the genomes represented at the ends. In constructing the phylogenetic tree, sequences of the 16S rRNA genes obtained in this study, amplified from sheep cheese samples, were used.
A large clade was formed with species from the LAB genera, such as Streptococcus (pink), Lactobacillus, Faecalibacterium, Lentilactobaccilus, Lacticaseibacillus, and Lactococcus (turquoise green and lilac), sharing a common ancestor, as they share the same order as the Lactobacillales. Well-defined groupings were observed, such as Pseudomonas (gray), Lactococcus (yellow), Streptococcus (pink), Lactobacillus (lilac), Staphylococcus (light green), and Acinetobacter (dark green) (Figure 3).
Genetic proximity that drew attention was that of the light green clade, consisting of pathogenic bacteria of the genera Alloiococcus, Staphylococcus, Macrococcus, and Brochothrix, with lactic acid bacteria (red, lilac, and turquoise green clades), mainly of the genera Lacticaseibacillus, Companilactobacillus, Limosilactobacillus, Lactiplantibacillus, Lentilactobacillus, and Loigolactobacillus (Figure 3). Additionally, a large clade in blue was generated with sequences that were not identified and was named Bacteria, which shares the same common ancestor as Streptococcus.
Due to the high abundance of LABs in the bacterial population of cheeses and because they are responsible for the positive characteristics of dairy products, this group is always in evidence in studies and research. However, it is important to investigate in more detail the bacteria that have the potential to cause disease, whether those commonly investigated, which are identified in food quality control legislation, emerging pathogens, and even species that are harmful to health milk technology.
Alpha diversity data were estimated using the Chao1, Simpson, and Shannon indices (Figure 4), given that the milk matrix supports a highly diverse microbiota due to its nutrient-rich nature [79]. From the analysis of bacterial population diversity in the cheeses, it is observed that the mean Chao1 index value is 53.4, ranging from 23.3 to 86.6 for samples ST056 and VF712, respectively. The samples RA127, VF712, and PT570 showed greater species diversity. The mean Simpson index was 0.350, and the Shannon index was 0.900. It can be seen in Figure 4 that 8 out of 13 samples had Chao1 index values above 50, indicating high species diversity. However, the same pattern does not hold for the Simpson and Shannon indices, indicating that the bacterial population does not maintain uniformity/richness and species abundance dominance. These findings reinforce the premise of previous studies that state the origin of the cheese influences the indices of richness, uniformity, and abundance of bacterial species [78].

Figure 4.
Alpha diversity indices of samples of sheep’s cheese sold in the southern region of Brazil. Chao-1 diversity index: qualitative index based on the richness of species present in the sample; Shannon index: considers the uniformity in the abundance of species; Simpson index: based on the dominance of species abundance.
Interestingly, the PT570 sample presented the highest values in all three indices analyzed, indicating it is a cheese with high diversity, uniformity/richness, and dominance of species abundance among all those analyzed. This cheese is produced with pasteurized milk, has a maturation period of 45 days (the youngest among the samples), and has an average moisture content of 27%. Wang et al. [80], in their study on Monascus cheese, also identified that there was higher alpha diversity in cheese samples with 10 days of maturation compared to those with 90 days, according to the Simpson index. The significant presence of Streptococcus spp. and Lactobacillus spp. is justified in Pecorino-type cheeses, largely due to the use of starter cultures predominantly composed of these two genera, which are also linked to flavor and texture characteristics [78]. In the sample in question, over 50% of the bacterial composition is of the genus Streptococcus, 10% of Lactobacillus, and, unexpectedly, 25% of the Pseudomonas.
Another important observation is the behavior identified in sample QJ188, which shows a high diversity of species according to the Chao1 index; however, the bacterial population has low uniformity and species dominance. This sample is characterized by the marked presence of LAB, which can be attributed to its short ripening time (60 days), being composed of 98% of the species Lactococcus lactis, a species used as a starter culture. Endres [78], in her study on sheep cheese produced in southern Brazil, identified the use of Lactobacillus helveticus as a starter culture and high levels of Streptococcus in colonial or fresh cheeses. A similar but less pronounced behavior is observed in sample RA127 (Figure 4).
The samples PT570, PR551, and PT211 exhibited the highest uniformity/richness (Shannon) and dominance (Simpson) of species abundance. It is interesting to highlight the profile of sample PR551, produced with unpasteurized milk, which presented the second highest Shannon and Simpson indices, corroborating the idea that microbial diversity is greater when thermal treatment of the raw material does not occur [75,76]. Meanwhile, sample GP209, also produced from raw milk, did not exhibit the same behavior, indicating once again that distinctions in animal management, production processes, and the final microbial composition of the milk directly influence the final product [78,81,82].
In the PCA analysis, which accounts for 99.6% of the total variance (PCA1 and PCA2), we observed that sample VF712 is closely associated with the Chao1 index, whereas sample PR551 exhibited a correlation with the Simpson and Shannon indices. Conversely, the PT570 cheese was positioned in a quadrant distant from the other analyzed samples. There were no distinct groupings among the samples; however, an association was noted between cheeses QJ188, RA127, and PM697. Near the Simpson and Shannon indices are cheeses PT454, ST056, RE550, ET945, PT917, and PT211, all produced in the state of RS, suggesting a potential distinguishing characteristic of cheeses from other states. This difference may be related to the microbiota “terroir” [83].
3.2. Profile of Bacteria with Pathogenic Potential and Harmful to Milk Technology
To detail and direct the analyses, the bacteria that could make the cheese unsuitable for consumption were classified into two groups: BPP—bacteria with potential pathogenicity to human health and BHMT—bacteria harmful to milk technology. Within BPP are foodborne pathogens, which cause poisoning and/or pose risks to public health, while BHMT consists of those bacteria that are resistant to thermal processes, which impart undesirable characteristics to the final product, spoilage, and biofilm-forming bacteria, according to the grouping proposed by Remor et al. [19]. These groups represent 3.9% of the total bacterial abundance in cheese. In Figure 2B, we can see the high abundance of similarity of the sequences obtained in this work with the phylum Firmicutes (43.0%), followed by Proteobacteria, with emphasis on the class Gammaproteobacteria (47.2%), which includes families of potential human pathogens [44,74,76].
In Table 2, we observed that 15 genera were classified as BPP, with the Acinetobacter genus being the most abundant, accounting for approximately 62.7% of the group (BPP), followed by the Pseudomonas genus, with 34.7%. The BHMT group was made up of six genera, with the Macrococcus genus being exclusive to this group. Furthermore, these two genera (Acinetobacter and Pseudomonas), together with Staphylococcus, appeared most frequently in the samples, being present in 8, 12, and 10 cheeses, respectively. These results reinforce the similar profile between the two groups (BPP and BHMT) and the existence of a central microbiome composed of harmful bacteria in cheese [84].

Table 2.
Species bacteria with pathogenic potential (BPP) and bacteria harmful to milk technology (BHMT) detected in samples of sheep’s milk cheese sold in the southern region of Brazil (n = 13).
Acinetobacter was identified in significant quantities, along with Macrococcus and/or Streptococcus, in samples of Halloumi cheeses produced with fresh pasteurized goat’s and/or sheep’s milk [76]. This genus was also prevalent in Feta cheese samples with Protected Designation of Origin (PDO), with Acinetobacter baumannii, Acinetobacter soli, Acinetobacter johnsonii, and Acinetobacter ursingii being the main species [44]. Interestingly, the last two species cited by Papadakis et al. [44] were also identified in this study.
The species Acinetobacter albensis was identified in 6 samples, representing 53.1% of the total abundance (TA) (excluding LAB and bacteria beneficial to milk technology) and more than 58% in both groups (BPP and BHMT), being the species with the greatest representation. It is worth noting that only sample VF712 represents 99.5% of the 53.1% of the total abundance of this species. In this same sample, Acinetobacter sp. was also identified, only at the genus level, representing 4.5% of the total BPP abundance. Despite the characteristic of long maturation time, around 180 days, and production with pasteurized milk, sample VF712 was affected by the incisive presence of this genus. This fact can be observed in Figure 5, which illustrates that the PCA, with the proximity of the Acinetobacter genus to this sample, is present in the same quadrant (Figure 5). The VF712 sample also presented a different bacterial composition profile than the other cheeses, with a high relationship with the genera Pantoea, Acinetobacter, and Alloiococcus (Figure 5).
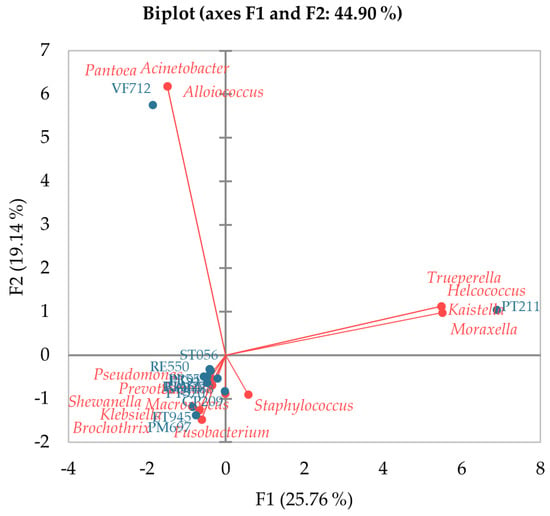
Figure 5.
Representation of Principal Component Analysis (PCA). The figure illustrates the PCA of the relationship between the number of sequences of BPP and BHMT genera with cheese samples. The active variables are represented in red (genera), while the active observations (cheese samples) are represented in blue.
The result reinforces the premise that these bacteria can be an environmental contaminant, as they are commonly found in the udder region, with a high incidence in raw milk [85], or are resistant to the conditions that the dairy product acquires throughout maturation. Kamimura et al. [77] described the psychotropic character of Acinetobacter and their ability to adapt to adversity, even when isolated on cloths used in the desilting process.
The genus Pseudomonas was the most prevalent taxon in studies carried out in food industry facilities. In analyses of the surfaces of dairy processing facilities, this bacterial group was present on more than 75% of the surfaces, which can be explained by the biofilm formation capacity of this contaminant [84], making its removal difficult and enabling continuous contamination. As it is one of the main contaminants of raw milk, the presence of Pseudomonas is due to several factors, such as temperature fluctuations in storage, herd health, and even the season, being more abundant in the months of November and December [85].
The two cheese samples produced with raw sheep’s milk (PR551 and GP209) have a very similar profile since both have an incidence of Pseudomonas sp. However, the first sample also has A. albensis in its composition of BPP and BHMT, while sample GP209 is composed of Kaistella haifensis, Staphylococcus sp., and Staphylococcus chromogenes. Corroborating the results mentioned regarding the high abundance of the genera Pseudomonas, Staphylococcus, Pasteurella, Enterobacter, Klebsiella, and Acinetobacter, a work conducted by Kamimura et al. [77] detected the presence of this group of bacteria in cheeses from Serra da Canastra. In samples referring to the floor microbiota, the main genera and families identified were Acinetobacter and Enterobacteriaceae, while in the microbiota on the shelves, the genus Staphylococcus predominated.
Of the sixteen bacterial taxa identified, only four were found in more than one sample, with the genus Pseudomonas being present in twelve of the thirteen samples. The samples with the greatest diversity of genera were PT211 (n = 7) and PM697, ET945, and RA127, with five genera each. However, they presented a low number of sequences, with 385, 157, 250, and 42 sequences, respectively. This result was confirmed in the PCA (Figure 5), in which the PT211 sample grouped separately with the genera Trueperella, Helcococcus, Moraxella, and Kaistella, distancing itself from the other cheeses. Samples PT570 (2 genera) and VF712 (4 genera) showed lower diversity and greater abundance of sequences, with 13,875 and 26,031, respectively.
The other bacteria were identified in isolated samples and may express specific problems in the cheese factory since their pathogenicity is linked to the environment and animals, and their occurrence in dairy products and foods, in general, is not common. Trueperella pyogenes is an example, with rare cases of contamination in humans, being an opportunistic animal pathogen associated with herd health and strongly related to the occurrence of mastitis and other infections [86]. Its detection occurred in a sample of cheese produced with pasteurized milk and aged for 90 days, showing that its presence was due to incorrect pasteurization or environmental cross-contamination.
The genus Klebsiella was identified in a cheese, with the species Klebsiella ornithinolytica (formerly known as Raoultella ornithinolytica) being the only one detected (Table 2). The presence of species of this genus has been reported mainly in water and the environment; however, species such as R. ornithinolytica and Klebsiella variicola have been considered hospital pathogens [87], emerging pathogens, and animal pathogens (mastitis) [88]. The species Alloicoccus otitis is another bacterial pathogen that causes otitis media [89], and its presence in sheep’s cheese has not yet been reported.
3.3. Correlation between Groups of Bacteria with Pathogenic and Harmful Potential for Milk Technology and Physicochemical Properties
It is scientifically known that the microbiota present in cheeses directly affects the sensory, organoleptic, and physicochemical characteristics of dairy products [13]. Furthermore, the nutritional composition and parameters such as moisture modulate microbial colonization and diversity [90], allowing for a distinction of flavors, odors, and characteristics among various types of cheese. From the Spearman’s correlation analysis, it was observed that the bacterial genera with pathogenic potential (BPP) and bacteria harmful to milk technology (BHMT) may have positive or negative associations with the PC properties of sheep’s milk cheeses (Figure 6). Notably, the genus Staphylococcus showed a positive correlation with carbohydrate content (blue) and a negative correlation with fat in dry extract (FDE) (red). The BHMT bacterial group also showed a positive association with carbohydrates, indicating that the presence of this group is higher in cheeses with higher carbohydrate content.
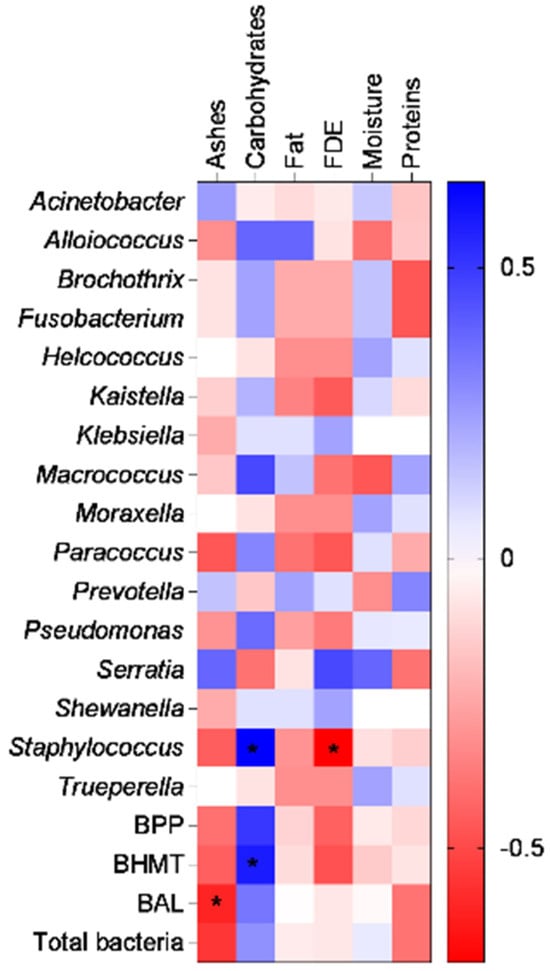
Figure 6.
Heat map demonstrating the correlation between bacterial groups and physicochemical parameters. BPP: abundance of bacteria with pathogenic potential and BHMT: abundance of bacteria specific to milk technology. The bluish color shows positive brightness, whitish shows without brightness, and reddish shows negative brightness. The correlation test used was Spearman’s non-parametric test and the asterisk indicates a significance of 95% (p < 0.05).
The FA profile of cheeses is directly related to the composition of the milk, which is influenced by herd management. Nutritional management, such as the type of forage, the proportion of concentrate, and supplementation with foods rich in polyunsaturated FAs, modulates the FA profile in milk [91,92,93,94]. Additionally, the influence of starter and adjunct cultures on the fat profile, especially in matured products [95,96], must be considered.
In Figure 7, the correlation between the bacterial groups and the FA profile in the analyzed cheeses is observed, and the significant associations were negative, meaning they were inversely proportional to the presence of a particular FA and the growth of the bacterial group. The BPP and BHMT groups are negatively associated with C20:1n9 (Gondoic acid) and C22:6n3 (Docosahexaenoic acid). Additionally, BHMT is also negatively correlated with C22:1n9 (Erucic acid). Studies have reported the effect of lipid complexes and isolated FAs against bacterial groups, including pathogenic ones, suggesting their use as a natural antibiotic, which is a promising alternative to traditional synthetic drugs [97,98,99,100,101,102]. Sukhikh et al. [102] investigated the antimicrobial effect of a purified lipid complex isolated from the microalgae Chlorella vulgaris and Arthrospira platensis against Escherichia coli, Bacillus subtilis, and Bacillus pumilus.
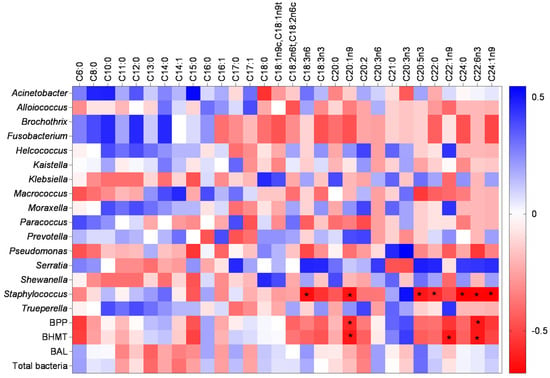
Figure 7.
Heat map demonstrating the correlation between bacterial groups and the fatty acid profile. BPP: abundance of bacteria with pathogenic potential and BHMT: abundance of bacteria specific to milk technology. The bluish color shows positive brightness, whitish shows without brightness, and reddish shows negative brightness. The correlation test used was Spearman’s non-parametric test and the asterisk indicates a significance of 95% (p < 0.05).
The genus Staphylococcus showed a negative correlation with seven different FAs: C18:3n6 (γ-Linolenic Acid), C20:1n9 (Gondoic Acid), C20:5n3 (Eicosapentaenoic Acid), C22:0 (Behenic Acid), C22:6n3 (Docosahexaenoic Acid), C24:0 (Lignoceric Acid), and C24:1n9 (Nervonic Acid). Linolenic acid in ethanolic extracts of C. vulgaris also showed antibacterial activity against Staphylococcus aureus and Salmonella typhi, the causative agent of typhoid fever [97,101]. Additionally, other researchers have verified the antimicrobial properties of eicosapentaenoic acid, stearidonic acid, and gamma-linolenic acid, with the latter two being inhibitors of oral pathogens such as Prevotella intermedia and Porphyromonas gingivalis, with minimum inhibitory concentrations of 39.06 and 9.76 μg/mL, respectively [99,100]. Inhibitions, at micromolar concentrations, of species such as Aggregatibacter actinomycetemcomitans, Candida albicans, Fusobacterium nucleatum subsp. vincenti, and Streptococcus mutans were also reported by Park et al. [100].
It is already scientifically known that the presence of VOCs can inhibit or even reduce microbial growth. In Figure 8, this inhibition profile was observed. The presence of various VOCs in cheese showed a negative association with the bacterial groups analyzed. Six bacterial groups—Staphylococcus, Pseudomonas, BPP, BHMT, LAB, and total bacteria—showed sensitivity (negative correlation) to the presence of four VOCs: Butyric acid and Ethanol (products of fermentative pathways of sugars such as glucose, galactose, and sucrose), Methyl Butyrate (a product of butyric acid fermentation), and Methyl Decanoate. [103]. Butyric acid directly influenced the Staphylococcus, BPP, and BHMT groups.
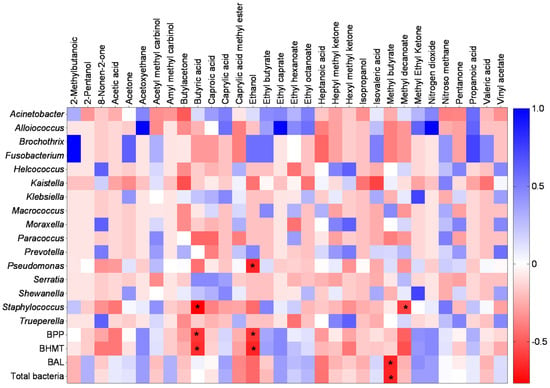
Figure 8.
Heat map demonstrating the correlation between bacterial groups and the profile of volatile organic compounds. BPP: abundance of bacteria with pathogenic potential and BHMT: abundance of bacteria specific to milk technology. The bluish color shows positive brightness, whitish shows without brightness, and reddish shows negative brightness. The correlation test used was Spearman’s non-parametric test and the asterisk indicates a significance of 95% (p < 0.05).
Interestingly, Butyric acid negatively influenced only the group of harmful and detrimental bacteria for dairy technology, showing a slight positive (non-significant) correlation with the group of LAB and total bacteria. The stimulation of the growth of butyric acid-producing microbiota may serve as another barrier to the development of undesirable groups, ensuring better quality control of cheeses. The presence of this VOC plays an important role in the flavor of many types of cheese, such as Camembert, Cheddar, Grana Padano, Gruyère, and Pecorino [104]. Species such as Lactobacillus paracasei, Staphylococcus equorum, Staphylococcus xylosus, and Enterococcus sp. are producers of butyric acid [105], which justifies the presence of this VOC in cheese samples, as the genus Lactobacillus was present, in some samples, in great abundance. However, the presence of butyric acid can impart negative characteristics and make the cheese unfit for consumption, such as rancid odor, unpleasant taste, and bloating [103,104]. The concentration of butyric acid capable of modifying the flavor of cheese varies according to the type; in Gouda, for example, this occurs at concentrations above 500 mg kg−1; whereas in Emmenthal, the flavor is modified whenever the ratio between propionic and butyric acids is less than 1 mg kg−1 [106].
3.4. Search for Resistance and Virulence Genes in Reference Genomes
Since ancient times, fermentation has been used as a method of food preservation because it considerably reduces the presence of potentially pathogenic microorganisms. However, recent studies have revealed the presence of different types of microorganisms, especially pathogenic bacteria, in these foods. The fermentation process can represent a significant problem for the health of consumers because it keeps microorganisms nearby and favors different gene transfer mechanisms between them, increasing the abundance of these genes within the community [107].
With Gegenees software, version 3.1, organisms belonging to the BPP and BHMT groups were used to evaluate the genomic similarity between them and with reference 16S rRNA from the NCBI database (Figure 9). When observing the heatmap, in general, one can notice more phylogenetically distant organisms, with the values marked closest to the red color. This profile was already expected due to the great diversity of genera selected for this investigation stage. However, some groups have attracted attention due to their high similarity. All sequences of the genes of the species identified in sheep’s cheese showed high similarity with the reference genes (85 to 100%), except for Pseudomonas weihenstephanensis, which obtained 57% similarity. Thus, the similarity analysis suggests that our sequences present similarity at the genomic level with their respective reference genomes (classified by NCBI), confirming the classification of the taxa. Based on these, the complete genomes of the reference species result that showed high similarity with the species identified in the cheeses were used for subsequent genomic analyses.
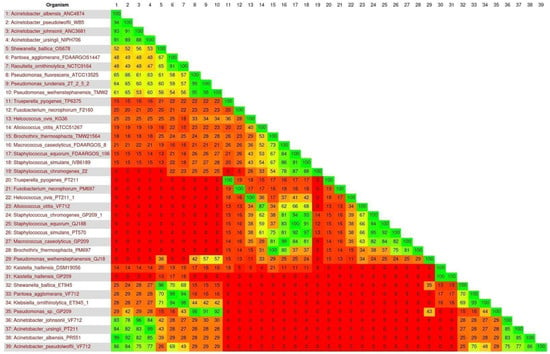
Figure 9.
Similarity analysis between species from the BPP and BHMT groups present in sheep’s cheese with reference organisms. In the heatmap, the same organisms described horizontally in each of the numbers are also allocated vertically. The greenish coloration demonstrates high similarity (values close to 100% similarity), the orange coloration shows a medium similarity and a redder coloration demonstrates low similarity between the genomes (values close to 0% similarity).
Complete reference genomes, obtained from NCBI databases, of the species identified in sheep’s cheese, were analyzed in the PanVita software, version 1, and a clustermap of resistance and virulence genes was generated to qualitatively and quantitatively verify the presence of these genes (Figure 10). In general, the resistance genes (Figure 10A) detected in the analysis using the CARD database totaled 146 genes detected in 14 species, of which 83 were different genes, divided into 6 different types of mechanisms acting on the inhibitory compound, which were efflux (n = 60), inactivation (n = 8), alteration of the target of action (n = 8), reduction in permeability (n = 6), protection (n = 2), and replacement of the antibiotic target (n = 2). Three genes (marA, ParR, and ramA) have efflux and permeability reduction functions simultaneously.
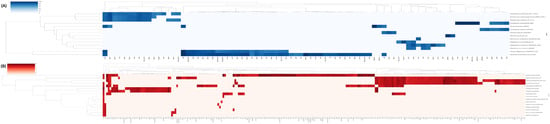
Figure 10.
Clustermap of resistance and virulence genes from the BPP and BHMT groups generated by PanVita software. (A) Resistance and (B) virulence genes. On the right, horizontally, the complete genomes of species from the BPP and BHMT groups are listed. At the bottom of each graph, the abbreviations of the names of the resistance/virulence genes found in the analysis are described. You can see colored squares with gradients of tones representing the number of resistance or virulence genes, according to intensity, in the genome.
Furthermore, the genes identified confer resistance to 28 antibiotics. The 3 main classes were fluoroquinolones, tetracyclines, and peptide antibiotics and disinfectant agents, with 28, 21, and 19 resistance genes, respectively. In a study conducted by Regecová et al. (2021), isolated strains of S. chromogenes showed the highest resistance to penicillin (95%), tetracycline (86%), and oxacillin (81%). Furthermore, through phenotypic manifestations and detection of the presence of the mecA gene, it was possible to verify that 30% of the isolates were resistant to methicillin. Additionally, 62% of the isolates were multidrug resistant.
According to Figure 10A, reference genomes of species detected in the BPP group presented a wide variety of mechanisms and genes, with a vast predominance focused on antibiotic efflux (n = 60) (Abau_AbaQ, abeM, acrB, acrD, adej, adek, adel, arlR, cpxA, CRP, Eclo_acrA, Ecol_mdfA, emrR, floR, H-NS, kdpE, Kpne_KpnE, Kpne_KpnF, Kpne_KpnG, Kpne_KpnH, leuO, LptD, marA, mdtA, mdtB, mdtC, mdtG, mdtH, MdtK, MexB, MexE, MexF, MexK, MexN, mgrA, msbA, norC, OpmH, OprM, OprN, oqxA, oqxB, Paer_CpxR, ParR, PmpM, ramA, RanA, RanB, rosA, rosB, rsmA, Saur_norA, sepA, tet(38), tetA(P), TolC, TriC, YajC, and YojI). The families of efflux pumps, as they are called, are located in the plasma membrane and use energy in different ways to translocate substances from the cells’ internal environment to the external environment and vice versa, playing a crucial role in the resistance of these cells bacteria [108]. They are often found conserved, suggesting maintenance of this type of antimicrobial resistance in evolution and consequently conferring some resistance against B-lactams, tetracyclines, macrolides, and quinolones, among others [109,110].
A second important group of genes found is resistance genes focused on altering the antibiotic target (ArnT, bacA, eptA, eptB, PmrF, sul2, and ugd) (Figure 10A). These types of resistance mechanisms can be considered central in this process since many antibiotics have a high affinity for growth inhibition or death, only with their primary target, and any change in this target makes the inhibitory compound inefficient, resulting in new resistance mechanisms [111].
Finally, a third large group of genes found in our analysis is related to antibiotic inactivation (APH(3″)-Ib, APH(6)-Id, FosA6, ORN-1, OXA-296, and Tet(X3)). This type of mechanism can damage or degrade the medicine and consequently reduce its efficiency. Through hydrolysis of the amide bond with the B-lactam ring or even by binding other tetracycline hydroxylase enzymes, these genes confer resistance to several classes, such as carbapenems and cephalosporins [111].
Considering the reference genome of the only species exclusive to the BHMT group, Macrococcus caseolyticus FDAARGOS 868, we can see the presence of a single norC gene with antibiotic efflux function. The species that presented the highest number of resistance genes were Raoultella ornithinolytica NCTC9164 with 41 genes, with the functions of efflux (n = 31), alteration of the target of action (n = 6), inactivation (n = 2), and reduction in permeability (n = 4), followed by Pantoea agglomerans FDAARGOS 1447 with 23 resistance genes (efflux—n = 20, alteration of the target of action—n = 2, and reduction in permeability—n = 1) (Figure 10A).
The genus Pseudomonas also presented a significant number of genes (n = 39) divided into 3 species: P. fluorescens_ATCC_13525 (n = 15), P. weihenstephanensis TMW2 1728 (n = 11), and P. lundensis_2T_2_5_2 (n = 13). Interestingly, a similar profile was found between the genes in the species of this genus. Resistance mechanisms are restricted to the functions of efflux and alteration of the target of action, the latter being expressed from the arnA gene in the three species. Regarding the efflux function, a similarity was observed between the Mex and Opr family genes. These families of efflux pumps (Mex and Opr, mainly) function in three states: Loose (L), Tight (T), and Open (O). Thus, they confer resistance to antibiotics through the electrochemical potential of protons [112]. A single gene found in this group, arnA, was correlated with the mechanism of antibiotic change, mainly the type of peptides (polymyxin).
In a recent study using in vivo methodology, isolated species of Staphylococcus were present in sheep’s cheese produced with raw milk, with resistance to multiple antimicrobial agents, 46% of which were multi-resistant. Furthermore, most isolates had at least one resistance gene in their genome [29]. In this in silico analysis, 16 resistance genes were detected in 3 species of Staphylococcus (S. equorum FDARGOS 1068, S. simulans IVB6189, and S. chromogenes 22). The genes arlR, mgrA, norA, Saur_norA, norC, sepA, tet(38) (efflux), dfrC (substitution of the antibiotic target), mphC (inactivation), and msrA (protection) are resistant to 7 classes of antibiotics (fluoroquinolone, cephalosporin, tetracycline, diaminopyrimidine, macrolide, streptogramin B, and streptogramin). The mphC and msrA genes were only identified in the species S. equorum FDARGOS 1068.
The species A. johnsonii ANC3681, A. ursingii NIPH706, A. otitis ATCC51267, B. thermosphacta D337, F. necrophorum subsp. Funduliforme F1260, and T. pyogenes TP6375 did not present resistance genes in the analysis and, therefore, are not represented in Figure 10.
With the clustermap of virulence genes, we can observe the correlation between the number of genes and their presence/absence in the reference genomes (Figure 10B). In general, 12 types of virulence mechanisms were identified in 247 different genes in the analysis (totaling 522 genes identified) with functions of adherence (n = 86), antimicrobial activity (n = 4), biofilm formation (n = 40), effector delivery system (n = 93), exoenzyme (n = 1), exotoxins (n = 1), immune modulation (n = 81), invasion (n = 5), motility (n = 127), nutritional factor/metabolic (n = 56), regulation (n = 18), and stress survival (n = 10).
According to Figure 10B, immune modulation and adherence processes were the main types found in the genomes of the BPP group. For immune modulation, the genes ACICU_RS00400, ACICU_RS00475, ACICU_RS00500, galE, galF, galU, gmhA/lpcA, gndA, KP1_RS17355, KP1_RS17345, KP1_RS17340, KP1_RS17240, KP1_RS17225, kdsA, KP1_RS17280, lpsA, lpxB, lpxC, lpxD, lpxL, pbpG, pgi, rfbA, rfbB, rfaD, rfaE, rfbD, rfbK1, tviB, ugd, and wcaJ were identified. For the adhesion process, the genes fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI, htpB, pilB, pilC, pilF, pilG, pilH, pilI, pilJ, pilM, pilP, pilQ pilR, pilT, pilU, rpoS, srtC-1/srtB, tufA, yagX/ecpC, yagW/ecpD, yagY/ecpB, and yagZ/ecpA were found.The process of immune modulation is considered one of the main types of virulence mechanisms used by microorganisms. Bacterial capsules and lipopolysaccharides stand out in this type of mechanism. Bacterial capsules in the cell wall provide a niche for the microorganism, favoring its response to stress and survival over time. Lipopolysaccharides, part of the structure of the microbial wall, act as an immunological barrier, controlling the passage of antibiotics and contributing to cellular integrity [113].
The virulence mechanism correlated with adherence is considered a crucial process, which can facilitate the fixation and internalization of the microorganism in the host’s epithelial cells, inducing several factors that can be apoptotic and lead to the death of host cells. These proteins are found mainly in the cell walls of microorganisms and, in addition to fixation, can facilitate transport between organisms, contributing to the success of pathogenesis [114].
This mechanism is frequently observed in bacteria of the genus Acinetobacter, considered a critical pathogen for human health [114]. This in silico work confirmed this characteristic, identifying that species of this genus (A. albensis ANC4874 and A. pseudolwoffii WB5) presented 12 different types of genes related to adherence, corresponding to 35.3% of the total identified (Figure 10B). Furthermore, this genus was the only one in our analysis to have the plcD gene, classified as an exotoxin in the VFDB database. This exotoxin helps the microorganism through lysis, contributing to invasion [115,116].
For the exclusive BHMT reference genome, Macrococcus caseolyticus FDAARGOS 868, the presence of five virulence genes was observed, two from the cap family (cap8E and cap8O) related to immune modulation, two from the clp family (clpP and clpC) with function of stress survival, and one, tufA, related to adherence.
Among the species that presented the highest number of virulence genes identified, the most notable are R. ornithinolytica NCTC9164, P. weihenstephanensis TMW2 1728 1, P. lundensis 2T_2_5_2, P. fluorescens ATCC 13525, and P. agglomerans FDAARGOS 1447 with 93, 89, 86, 79 and 57 genes, respectively. It is important to consider that the species R. ornithinolytica NCTC9164 and P. agglomerans FDAARGOS 1447 also had the highest number of genes related to antibiotic resistance. Regecová et al. [27] warn in their work about the need to investigate the presence of other species of bacteria in dairy products, not just those of recognized importance. Our survey shows that species of apparently minor importance can present a risk to consumers’ health and, eventually, become emerging pathogens in the future.
It is also important to highlight the presence of other virulence mechanisms, such as motility and biofilm formation. The presence of genes from the mot family, found in the genus Pseudomonas, are responsible for bacterial flagellar movement and, consequently, act in a chemotactic manner, interacting with other organisms [117]. On the other hand, we have the presence of flagella, represented by genes from the fli family, which, in addition to helping in the motility process, play a crucial role in the formation of biofilms [118]. Biofilms facilitate adhesion between bacteria, favoring the process of horizontal gene transfer, which can make the fermented food production process extremely delicate [108].
The species A. johnsonii ANC3681, A. ursingii NIPH706, A. otitis ATCC51267, and T. pyogenes TP6375 did not present virulence genes found in the analysis and, therefore, were not represented in Figure 10.
4. Conclusions
From this work, it was possible to characterize the microbiota of sheep’s cheese produced and/or sold in southern Brazil and correlate it with the PC, FA, and VOC parameters. By directing the results towards the detection of BPP and BHMT bacteria, it was possible to elucidate the current scenario of the microbiological quality of these products.
Even in compliance with legislation, the cheeses contained bacterial groups that are neglected in investigations because they are not important for the food industry, but could become emerging pathogens in the future or generate problems for the production sector. Taking into account the in silico analysis of complete reference genomes with high similarity to the isolates in this work, through genomics, it was possible to obtain complementary results that draw attention to the need for a more detailed investigation of these bacteria in cheeses. The high prospect of genes related to antibiotic resistance and virulence, even the production of toxins, serves as a warning for future investigations in dairy products.
We state that this study is exploratory in nature and highlight the importance of sequencing the complete genomes of these neglected species present in cheese to validate and confirm the presence of resistance and virulence genes. Furthermore, it is also necessary to use quantitative or semi-quantitative tools to quantify the abundance of these species and verify the expression of harmful genes in foods. The genus Staphyloccocus showed a significant negative correlation with the PC, FA, and VOC parameters. The bacterial groups BPP and BHMT also showed a negative correlation with some FA and VOC. In general, this study will guide future investigations focusing on bacteria unsuitable for cheese, especially sheep’s cheese. Work like this is necessary to provide a scientific basis for the particularities of these dairy products, also generating insights into possible complications and a better understanding of the problems that the sheep’s cheese production chain may face.
Author Contributions
Conceptualization, M.A.P.P., W.d.C.O. and N.S.P.d.S.R.; data curation, M.A.P.P., W.d.C.O. and A.G.F.; formal analysis, M.A.P.P., S.C.S.C., W.d.C.O. and A.G.F.; funding acquisition, M.A.P.P., W.d.C.O. and N.S.P.d.S.R.; investigation, W.d.C.O., M.A.P.P. and S.C.S.C.; methodology, M.A.P.P., S.C.S.C., M.B.P.P.O., W.d.C.O. and A.G.F.; project administration, N.S.P.d.S.R.; software, A.G.F.; supervision, N.S.P.d.S.R.; visualization, M.A.P.P., M.B.P.P.O. and W.d.C.O.; writing—original draft, M.A.P.P., W.d.C.O. and A.G.F.; writing—review and editing, M.A.P.P., W.d.C.O., A.G.F., M.B.P.P.O. and N.S.P.d.S.R. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Fundação de Amparo à Pesquisa do Rio Grande do Sul—FAPERGS (registration number 23/2551-0000170-5), grants funded: Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—CAPES (registration number 88882.461702/2019-01) e Conselho Nacional de Desenvolvimento Científico e Tecnológico—CNPq (registration number 150197/2023-3).
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors on request.
Acknowledgments
Sara C. Cunha acknowledges FCT for the 2022.07841.CEECIND/CP1724/CT0014 contract and also thanks FCT/MCTES—UIDP/50006/2020. The team’s researchers would like to thank Siomar de Castro Soares, professor and researcher at the Department of Microbiology, Immunology and Parasitology, Laboratory of Immunology and Bioinformatics, Federal University of Triângulo Mineiro, for all his support in the bioinformatics analyses.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- FAOSTAT Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org/faostat/en/#data/QCL/visualize (accessed on 23 July 2024).
- Bianchi, A.E.; Monteiro, A.L.G.; Morais, O.R.d.; Beltrão, R.B.F.; Debortoli, E.C. Caracterização dos sistemas produtivos de ovinos de leite no Brasil. MilkPoint. 2016, Seção Radar Técnico, Ovinos e Caprinos, 1-5. Available online: http://www.infoteca.cnptia.embrapa.br/infoteca/handle/doc/1073171 (accessed on 23 July 2024).
- Barłowska, J.; Szwajkowska, M.; Litwińczuk, Z.; Król, J. Nutritional Value and Technological Suitability of Milk from Various Animal Species Used for Dairy Production. Compr. Rev. Food Sci. Food Saf. 2011, 10, 291–302. [Google Scholar] [CrossRef]
- Bittante, G.; Amalfitano, N.; Bergamaschi, M.; Patel, N.; Haddi, M.-L.; Benabid, H.; Pazzola, M.; Vacca, G.M.; Tagliapietra, F.; Schiavon, S. Composition and Aptitude for Cheese-Making of Milk from Cows, Buffaloes, Goats, Sheep, Dromedary Camels, and Donkeys. J. Dairy Sci. 2022, 105, 2132–2152. [Google Scholar] [CrossRef] [PubMed]
- Dalmina, E.M.; Malta, D.S.; Silva, F.C.; Tribst, A.A.L.; Rigo, E.; Cavalheiro, D. Sodium Reduction in “Requeijão Cremoso” Processed Cheese Made from Fresh and Refrigerated Sheep Milk. J. Food Process. Preserv. 2022, 46, e16418. [Google Scholar] [CrossRef]
- Hassan, F.A.; Abbas, H.M.; Abd El-Gawad, M.A.; Enab, A.K. Goats Dairy Products as a Potentially Functional Food. Life Sci. J. 2014, 11, 648–657. [Google Scholar]
- Ramos, T.M.; Gajo, A.A.; Pinto, S.M.; Abreu, L.R.; Pinheiro, A.C. Perfil de textura de Labneh (iogurte grego). Rev. Inst. Laticínios Cândido Tostes 2009, 64, 8–12. [Google Scholar]
- Robinson, R.K.; Tamime, A.Y. Feta & Related Cheeses; CRC Press: Boca Raton, FL, USA, 1996; ISBN 978-0-7476-0077-0. [Google Scholar]
- Furtado, M.M. Quesos Típicos de Latinoamérica; Danisco: São Paulo, Brazil, 2005. [Google Scholar]
- Addis, M.; Piredda, G.; Pes, M.; Di Salvo, R.; Scintu, M.F.; Pirisi, A. Effect of the Use of Three Different Lamb Paste Rennets on Lipolysis of the PDO Pecorino Romano Cheese. Int. Dairy J. 2005, 15, 563–569. [Google Scholar] [CrossRef]
- Nespolo, C.R.; Bianchi, A.E.; Queirós, A.A.E.; Farinatti, L.H.E. A Produção de Leite Ovino e Seus Derivados No Oeste Catarinense: Uma Alternativa Para o Produtor e Para o Consumidor. In Anais do Simpósio de Segurança Alimentar; FAURGS: Gramado, Brazil, 2012; Volume 4. [Google Scholar]
- Pedroso, M.A.P.; Nunes, L.S.; Uliana, G.C.; Richards, N.S.P.d.S. Estratégias e Inovações No Mercado Lácteo: Uma Análise Dos Produtos de Leite de Ovelha No Sul Do Brasil. Fazer Ciência 2023, 157, 55–60. [Google Scholar]
- Tilocca, B.; Costanzo, N.; Morittu, V.M.; Spina, A.A.; Soggiu, A.; Britti, D.; Roncada, P.; Piras, C. Milk Microbiota: Characterization Methods and Role in Cheese Production. J. Proteom. 2020, 210, 103534. [Google Scholar] [CrossRef] [PubMed]
- Paszczyk, B.; Łuczyńska, J. The Comparison of Fatty Acid Composition and Lipid Quality Indices in Hard Cow, Sheep, and Goat Cheeses. Foods 2020, 9, 1667. [Google Scholar] [CrossRef]
- Fusieger, A.; Martins, M.C.F.; de Freitas, R.; Nero, L.A.; de Carvalho, A.F. Technological Properties of Lactococcus Lactis Subsp. Lactis Bv. Diacetylactis Obtained from Dairy and Non-Dairy Niches. Braz. J. Microbiol. 2020, 51, 313–321. [Google Scholar] [CrossRef]
- Bovolenta, S.; Romanzin, A.; Corazzin, M.; Spanghero, M.; Aprea, E.; Gasperi, F.; Piasentier, E. Volatile Compounds and Sensory Properties of Montasio Cheese Made from the Milk of Simmental Cows Grazing on Alpine Pastures. J. Dairy Sci. 2014, 97, 7373–7385. [Google Scholar] [CrossRef] [PubMed]
- Pillonel, L.; Ampuero, S.; Tabacchi, R.; Bosset, J. Analytical Methods for the Determination of the Geographic Origin of Emmental Cheese: Volatile Compounds by GC/MS-FID and Electronic Nose. Eur. Food Res. Technol. 2003, 216, 179–183. [Google Scholar] [CrossRef]
- Possas, A.; Bonilla-Luque, O.M.; Valero, A. From Cheese-Making to Consumption: Exploring the Microbial Safety of Cheeses through Predictive Microbiology Models. Foods 2021, 10, 355. [Google Scholar] [CrossRef] [PubMed]
- Remor, A.; de Vasconcelos, T.C.B.; Belo, V.S.; Zanella, R.; dos Santos, E.D.; Dickel, E.L. Metagenomic investigation in colonial cheese produced with pasteurized milk and sold in the state of Rio Grande do Sul, Brazil. Veterinária Zootec. 2021, 28, 1–9. [Google Scholar] [CrossRef]
- Amato, E.; Filipello, V.; Gori, M.; Lomonaco, S.; Losio, M.N.; Parisi, A.; Huedo, P.; Knabel, S.J.; Pontello, M. Identification of a Major Listeria Monocytogenes Outbreak Clone Linked to Soft Cheese in Northern Italy—2009–2011. BMC Infect. Dis. 2017, 17, 342. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.D.; Pedroso, S.H.S.P.; Sandes, S.H.C.; Silva, G.O.; Luiz, K.C.M.; Dias, R.S.; Filho, R.A.T.; Figueiredo, H.C.P.; Santos, S.G.; Nunes, A.C.; et al. Virulence Factors and Antimicrobial Resistance of Staphylococcus aureus Isolated from the Production Process of Minas Artisanal Cheese from the Region of Campo Das Vertentes, Brazil. J. Dairy Sci. 2020, 103, 2098–2110. [Google Scholar] [CrossRef] [PubMed]
- Alotaibi, K.; Khan, A.A. Prevalence and Molecular Characterization of Shiga Toxin-Producing Escherichia coli from Food and Clinical Samples. Pathogens 2023, 12, 1302. [Google Scholar] [CrossRef] [PubMed]
- Withenshaw, S.M.; Smith, R.P.; Davies, R.; Smith, A.E.O.; Gray, E.; Rodgers, J. A Systematized Review and Qualitative Synthesis of Potential Risk Factors Associated with the Occurrence of Non-O157 Shiga Toxin-Producing Escherichia coli (STEC) in the Primary Production of Cattle. Compr. Rev. Food Sci. Food Saf. 2022, 21, 2363–2390. [Google Scholar] [CrossRef]
- Pinedo, L.C.; Mughini-Gras, L.; Franz, E.; Hald, T.; Pires, S.M. Sources and Trends of Human Salmonellosis in Europe, 2015–2019: An Analysis of Outbreak Data. Int. J. Food Microbiol. 2022, 379, 109850. [Google Scholar] [CrossRef]
- Primavilla, S.; Roila, R.; Rocchegiani, E.; Blasi, G.; Petruzzelli, A.; Gabucci, C.; Ottaviani, D.; Di Lullo, S.; Branciari, R.; Ranucci, D.; et al. Assessment of the Microbiological Safety and Hygiene of Raw and Thermally Treated Milk Cheeses Marketed in Central Italy between 2013 and 2020. Life 2023, 13, 2324. [Google Scholar] [CrossRef]
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. High-Throughput Sequencing for Detection of Subpopulations of Bacteria Not Previously Associated with Artisanal Cheeses. Appl. Environ. Microbiol. 2012, 78, 5717–5723. [Google Scholar] [CrossRef] [PubMed]
- Regecová, I.; Výrostková, J.; Zigo, F.; Gregová, G.; Kováčová, M. Detection of Antimicrobial Resistance of Bacteria Staphylococcus chromogenes Isolated from Sheep’s Milk and Cheese. Antibiotics 2021, 10, 570. [Google Scholar] [CrossRef] [PubMed]
- Danquah, C.A.; Minkah, P.A.B.; Duah-Junior, I.O.; Amankwah, K.B.; Somuah, S.O. Antimicrobial Compounds from Microorganisms. Antibiotics 2022, 11, 285. [Google Scholar] [CrossRef] [PubMed]
- Endres, C.M.; Moreira, E.; de Freitas, A.B.; Castel, A.P.D.; Graciano, F.; Mann, M.B.; Frazzon, A.P.G.; Mayer, F.Q.; Frazzon, J. Evaluation of Enterotoxins and Antimicrobial Resistance in Microorganisms Isolated from Raw Sheep Milk and Cheese: Ensuring the Microbiological Safety of These Products in Southern Brazil. Microorganisms 2023, 11, 1618. [Google Scholar] [CrossRef] [PubMed]
- Dacres, H.; Weihs, F.; Wang, J.; Anderson, A.; Trowell, S.C. Bioluminescence Resonance Energy Transfer Biosensor for Measuring Activity of a Protease Secreted by Pseudomonas fluorescens Growing in Milk. Anal. Chim. Acta 2023, 1270, 341401. [Google Scholar] [CrossRef]
- Otero-Asman, J.R.; Sánchez-Jiménez, A.; Bastiaansen, K.C.; Wettstadt, S.; Civantos, C.; García-Puente, A.; Bitter, W.; Llamas, M.A. The Prc and CtpA Proteases Modulate Cell-Surface Signaling Activity and Virulence in Pseudomonas aeruginosa. Science 2023, 26, 107216. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Ye, A.; Jiang, N. The Role of Bacteria in Gallstone Formation. Folia Microbiol. 2024, 69, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Parija, S.C. Pseudomonas, Burkholderia and Acinetobacter. In Textbook of Microbiology and Immunology; Parija, S.C., Ed.; Springer Nature: Singapore, 2023; pp. 553–561. ISBN 978-981-19331-5-8. [Google Scholar]
- Leonard, S.R.; Mammel, M.K.; Lacher, D.W.; Elkins, C.A. Application of Metagenomic Sequencing to Food Safety: Detection of Shiga Toxin-Producing Escherichia coli on Fresh Bagged Spinach. Appl. Environ. Microbiol. 2015, 81, 8183–8191. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Waldron, L.; Ballarini, A.; Narasimhan, V.; Jousson, O.; Huttenhower, C. Metagenomic Microbial Community Profiling Using Unique Clade-Specific Marker Genes. Nat. Methods 2012, 9, 811–814. [Google Scholar] [CrossRef]
- Erhardt, M.M.; Oliveira, W.d.C.; Fröder, H.; Marques, P.H.; Oliveira, M.B.P.P.; Richards, N.S.P.d.S. Lactic Bacteria in Artisanal Cheese: Characterization through Metagenomics. Fermentation 2023, 9, 41. [Google Scholar] [CrossRef]
- Keenum, I.; Wind, L.; Ray, P.; Guron, G.; Chen, C.; Knowlton, K.; Ponder, M.; Pruden, A. Metagenomic Tracking of Antibiotic Resistance Genes through a Pre-Harvest Vegetable Production System: An Integrated Lab-, Microcosm- and Greenhouse-Scale Analysis. Environ. Microbiol. 2022, 24, 3705–3721. [Google Scholar] [CrossRef]
- Moon, S.H.; Udaondo, Z.; Abram, K.Z.; Li, X.; Yang, X.; DiCaprio, E.L.; Jun, S.-R.; Huang, E. Isolation of AmpC- and Extended Spectrum β-Lactamase-Producing Enterobacterales from Fresh Vegetables in the United States. Food Control 2022, 132, 108559. [Google Scholar] [CrossRef]
- Onalenna, O.; Rahube, T.O. Assessing Bacterial Diversity and Antibiotic Resistance Dynamics in Wastewater Effluent-Irrigated Soil and Vegetables in a Microcosm Setting. Heliyon 2022, 8, e09089. [Google Scholar] [CrossRef]
- Valentino, V.; Sequino, G.; Cobo-Díaz, J.F.; Álvarez-Ordóñez, A.; De Filippis, F.; Ercolini, D. Evidence of Virulence and Antibiotic Resistance Genes from the Microbiome Mapping in Minimally Processed Vegetables Producing Facilities. Food Res. Int. 2022, 162, 112202. [Google Scholar] [CrossRef]
- Yasir, M.; Al-Zahrani, I.A.; Bibi, F.; Abd El Ghany, M.; Azhar, E.I. New Insights of Bacterial Communities in Fermented Vegetables from Shotgun Metagenomics and Identification of Antibiotic Resistance Genes and Probiotic Bacteria. Food Res. Int. 2022, 157, 111190. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Li, L.; Xu, Y.; Li, H.; Yu, Y.; Xu, Z. Metagenomics Reveals the Microbial Community Responsible for Producing Biogenic Amines During Mustard [Brassica Juncea (L.)] Fermentation. Front. Microbiol. 2022, 13, 824644. [Google Scholar] [CrossRef] [PubMed]
- Yurt, M.N.Z.; Ersoy Omeroglu, E.; Tasbasi, B.B.; Acar, E.E.; Altunbas, O.; Ozalp, V.C.; Sudagidan, M. Bacterial and Fungal Microbiota of Mould-Ripened Cheese Produced in Konya. Int. J. Dairy Technol. 2023, 76, 627–637. [Google Scholar] [CrossRef]
- Papadakis, P.; Konteles, S.; Batrinou, A.; Ouzounis, S.; Tsironi, T.; Halvatsiotis, P.; Tsakali, E.; Van Impe, J.F.M.; Vougiouklaki, D.; Strati, I.F.; et al. Characterization of Bacterial Microbiota of P.D.O. Feta Cheese by 16S Metagenomic Analysis. Microorganisms 2021, 9, 2377. [Google Scholar] [CrossRef]
- Masenya, K.; Manganyi, M.C.; Dikobe, T.B. Exploring Cereal Metagenomics: Unravelling Microbial Communities for Improved Food Security. Microorganisms 2024, 12, 510. [Google Scholar] [CrossRef]
- Pérez-Cobas, A.E.; Gomez-Valero, L.; Buchrieser, C. Metagenomic Approaches in Microbial Ecology: An Update on Whole-Genome and Marker Gene Sequencing Analyses. Microb. Genom. 2020, 6, e000409. [Google Scholar] [CrossRef]
- Taş, N.; de Jong, A.E.; Li, Y.; Trubl, G.; Xue, Y.; Dove, N.C. Metagenomic Tools in Microbial Ecology Research. Curr. Opin. Biotechnol. 2021, 67, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Gobbetti, M.; Di Cagno, R.; Calasso, M.; Neviani, E.; Fox, P.F.; De Angelis, M. Drivers That Establish and Assembly the Lactic Acid Bacteria Biota in Cheeses. Trends Food Sci. Technol. 2018, 78, 244–254. [Google Scholar] [CrossRef]
- Lemes, M.R.; Rodrigues, T.C.V.; Jaiswal, A.K.; Tiwari, S.; Sales-Campos, H.; Andrade-Silva, L.E.; Oliveira, C.J.F.; Azevedo, V.; Rodrigues, V.; Soares, S.C.; et al. In Silico Designing of a Recombinant Multi-Epitope Antigen for Leprosy Diagnosis. J. Genet. Eng. Biotechnol. 2022, 20, 128. [Google Scholar] [CrossRef] [PubMed]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Durkin, A.S.; et al. Genome Analysis of Multiple Pathogenic Isolates of Streptococcus agalactiae: Implications for the Microbial “Pan-Genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef]
- Barh, D.; Tiwari, S.; Jain, N.; Ali, A.; Santos, A.R.; Misra, A.N.; Azevedo, V.; Kumar, A. In Silico Subtractive Genomics for Target Identification in Human Bacterial Pathogens. Drug Dev. Res. 2011, 72, 162–177. [Google Scholar] [CrossRef]
- Lux, T.M.; Lee, R.; Love, J. Genome-Wide Phylogenetic Analysis of the Pathogenic Potential of Vibrio furnissii. Front. Microbiol. 2014, 5, 435. [Google Scholar] [CrossRef]
- IAL Instituto Adolfo Lutz. Normas Analíticas Do Instituto Adolfo Lutz, 3rd ed.; IMESP: São Paulo, Brazil, 1985; Volume 1. [Google Scholar]
- IAL Instituto Adolfo Lutz. Métodos Físico-Químicos Para Análise de Alimentos; ANVISA: São Paulo, Brazil, 2008. [Google Scholar]
- Tontisirin, K.; MacLean, W.C.; Warwick, P. Food Energy: Methods of Analysis and Conversion Factors: Report of a Technical Workshop, Rome, 3–6 December 2002; FAO: Rome, Italy, 2003; ISBN 92-5-105014-7. [Google Scholar]
- Bligh, E.; Dyer, W. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Hartman, L.; Lago, R.C. Rapid Preparation of Fatty Acid Methyl Esters from Lipids. Lab. Pract. 1973, 22, 475–476. [Google Scholar]
- Visentainer, J.V. Aspectos analíticos da resposta do detector de ionização em chama para ésteres de ácidos graxos em biodiesel e alimentos. Quím. Nova 2012, 35, 274–279. [Google Scholar] [CrossRef]
- Zianni, R.; Mentana, A.; Tomaiuolo, M.; Campaniello, M.; Iammarino, M.; Centonze, D.; Palermo, C. Volatolomic Approach by HS-SPME/GC–MS and Chemometric Evaluations for the Discrimination of X-Ray Irradiated Mozzarella Cheese. Food Chem. 2023, 423, 136239. [Google Scholar] [CrossRef]
- NIST NIST V2.2 (National Institute of Standards and Technology, USA) Library Database. Available online: https://webbook.nist.gov/chemistry/name-ser/ (accessed on 16 June 2024).
- DePaula, J.; Cunha, S.C.; Cruz, A.; Sales, A.L.; Revi, I.; Fernandes, J.; Ferreira, I.M.P.L.V.O.; Miguel, M.A.L.; Farah, A. Volatile Fingerprinting and Sensory Profiles of Coffee Cascara Teas Produced in Latin American Countries. Foods 2022, 11, 3144. [Google Scholar] [CrossRef]
- Galvan-Lima, Â.; Cunha, S.C.; Martins, Z.E.; Soares, A.G.; Ferreira, I.M.P.L.V.O.; Farah, A. Headspace Volatolome of Peel Flours from Citrus Fruits Grown in Brazil. Food Res. Int. 2021, 150, 110801. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Wickham, H. Reshaping Data with the Reshape Package. J. Stat. Softw. 2007, 21, 1–20. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package 2024. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 23 July 2024).
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Mori, H.; Maruyama, T.; Yano, M.; Yamada, T.; Kurokawa, K. VITCOMIC2: Visualization Tool for the Phylogenetic Composition of Microbial Communities Based on 16S rRNA Gene Amplicons and Metagenomic Shotgun Sequencing. BMC Syst. Biol. 2018, 12, 30. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Agren, J.; Sundström, A.; Håfström, T.; Segerman, B. Gegenees: Fragmented Alignment of Multiple Genomes for Determining Phylogenomic Distances and Genetic Signatures Unique for Specified Target Groups. PLoS ONE 2012, 7, e39107. [Google Scholar] [CrossRef]
- Rodrigues, D.L.N.; Ariute, J.C.; Rodrigues da Costa, F.M.; Benko-Iseppon, A.M.; Barh, D.; Azevedo, V.; Aburjaile, F. PanViTa: Pan Virulence and resistance Analysis. Front. Bioinform. 2023, 3, 1070406. [Google Scholar] [CrossRef] [PubMed]
- Bennato, F.; Di Domenico, M.; Ianni, A.; Di Gialleonardo, L.; Cammà, C.; Martino, G. Grape Pomace in Ewes Diet Affects Metagenomic Profile, Volatile Compounds and Biogenic Amines Contents of Ripened Cheese. Fermentation 2022, 8, 598. [Google Scholar] [CrossRef]
- Kamilari, E.; Anagnostopoulos, D.A.; Papademas, P.; Efthymiou, M.; Tretiak, S.; Tsaltas, D. Snapshot of Cyprus Raw Goat Milk Bacterial Diversity via 16S rDNA High-Throughput Sequencing; Impact of Cold Storage Conditions. Fermentation 2020, 6, 100. [Google Scholar] [CrossRef]
- Kamilari, E.; Anagnostopoulos, D.A.; Papademas, P.; Kamilaris, A.; Tsaltas, D. Characterizing Halloumi Cheese’s Bacterial Communities through Metagenomic Analysis. LWT 2020, 126, 109298. [Google Scholar] [CrossRef]
- Kamimura, B.A.; Cabral, L.; Noronha, M.F.; Baptista, R.C.; Nascimento, H.M.; Sant’Ana, A.S. Amplicon Sequencing Reveals the Bacterial Diversity in Milk, Dairy Premises and Serra Da Canastra Artisanal Cheeses Produced by Three Different Farms. Food Microbiol. 2020, 89, 103453. [Google Scholar] [CrossRef]
- Endres, C.M.; Castro, Í.M.S.; Trevisol, L.D.; Severo, J.M.; Mann, M.B.; Varela, A.P.M.; Frazzon, A.P.G.; Mayer, F.Q.; Frazzon, J. Molecular Characterization of the Bacterial Communities Present in Sheep’s Milk and Cheese Produced in South Brazilian Region via 16S rRNA Gene Metabarcoding Sequencing. LWT 2021, 147, 111579. [Google Scholar] [CrossRef]
- Porcellato, D.; Aspholm, M.; Skeie, S.B.; Monshaugen, M.; Brendehaug, J.; Mellegård, H. Microbial Diversity of Consumption Milk during Processing and Storage. Int. J. Food Microbiol. 2018, 266, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.; Qiu, S.; Wang, B.; Zeng, H. Metagenomic and Flavoromic Profiling Reveals the Correlation between the Microorganisms and Volatile Flavor Compounds in Monascus-Fermented Cheese. Food Res. Int. 2024, 188, 114483. [Google Scholar] [CrossRef]
- Araújo-Rodrigues, H.; Tavaria, F.K.; dos Santos, M.T.P.G.; Alvarenga, N.; Pintado, M.M. A Review on Microbiological and Technological Aspects of Serpa PDO Cheese: An Ovine Raw Milk Cheese. Int. Dairy J. 2020, 100, 104561. [Google Scholar] [CrossRef]
- Bencini, R.; Pulina, G. The Quality of Sheep Milk: A Review. Aust. J. Exp. Agric. 1997, 37, 485–504. [Google Scholar] [CrossRef]
- Felder, D.; Burns, D.; Chang, D. Defining Microbial Terroir: The Use of Native Fungi for the Study of Traditional Fermentative Processes. Int. J. Gastron. Food Sci. 2012, 1, 64–69. [Google Scholar] [CrossRef]
- Xu, Z.S.; Ju, T.; Yang, X.; Gänzle, M. A Meta-Analysis of Bacterial Communities in Food Processing Facilities: Driving Forces for Assembly of Core and Accessory Microbiomes across Different Food Commodities. Microorganisms 2023, 11, 1575. [Google Scholar] [CrossRef]
- Yuan, H.; Han, S.; Zhang, S.; Xue, Y.; Zhang, Y.; Lu, H.; Wang, S. Microbial Properties of Raw Milk throughout the Year and Their Relationships to Quality Parameters. Foods 2022, 11, 3077. [Google Scholar] [CrossRef]
- Rzewuska, M.; Kwiecień, E.; Chrobak-Chmiel, D.; Kizerwetter-Świda, M.; Stefańska, I.; Gieryńska, M. Pathogenicity and Virulence of Trueperella pyogenes: A Review. Int. J. Mol. Sci. 2019, 20, 2737. [Google Scholar] [CrossRef]
- Seng, P.; Boushab, B.M.; Romain, F.; Gouriet, F.; Bruder, N.; Martin, C.; Paganelli, F.; Bernit, E.; Treut, Y.P.L.; Thomas, P.; et al. Emerging Role of Raoultella ornithinolytica in Human Infections: A Series of Cases and Review of the Literature. Int. J. Infect. Dis. 2016, 45, 65–71. [Google Scholar] [CrossRef]
- Rodríguez-Medina, N.; Barrios-Camacho, H.; Duran-Bedolla, J.; Garza-Ramos, U. Klebsiella variicola: An Emerging Pathogen in Humans. Emerg. Microbes Infect. 2019, 8, 973–988. [Google Scholar] [CrossRef]
- Khoramrooz, S.S.; Mirsalehian, A.; Emaneini, M.; Jabalameli, F.; Aligholi, M.; Saedi, B.; Bazargani, A.; Taherikalani, M.; Borghaei, P.; Razmpa, E. Frequency of Alloicoccus otitidis, Streptococcus pneumoniae, Moraxella catarrhalis and Haemophilus influenzae in Children with Otitis Media with Effusion (OME) in Iranian Patients. Auris Nasus Larynx 2012, 39, 369–373. [Google Scholar] [CrossRef]
- Lima, B.B.; Leal, M.C. Parâmetros Indicadores de Qualidade de Queijos Artesanais Comercializados Em Castro-PR; Universidade Federal do Paraná: Curitiba, Brazil, 2017. [Google Scholar]
- Kliem, K.E.; Humphries, D.J.; Grandison, A.S.; Morgan, R.; Livingstone, K.M.; Givens, D.I.; Reynolds, C.K. Effect of a Whey Protein and Rapeseed Oil Gel Feed Supplement on Milk Fatty Acid Composition of Holstein Cows. J. Dairy Sci. 2019, 102, 288–300. [Google Scholar] [CrossRef]
- Margalho, L.P.; Kamimura, B.A.; Pimentel, T.C.; Balthazar, C.F.; Araujo, J.V.A.; Silva, R.; Conte-Junior, C.A.; Raices, R.S.L.; Cruz, A.G.; Sant’Ana, A.S. A Large Survey of the Fatty Acid Profile and Gross Composition of Brazilian Artisanal Cheeses. J. Food Compos. Anal. 2021, 101, 103955. [Google Scholar] [CrossRef]
- Mitani, T.; Asakuma, S.; Shinoda, Y.; Ueda, Y.; Aoki, Y.; Oshita, T. Effects of Ear Corn Silage Supplementation on Milk Production and Milk Fatty Acid Profiles in Grazing Dairy Farms. Anim. Sci. J. 2020, 91, e13454. [Google Scholar] [CrossRef]
- Nudda, A.; Cannas, A.; Correddu, F.; Atzori, A.S.; Lunesu, M.F.; Battacone, G.; Pulina, G. Sheep and Goats Respond Differently to Feeding Strategies Directed to Improve the Fatty Acid Profile of Milk Fat. Animals 2020, 10, 1290. [Google Scholar] [CrossRef]
- Silva, H.L.A.; Balthazar, C.F.; Esmerino, E.A.; Vieira, A.H.; Cappato, L.P.; Neto, R.P.C.; Verruck, S.; Cavalcanti, R.N.; Portela, J.B.; Andrade, M.M.; et al. Effect of Sodium Reduction and Flavor Enhancer Addition on Probiotic Prato Cheese Processing. Food Res. Int. 2017, 99, 247–255. [Google Scholar] [CrossRef]
- Silva, H.L.A.; Balthazar, C.F.; Esmerino, E.A.; Neto, R.P.C.; Rocha, R.S.; Moraes, J.; Cavalcanti, R.N.; Franco, R.M.; Tavares, M.I.B.; Santos, J.S.; et al. Partial Substitution of NaCl by KCl and Addition of Flavor Enhancers on Probiotic Prato Cheese: A Study Covering Manufacturing, Ripening and Storage Time. Food Chem. 2018, 248, 192–200. [Google Scholar] [CrossRef]
- Ahmad, M.T.; Shariff, M.; Md. Yusoff, F.; Goh, Y.M.; Banerjee, S. Applications of Microalga Chlorella vulgaris in Aquaculture. Rev. Aquac. 2020, 12, 328–346. [Google Scholar] [CrossRef]
- Mostafa, S.S.M. Microalgal Biotechnology: Prospects and Applications. In Plant Science; IntechOpen: London, UK, 2012; pp. 275–314. ISBN 978-953-51-0905-1. [Google Scholar]
- Nassar, M.; Jaffery, A.; Ibrahim, B.; Baraka, B.; Abosheaishaa, H. The Multidimensional Benefits of Eicosapentaenoic Acid: From Heart Health to Inflammatory Control. Egypt. J. Intern. Med. 2023, 35, 81. [Google Scholar] [CrossRef]
- Park, N.-H.; Choi, J.-S.; Hwang, S.-Y.; Kim, Y.-C.; Hong, Y.-K.; Cho, K.K.; Choi, I.S. Antimicrobial Activities of Stearidonic and Gamma-Linolenic Acids from the Green Seaweed Enteromorpha Linza against Several Oral Pathogenic Bacteria. Bot. Stud. 2013, 54, 39. [Google Scholar] [CrossRef]
- Ru, I.T.K.; Sung, Y.Y.; Jusoh, M.; Wahid, M.E.A.; Nagappan, T. Chlorella vulgaris: A Perspective on Its Potential for Combining High Biomass with High Value Bioproducts. Appl. Phycol. 2020, 1, 2–11. [Google Scholar] [CrossRef]
- Sukhikh, S.; Prosekov, A.; Ivanova, S.; Maslennikov, P.; Andreeva, A.; Budenkova, E.; Kashirskikh, E.; Tcibulnikova, A.; Zemliakova, E.; Samusev, I.; et al. Identification of Metabolites with Antibacterial Activities by Analyzing the FTIR Spectra of Microalgae. Life 2022, 12, 1395. [Google Scholar] [CrossRef]
- Perry, K.S.P. Queijos: Aspectos químicos, bioquímicos e microbiológicos. Química Nova 2004, 27, 293–300. [Google Scholar] [CrossRef]
- Curioni, P.M.G.; Bosset, J.O. Key Odorants in Various Cheese Types as Determined by Gas Chromatography-Olfactometry. Int. Dairy J. 2002, 12, 959–984. [Google Scholar] [CrossRef]
- Cardoso, M.G.B. Microbiota bacteriana e compostos voláteis de queijo Minas Artesanal da região da Serra da Canastra; Universidade Federal de Lavras: Lavras, Brazil, 2022. [Google Scholar]
- Furtado, M.M. O Estufamento Tardio Dos Queijos: Características e Prevenção, Uma Revisão. Rev. Inst. Laticínios Cândido Tostes 1985, 40, 3–39. [Google Scholar]
- Vinayamohan, P.G.; Viju, L.S.; Joseph, D.; Venkitanarayanan, K. Fermented Foods as a Potential Vehicle of Antimicrobial-Resistant Bacteria and Genes. Fermentation 2023, 9, 688. [Google Scholar] [CrossRef]
- Gaurav, A.; Bakht, P.; Saini, M.; Pandey, S.; Pathania, R. Role of Bacterial Efflux Pumps in Antibiotic Resistance, Virulence, and Strategies to Discover Novel Efflux Pump Inhibitors. Microbiology 2023, 169, 001333. [Google Scholar] [CrossRef] [PubMed]
- Naidu, V.; Bartczak, A.; Brzoska, A.J.; Lewis, P.; Eijkelkamp, B.A.; Paulsen, I.T.; Elbourne, L.D.H.; Hassan, K.A. Evolution of RND Efflux Pumps in the Development of a Successful Pathogen. Drug Resist. Updates 2023, 66, 100911. [Google Scholar] [CrossRef] [PubMed]
- Saini, N.; Aamir, M.; Singh, V.K.; Deepak, B.; Mona, S. Unveiling the Microbial Diversity and Functional Dynamics of Shiv Kund, Sohna Hot Spring, India through a Shotgun Metagenomics Approach. Arch. Microbiol. 2023, 205, 323. [Google Scholar] [CrossRef] [PubMed]
- Darby, E.M.; Trampari, E.; Siasat, P.; Gaya, M.S.; Alav, I.; Webber, M.A.; Blair, J.M.A. Molecular Mechanisms of Antibiotic Resistance Revisited. Nat. Rev. Microbiol. 2023, 21, 280–295. [Google Scholar] [CrossRef]
- Gervasoni, S.; Mehla, J.; Bergen, C.R.; Leus, I.V.; Margiotta, E.; Malloci, G.; Bosin, A.; Vargiu, A.V.; Lomovskaya, O.; Rybenkov, V.V.; et al. Molecular Determinants of Avoidance and Inhibition of Pseudomonas aeruginosa MexB Efflux Pump. mBio 2023, 14, e01403-23. [Google Scholar] [CrossRef]
- Zhou, J.-X.; Feng, D.-Y.; Li, X.; Zhu, J.-X.; Wu, W.-B.; Zhang, T. Advances in Research on Virulence Factors of Acinetobacter baumannii and Their Potential as Novel Therapeutic Targets. J. Appl. Microbiol. 2023, 134, lxac089. [Google Scholar] [CrossRef]
- Shadan, A.; Pathak, A.; Ma, Y.; Pathania, R.; Singh, R.P. Deciphering the Virulence Factors, Regulation, and Immune Response to Acinetobacter baumannii Infection. Front. Cell. Infect. Microbiol. 2023, 13, 1053968. [Google Scholar] [CrossRef]
- Fursova, N.K.; Fursov, M.V.; Astashkin, E.I.; Fursova, A.D.; Novikova, T.S.; Kislichkina, A.A.; Sizova, A.A.; Fedyukina, G.N.; Savin, I.A.; Ershova, O.N. Multidrug-Resistant and Extensively Drug-Resistant Acinetobacter baumannii Causing Nosocomial Meningitis in the Neurological Intensive Care Unit. Microorganisms 2023, 11, 2020. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Zhao, D.; Huang, N.; Liu, S.; Zheng, J.; Cao, J.; Zeng, W.; Zheng, X.; Wang, L.; Zhou, T.; et al. Clinical Impact of the Type VI Secretion System on Clinical Characteristics, Virulence and Prognosis of Acinetobacter baumannii during Bloodstream Infection. Microb. Pathog. 2023, 182, 106252. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.A.; Saha, P. Chemotactic Response of P-Nitrophenol Degrading Pseudomonas asiatica Strain PNPG3 through Phenotypic and Genome Sequence-Based in Silico Studies. 3 Biotech 2023, 13, 408. [Google Scholar] [CrossRef]
- He, L.; Zhao, L.; Li, Q.; Huang, L.; Qin, Y.; Zhuang, Z.; Wang, X.; Huang, H.; Zhang, J.; Zhang, J.; et al. Flagellar Gene fliP Contributes to the Virulence of Pseudomonas plecoglossicida by Regulating Its Motility. Aquaculture 2023, 576, 739874. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).